Abstract
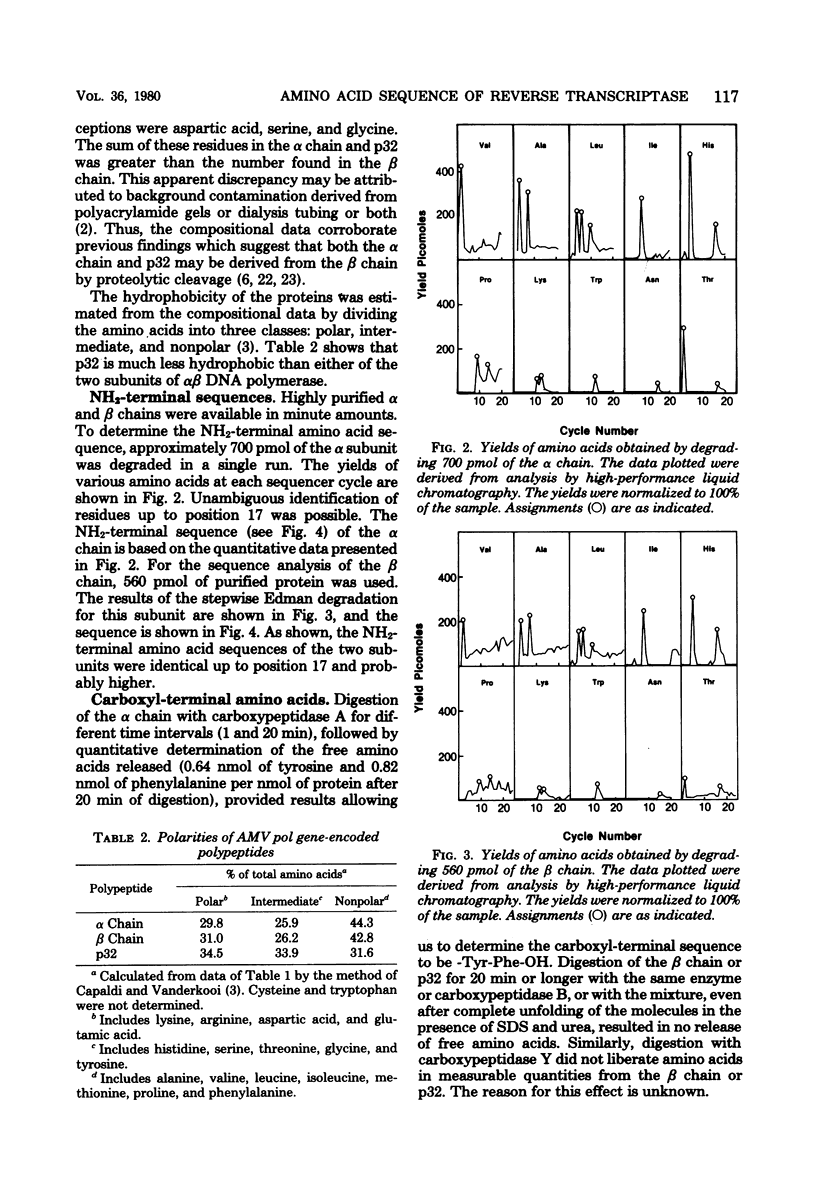
The NH2-terminal amino acid sequences of the α and β chains of avian myeloblastosis αβ DNA polymerase were determined by using microsequence analysis in the subnanomole range and were found to be identical up to 17 residues. The common sequence was as follows: Thr-Val-Ala-Leu-His-Leu-Ala-Ile-Pro-Leu-Lys-Trp-Lys-Pro-Asn-His-Thr-. This result provides convincing chemical evidence that the α chain is derived from the NH2-terminal region of the β chain by proteolytic cleavage, whereas the amino acid composition for these α and β subunits and p32 DNA endonuclease suggests that the latter is derived from the carboxyl-terminal region of the β chain.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer S. H., Noyes A. N., Boyer M. L., Marr K. Hemoglobin 3 chains in apes. Primary structures and the presumptive nature of back mutation in a normally silent gene. J Biol Chem. 1973 Feb 10;248(3):992–1003. [PubMed] [Google Scholar]
- Brown W. E., Howard G. C. Amino acid composition of proteins eluted from polyacrylamide gels: background considerations. Anal Biochem. 1980 Jan 15;101(2):294–298. doi: 10.1016/0003-2697(80)90189-x. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- Eisenman R. N., Mason W. S., Linial M. Synthesis and processing of polymerase proteins of wild-type and mutant avian retroviruses. J Virol. 1980 Oct;36(1):62–78. doi: 10.1128/jvi.36.1.62-78.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Verma I. M. Studies on the reverse transcriptase of RNA tumor viruses. Structural relatedness of two subunits of avian RNA tumor viruses. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4991–4994. doi: 10.1073/pnas.71.12.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979 Mar 10;254(5):1606–1613. [PubMed] [Google Scholar]
- Grandgenett D. P. Dissociation of alpha beta DNA polymerase of avian myeloblastosis virus by dimethyl sulfoxide. J Virol. 1976 Mar;17(3):950–961. doi: 10.1128/jvi.17.3.950-961.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P., Gerard G. F., Green M. A single subunit from avian myeloblastosis virus with both RNA-directed DNA polymerase and ribonuclease H activity. Proc Natl Acad Sci U S A. 1973 Jan;70(1):230–234. doi: 10.1073/pnas.70.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Hayashi R., Moore S., Stein W. H. Carboxypeptidase from yeast. Large scale preparation and the application to COOH-terminal analysis of peptides and proteins. J Biol Chem. 1973 Apr 10;248(7):2296–2302. [PubMed] [Google Scholar]
- Henderson L. E., Copeland T. D., Oroszlan S. Separation of amino acid phenylthiohydantoins by high-performance liquid chromatography on phenylalkyl support. Anal Biochem. 1980 Feb;102(1):1–7. doi: 10.1016/0003-2697(80)90307-3. [DOI] [PubMed] [Google Scholar]
- Henderson L. E., Copeland T. D., Smythers G. W., Marquardt H., Oroszlan S. Amino-terminal amino acid sequence and carboxyl-terminal analysis of Rauscher murine leukemia virus glycoproteins. Virology. 1978 Mar;85(1):319–322. doi: 10.1016/0042-6822(78)90437-3. [DOI] [PubMed] [Google Scholar]
- Hizi A., McCrae M. A., Joklik W. K. Studies on the amino acid sequence content of proteins specified by the gag and pol genes of avian sarcoma virus B77. Virology. 1978 Aug;89(1):272–284. doi: 10.1016/0042-6822(78)90059-4. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. Direct microsequence analysis of polypeptides using an improved sequenator, a nonprotein carrier (polybrene), and high pressure liquid chromatography. Biochemistry. 1978 May 30;17(11):2124–2133. doi: 10.1021/bi00604a016. [DOI] [PubMed] [Google Scholar]
