Abstract
miRNAs are small non-coding RNAs that inhibit translation and promote mRNA decay. The levels of mature miRNAs are the result of different rates of transcription, processing, and turnover. The non-canonical polymerase Gld2 has been implicated in the stabilization of miR-122 possibly by catalyzing 3′ monoadenylation, however, there is little evidence that this relationship is one of cause and effect. Here, we biochemically characterize Gld2 involvement in miRNA monoadenylation and its effect on miRNA stability. We find that Gld2 directly monoadenylates and stabilizes specific miRNA populations in human fibroblasts and that sensitivity to monoadenylation-induced stability depends on nucleotides in the miRNA 3′ end. These results establish a novel mechanism of miRNA stability and resulting post-transcriptional gene regulation.
INTRODUCTION
MicroRNAs (miRNAs) are 21-24 nucleotide RNAs involved in the post-transcriptional regulation of virtually all biological processes (Ambros, 2004). Through Watson-Crick base-pairing with their 5′ (seed) nucleotides to 3′ untranslated regions (3′ UTRs), they inhibit translation, induce de-adenylation and destruction, or otherwise abrogate the expression of mRNAs (Nottrott et al., 2006; Guo et al., 2010; Bazzini et al., 2012). miRNAs are processed from primary transcripts (pri-miRNAs) into pre-miRNA stem-loop structures in the nucleus by Drosha; after export to the cytoplasm, the pre-miRNAs are further processed into imperfect miRNA duplexes by the RNAse III enzyme Dicer. Finally, the inactive (passenger) strand is destroyed and the mature (guide strand) miRNA is loaded into what becomes an active Argonaute 2 (Ago2)-containing RNA-induced silencing complex (RISC). Thus, the amounts of mature miRNAs are the result of transcription, processing, and turnover (Ambros, 2004; Bartel, 2004). In addition, various RNA binding proteins such as hnRNP A1 (Guil and Cáceres, 2007), KSRP (Trabucchi et al., 2009), and TDP-43 (Buratti et al., 2010; Kawahara and Mieda-Sato, 2012) have been shown to modulate the biogenesis of specific miRNAs. Several miRNA 3′ modifications have been implicated in the regulation of miRNA turnover (Li et al., 2005; Horwich et al., 2007) and recently, high-throughput sequencing studies detected nucleotide additions on miRNA 3′ termini in animal cells. These additional one or very rarely two nucleotides are not found in genomic sequences and are termed non-templated additions. One function of these extra nucleotides is to modulate miRNA efficacy to enter into RISC (Burroughs et al., 2010), which in turn could modify their stability or ability to regulate translation. The non-templated 3′ nucleotide additions occur only on specific miRNAs and are cell type, developmental, or disease state-specific, suggesting an essential role in many biological processes (Wyman et al., 2011).
Although the importance of regulated miRNA stability seems self-evident, the mechanism(s) involved are generally unknown. miR-382, a miRNA that contributes to HIV-1 pro-virus latency, is particularly unstable; mutational analysis has demonstrated that substitutions in the last seven nucleotides increase its stability (Bail et al., 2010). Similarly, stability of the miR-16 family is dynamically regulated throughout the cell cycle and the seed region and 3′ nucleotides of one of them, miR-503, are particularly important for controlling its steady state levels (Rissland et al., 2011). Recent evidence suggests that non-templated 3′ monoadenylation might be a determinant of miRNA stability; however, there is no direct evidence that this is the case. In mouse liver and neonatal human fibroblasts, removal or depletion of Gld2 (also called PAPD4 or TUTase2) results in a marked down-regulation of mature miR-122, but not its precursor (Katoh et al., 2009; Burns et al., 2011). Gld2 was first characterized in C. elegans as a cytoplasmic non-canonical poly(A) polymerase involved in germline development (Wang et al., 2002); its most well characterized function is to polyadenylate mRNAs in oocytes and neurons, thereby stimulating translation. In these cases, Gld2 is tethered to the mRNA 3′ end by an RNA binding protein such as CPEB or Gld3 (Barnard et al., 2004; Kim and Richter, 2006; Udagawa et al., 2012;Wang et al., 2002). In mouse liver and human fibroblasts, Gld2 is thought to catalyze a 3′ monoadenylation reaction, thereby stabilizing miR-122 (Katoh et al., 2009; Burns et al., 2011). In the fibroblasts, direct or indirect Gld2-stimulated monoadenylation and stabilization of miR-122 elicits a down-regulation of CPEB mRNA expression, which in turn tempers CPEB’s regulation of p53 mRNA polyadenylation-induced translation (Burns et al., 2011).
In the present study, we have analyzed the involvement of Gld2-catalyzed monoadenylation in miRNA stability. We demonstrate that Gld2 adds a single nucleotide to the 3′ end of specific miRNAs, show directly that monoadenylation stabilizes and prolongs the activity of some but not all miRNAs, and present data indicating that sensitivity to monoadenylation-induced stability depends on nucleotides at the 3′ end of the miRNA. Finally, we present evidence that mature miRNA stability is the product of a complex combinatorial control.
RESULTS
Gld2 monoadenylates small RNAs
To investigate Gld2 monoadenylation activity, Flag-tagged Gld2 (WT or a catalytically inactive mutant form, D215A) was ectopically expressed in human primary foreskin fibroblasts (Figure 1A) followed by Flag immunoprecipitation and incubation with single-stranded miRNAs in the presence of α-32P-ATP. The RNA was then extracted and analyzed by PAGE and phosphorimaging. Figure 1B shows that Gld2 monoadenylated miR-122, let-7a, and miR-134 to similar extents. Cells that did not express ectopic Gld2, or expressed the inactive D215A mutant form did not adenylate the RNAs. To assess whether other RNAs can be monoadenylated by Gld2, random sequence RNA 18-, 21-, and 24-mers were tested in assays identical to those described above; in each case, Gld2 monoadenylated the RNAs (Supplementary Figure 1). These data show that Gld2 monoadenylates miRNAs but at least in vitro, does not distinguish between them and other small RNAs.
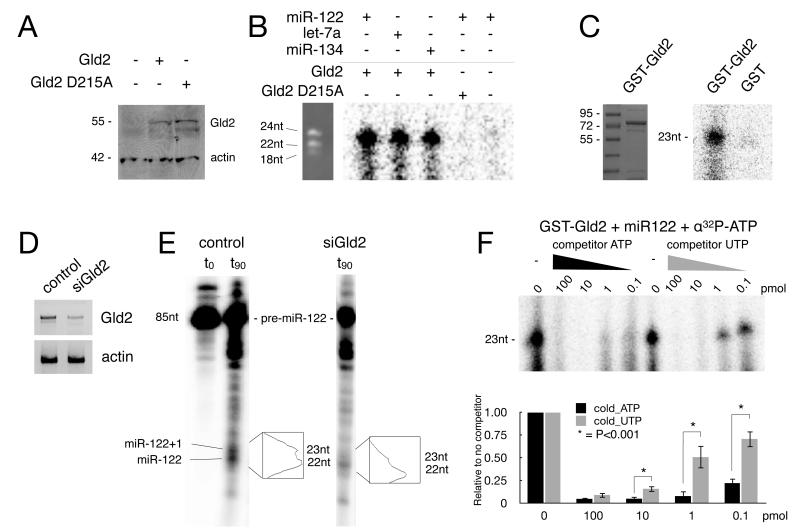
Figure 1. Gld2 monoadenylates miRNAs.