- Kacian D. L., Watson K. F., Burny A., Spiegelman S. Purification of the DNA polymerase of avian myeloblastosis virus. Biochim Biophys Acta. 1971 Sep 24;246(3):365–383. doi: 10.1016/0005-2787(71)90773-8. [DOI] [PubMed] [Google Scholar]
- Klapper D. G., Wilde C. E., 3rd, Capra J. D. Automated amino acid sequence of small peptides utilizing Polybrene. Anal Biochem. 1978 Mar;85(1):126–131. doi: 10.1016/0003-2697(78)90282-8. [DOI] [PubMed] [Google Scholar]
- Oroszlan S., Copeland T. D., Henderson L. E., Stephenson J. R., Gilden R. V. Amino-terminal sequence of bovine leukemia virus major internal protein: homology with mammalian type C virus p30 structural proteins. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2996–3000. doi: 10.1073/pnas.76.6.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroszlan S., Copeland T., Summers M. R., Smythers G., Gilden R. V. Amino acid sequence homology of mammalian type C RNA virus major internal proteins. J Biol Chem. 1975 Aug 25;250(16):6232–6239. [PubMed] [Google Scholar]
- Oroszlan S., Henderson L. E., Stephenson J. R., Copeland T. D., Long C. W., Ihle J. N., Gilden R. V. Amino- and carboxyl-terminal amino acid sequences of proteins coded by gag gene of murine leukemia virus. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1404–1408. doi: 10.1073/pnas.75.3.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettenmier C. W., Karess R. E., Anderson S. M., Hanafusa H. Tryptic peptide analysis of avian oncovirus gag and pol gene products. J Virol. 1979 Oct;32(1):102–113. doi: 10.1128/jvi.32.1.102-113.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho H. M., Grandgenett D. P., Green M. Sequence relatedness between the subunits of avian myeloblastosis virus reverse transcriptase. J Biol Chem. 1975 Jul 10;250(13):5278–5280. [PubMed] [Google Scholar]
- Schiff R. D., Grandgenett D. P. Virus-coded origin of a 32,000-dalton protein from avian retrovirus cores: structural relatedness of p32 and the beta polypeptide of the avian retrovirus DNA polymerase. J Virol. 1978 Oct;28(1):279–291. doi: 10.1128/jvi.28.1.279-291.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal G., Loeb L. A. On the fidelity of DNA replication. Enzyme activities associated with DNA polymerases from RNA tumor viruses. J Biol Chem. 1976 Feb 25;251(4):975–981. [PubMed] [Google Scholar]
- Shealy D. J., Rueckert R. R. Proteins of Rous-associated virus 61, an avian retrovirus: common precursor for glycoproteins gp85 and gp35 and use of pactamycin to map translational order of proteins in the gag, pol, and env genes. J Virol. 1978 May;26(2):380–388. doi: 10.1128/jvi.26.2.380-388.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Modak M. J. Enzymatic activities associated with avian and murine retroviral DNA polymerases. Catalysis of and active site involvement in pyrophosphate exchange and pyrophosphorolysis reactions. J Biol Chem. 1980 Mar 10;255(5):2000–2004. [PubMed] [Google Scholar]
- Tarr G. E., Beecher J. F., Bell M., McKean D. J. Polyquarternary amines prevent peptide loss from sequenators. Anal Biochem. 1978 Feb;84(2):622–7?0=ENG. doi: 10.1016/0003-2697(78)90086-6. [DOI] [PubMed] [Google Scholar]