(A) Western blot of Flag-Gld2 and catalytically inactive Flag-D125A-Gld2 ectopically expressed in fibroblasts. Actin served as a loading control. (B) Immunoprecipitated Flag-Gld2 and Flag-D215A-Gld2 were used in monoadenylation assays with miR-122, let-7a, and miR-134 in the presence of α-32P-ATP. A Flag immunoprecipitate from mock-transfected cells was used as a control. RNA molecular weight markers are shown on the left. (C) Coomassie blue stained gel of recombinant Gld2 (left) and GST-Gld2 used in monoadenylation reactions with miR122 and α32-ATP (right). (D) Semi-quantitative RT-PCR of Gld2 and actin RNAs in control and Gld2 siRNA-depleted fibroblasts. The amount of depletion of Gld2 was ~75%. (E) In vitro-transcribed 32P-pre-miR122 was added to control or Gld2-depleted fibroblast extracts (t0) and incubated for 90 min. The pre-miRNA was processed to mature (22 nucleotides) and mature monoadenylated (23 nucleotides) forms in the control extract, but only the mature form (22 nucleotides) in the Gld2-depleted extracts. (F) Recombinant Gld2 was used in an in vitro adenylation reaction with miR-122 and varying amounts of competitor ATP and UTP. The histogram shows the quantification of 3 replicate experiments (the bars refer to SEM and the asterisk refers to statistical significance, Student’s t test, p<0.001).
miRNAs are extensively modified on their 3′ ends (Burroughs et al., 2010; Wyman et al., 2011), and thus it was formally possible that a Gld2 co-precipitating protein monoadenylated the miRNAs, or that the miRNAs were polyadenylated and then trimmed to a single adenylate residue by a deadenylating enzyme (Ameres et al., 2010; Han et al., 2011; Liu et al., 2011). Consequently, highly purified recombinant GST-Gld2 (Figure 1C, left panel) was incubated with miR-122 and α32P-ATP followed by analysis for monoadenylation. Figure 1C (right panel) shows that recombinant GST-Gld2, but not GST, catalyzed the addition of a single adenylate residue to miR-122. These data indicate not only that Gld2 monoadenylates miRNA, but that it does not require the tethering that is necessary for the enzyme to polyadenylate mRNA (Barnard et al., 2004; Wang et al., 2002).
We also investigated whether Gld2 was required for pre-miRNA processing as well as monoadenylation. Extracts from human primary fibroblasts depleted of Gld2 (Figure 1D) were incubated with 5′ 32P-ATP-labeled pre-miR-122 for 90 minutes, followed by RNA extraction and analysis by denaturing PAGE. Control extracts processed, albeit inefficiently, the labeled pre-miRNA into two distinct mature miRNA species, 22 and 23 nucleotides in length. Conversely, the Gld2-depleted extracts produced mostly a single band of 22 nucleotides (Figure 1E). These results indicate that Gld2 does not affect pre-miR-122 processing, but that it is necessary for the 3′ end monoadenylation that increases the length of the mature miRNA from 22 to 23 nucleotides.
It was formally possible that the additional 3′ nucleotide in the previous assay was not necessarily an adenylate residue. Because deep-sequencing studies found that the most prevalent miRNA additions were either one adenylate or one uridylate residue (Burroughs et al., 2010; Wyman et al., 2011), we determined whether the enzyme can monouridylate as well as monoadenylate miRNAs. Recombinant Gld2 was incubated with miR-122, α-32P-ATP, and increasing amounts of radioinert ATP or UTP. The radioinert ATP competed more effectively with the α-32P-ATP than radioinert UTP at all concentrations except the highest one (Student t test, p<0.001) (Fig. 1F). Therefore, ATP is the preferred nucleoside triphosphate for Gld2 to modify miRNA 3′ ends; monouridylation may be catalyzed by a different enzyme.
Monoadenylation promotes miRNA stability
Experiments using Gld2 knockout mouse liver (Katoh et al., 2009) and Gld2-depleted human primary fibroblasts (Burns et al., 2011) suggest that 3′ monoadenylation stabilizes miR-122, however, there is no direct evidence that this relationship is one of cause and effect. To assess this possibility, human fibroblasts were transfected with a radiolabeled miR-122 pre-annealed duplex (see scheme in Figure 2D) that lacked or contained a 3′ adenosine on the leading strand. Such duplexes resemble Dicer products with 5′ phosphate and 3′ hydroxyl groups on both strands and after removal and destruction of the passenger strand feed the leading strand into RISC (MacRae and Doudna, 2007). RNA was then extracted at different times and examined by denaturing PAGE. To minimize the dilution of the miRNA with cell doublings, we plated transfected cells at confluency and loaded the same amount of total RNA onto the gel for each time point. 3′ monoadenylated miR-122 had almost twice the half-life (1.8-fold) compared to the non-monoadenylated form (~54 and ~30 hours, respectively) (Figure 2A). We also incubated fibroblast cell extracts with the same miRNAs and measured their stabilities. Although the miRNAs degraded more rapidly than they did in intact cells, there was still a stabilizing effect of the monoadenylate residue on miR-122 (~1.6 fold, ~2.4 versus ~1.5 hours) (Figure 2B). To assess whether 3′ monoadenylation can stabilize any miRNA, we performed the same in vitro assay with let-7a, which was selected at random. Let-7a, which was very stable in the extracts compared to miR-122 (t1/2 of ~3 versus 1.5 hours) was unaffected by 3′ monoadenylation (Figure 2C). These results show directly that 3′ monoadenylation increases the stability of miR-122 but has little effect on the inherently stable miRNAs such as let-7a.
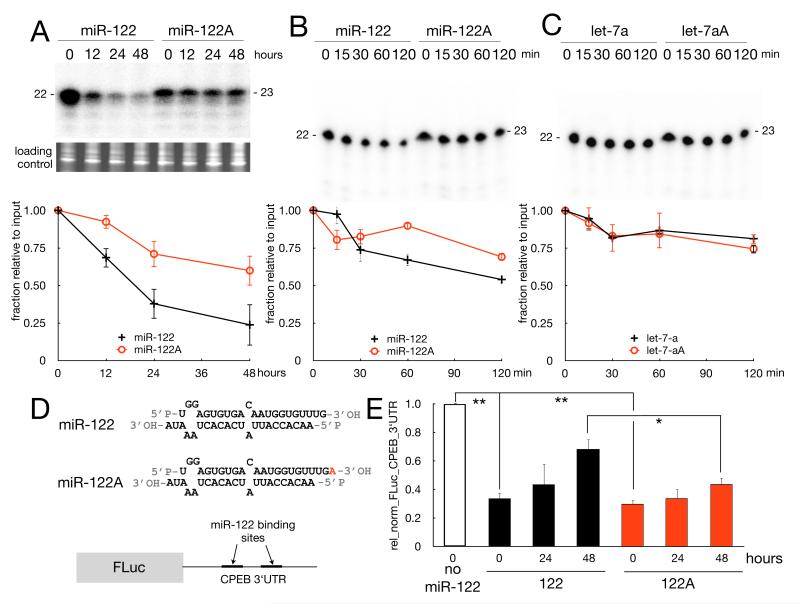
Figure 2. Monoadenylation stabilizes miR-122.
(A) Human primary fibroblasts were transfected with 32P-labeled miR-122 or miR-122A (monoadenylated) duplexes (depicted in panel D). Small RNAs were extracted at the times indicated and resolved by urea-PAGE and quantified by phosphorimaging (2 replicates, the bars represent SD). Ethidium bromide-staining of total small RNAs at each time point is shown as loading control. (B and C) 32P-labeled miR-122 and miR-122A (B) or let-7a and let-7aA (C) duplexes were incubated with extracts from human primary fibroblast for times indicated. The RNA was then extracted and resolved and quantified by urea-PAGE and phosphorimaging (2 replicates, bars refer to SD). (E) Plasmids encoding firefly luciferase mRNA appended with the CPEB 3′ UTR containing two miR-122 binding sites (illustrated in D) and Renilla luciferase were transfected into human fibroblasts alone or with miR-122 or miR-122A duplexes. Extracts prepared at the times indicated were analyzed for bioluminescence; firefly activity was normalized to that of Renilla activity. The values indicated are relative to control (no miR-122) at (t0). The error bars are SD of 3 replicates (Student t-test, *=p<0.01, **=p<0.001).
We next examined whether the monoadenylation-enhanced stability of miR-122 affects target mRNA expression. Human fibroblasts were transfected with an imperfect duplex (same as before) containing adenylated or non-adenylated miR-122 together with plasmids encoding Renilla luciferase mRNA (as a control) and firefly luciferase mRNA appended with the CPEB 3′UTR, which harbors two miR-122 sites (Figure 2D) (Burns et al., 2011). Although both non-adenylated and monoadenylated miR-122 inhibited reporter expression to about the same extent (~3 fold) at time 0 (24 hours post-transfection), by 48 hours, the monoadenylated form was about two fold more effective in inhibiting reporter expression compared to the nonadenylated form (Figure 2E). These data indicate that the enhanced stability of monoadenylated miRNA results in reduced target mRNA expression.
Gld2 depletion reduces the 3′ monoadenylation and stability of specific miRNAs
To determine whether miRNA monoadenylation and/or stability is mediated by Gld2 in vivo, we deep-sequenced libraries of miRNAs from control and Gld2-depleted fibroblasts. Of the 574 distinct miRNAs sequenced, 8.7% (n=50) contained non-templated 3′ monoadenylate residues, which generally agrees with other studies (Burroughs et al., 2010). Interestingly, 5.4% of miRNAs (n=31) contained a single 3′ uridylate residue (n=31), and 3.5% (n=20) contained either an adenylate or uridylate residue (Figure 3A). We were unable to estimate either the fraction of miRNAs that have 2 or more adenosines at their 3′ ends, or shorter species (e.g., 19 or 20 nucleotides) that harbored 3′ non-templated adenosines because the discrepancy rate between mature miRNA and genomic sequence at these positions fell within the average mutation rate (less than 0.05%) found at other miRNA nucleotide positions, making potential non-templated additions indistinguishable from sequencing errors. Upon Gld2 knockdown, 86% (n=43 out of 50) of the miRNAs that were monoadenylated in the control experienced a reduction of the monoadenylated species (on average, a 2.1-fold decrease). As a comparison, the uridylated miRNAs experienced an almost equal increase or decrease (n=14 and 16, respectively, out of 30) (on average a 1.07-fold reduction), indicating the importance of Gld2 for monoadenylation but not monouridylation (Figure 3B). Surprisingly, the percent of each miRNA species that was monoadenylated was low, and ranged from <1% to 8.4%. Even so, depletion of Gld2 causes a significant reduction (Student t test, p=0.016) in the percent of these monoadenylated species from a mean value of 2.8% to 1.5%(Figure 3C).
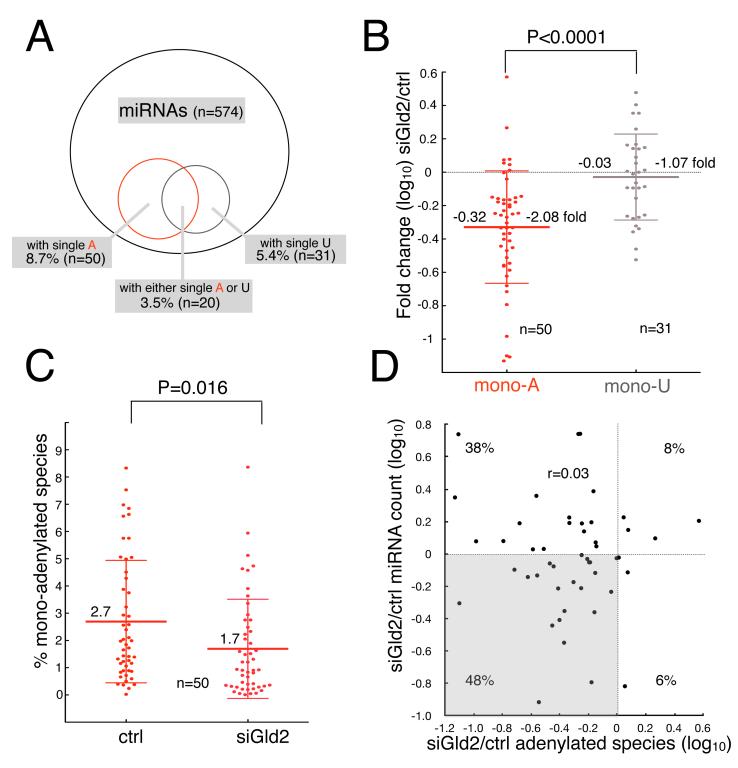
Figure 3. Gld2 depletion elicits decreased miRNA monoadenylation.
(A) Venn diagram and the percent of monoadenylated (A) or monouridylated (U) species for 574 sequenced miRNA species in human primary fibroblasts. (B) Scatter plot of the fold change (log10) of monoadenylated or monouridylated species for each miRNA. The mean reduction of monoadenylated species was 2.08 fold and of monouridylated species was 1.07 fold. The mean is indicated by the large horizontal bars and standard deviations are indicated by the smaller bars (Student’s t-test, p<0.0001). (C) Scatter plot of the percentage of monoadenylated species for each miRNA in non-targeting or Gld2 siRNA-treated human primary fibroblasts. The large horizontal bars represent mean values and the smaller bars depict standard deviations (Student’s t-test, p=0.016). (D) Scatter plot of the change (log10) in monoadenylated miRNA species versus sequencing read counts in control and Gld2-depleted fibroblasts. The shaded quadrant highlights miRNAs that have fewer monoadenylated species and whose read count decreased upon Gld2 depletion.
A plot of the ratio of the number of miRNA reads in Gld2-depleted cells to control cells versus the ratio of the monoadenylated miRNA species in Gld2-depleted to controls cells shows a Spearman r coefficient of 0.03, which implies no correlation between the two parameters (Figure 3D). This observation reveals that not all the miRNAs with fewer monoadenylated species after Gld2 depletion decrease in amount. Indeed, it may be that only discrete miRNA sub-populations are monoadenylation-sensitive, and/or that Gld2 depletion causes a change in miRNA gene transcription or processing of sufficient magnitude to override the monoadenylation stabilization effect. To assess these possibilities, we selected 14 miRNAs that decreased in amount as assessed by deep-seq read count (and confirmed by RT-qPCR, not shown) and experienced a reduction in monoadenylation upon Gld2 depletion (shaded in the lower left quadrant of Figure 3D) and measured miRNA precursor levels (both pri- and pre-); these values were then compared to mature miRNA amounts and the fraction that was monoadenylated. We found that miRNAs fell into two categories. The first category is exemplified by miR-34a and all the miRNAs in the gray box in Figure 4A. Here, Gld2 depletion that resulted in a decrease of miRNA amounts was accompanied by similar reductions of miRNA precursors (compare light gray bars of mature miRNAs to dark gray bars of miRNA precursors). Using miRNA-34a as an example, Gld2 depletion elicited a 75% reduction in monoadenylated species and a 25% reduction in the amount of mature miRNA. However, Gld2 depletion also elicited a reduction of miR34a precursor by about 20%. Thus, the nearly parallel reduction of miR-34a precursor with mature miR-34a indicates that Gld2 depletion most likely indirectly affected precursor levels by, for example, altering the translation of mRNA encoding an essential transcription or processing factor. Therefore, Gld2 depletion probably had little effect on mature miR-34a stability.
Figure 4. Monoadenylation stabilizes specific miRNAs.
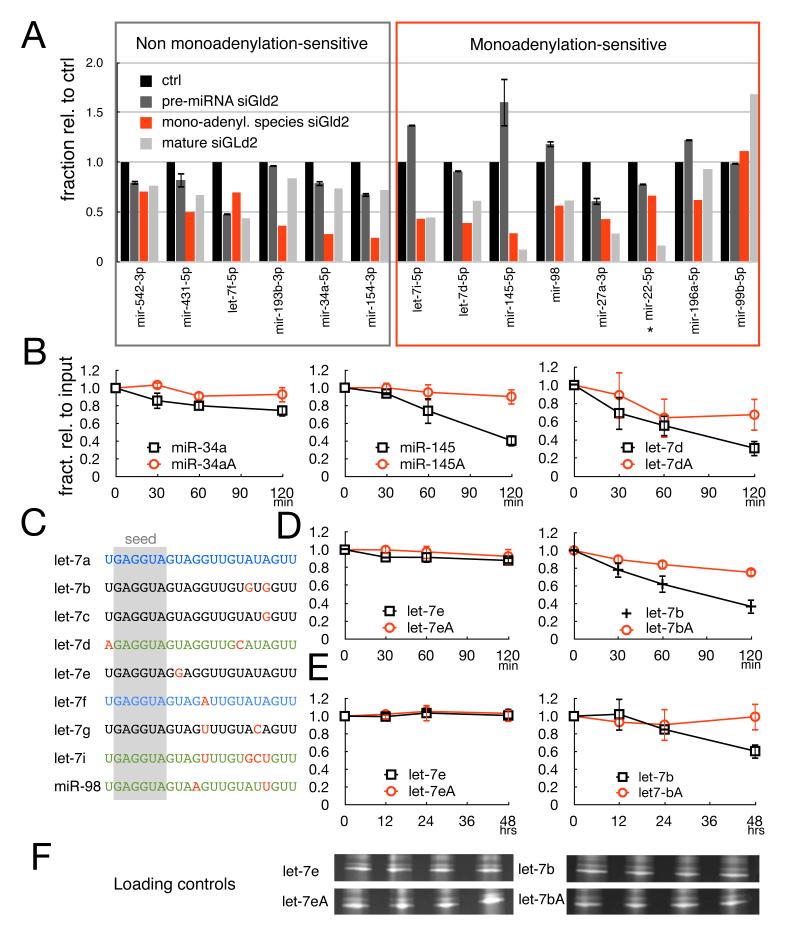
(A) Histogram comparing the changes from control levels (black bar, set at 1 in all cases) of pre-miRNAs (dark gray), mature miRNAs (light gray) and monoadenylated species (red) upon Gld2 depletion. Pre-miRNA levels were quantified by RT-qPCR and are presented as mean + SEM from 3 independent experiments. Mature miRNA levels and monoadenylated species were computed from deep-sequencing data as described in the Experimental methods. Boxed in gray are miRNAs whose changes in mature levels upon Gld-2 depletion are parallel to changes in their precursor forms. Boxed in red are miRNAs whose change in mature levels by Gld2 depletion is not reflected by parallel changes in their precursor forms. (B) Quantification of in vitro miRNA stability assays (performed and quantified as in Figure 2B) of miR-34a, miR-145, let-7d, and their corresponding monoadenylated forms. (C) Alignment of the let-7 family members; nucleotides in red are divergent from those in let-7a; miRNAs in blue are family members that are not stabilized by monoadenylation; miRNAs in green are family members that are stabilized by monoadenylation. (D) In vitro miRNA stability assays of let-7e and let-7b and their corresponding monoadenylated forms. (E) In vivo miRNA stability assays of let-7e and let-7b and their corresponding monoadenylated forms. (F) Loading controls (ethidium bromide-staining of small RNAs) corresponding to same samples denoted in panel E.
Conversely, for other miRNAs such as miR-145 and let-7d (and for all the ones in the red box in Figure 4A), mature miRNA levels reduced upon Gld2 depletion did not reflect a change in transcription and/or processing. Consider miR-145 as a specific example: Gld2 depletion elicited a ~90% reduction in mature miRNA, yet precursor levels not only did not decrease, they increased ~1.6-fold. Thus in this case, the reduction of mature miR-145 levels was accompanied only by a parallel decrease in monoadenylated miR-145, suggesting that the loss of monoadenylation was responsible for instability of the mature miRNA.
To investigate whether monoadenylation indeed mediates stability of miR-145 and let-7d, but not miR-34a, we performed in vitro assays as in Figure 2B. miR-34a was inherently stable and the addition of a 3′ monoadenylate residue had little effect on half-life. On the other hand, miR-145 and let-7d were inherently unstable, and 3′ monoadenylation enhanced their stabilities (~3.5 and ~2 fold, respectively). Taken together, these results indicate that some but not all populations of miRNAs are stabilized by 3′ monoadenylation.
Monoadenylation-mediated stabilization of specific miRNAs depends on nucleotides in the 3′ end
What distinguishes stable from unstable miRNAs, and does monoadenylation increase the half-life of the ones that are unstable? The 9 let-7 family members offer an interesting group of miRNAs with which to address these questions. The data in Figure 2C show that let-7a is stable and is unaffected by the addition of a 3′ monoadenylate residue. The data in Figures 4A and B demonstrate that let-7i, let-7d, and miR-98 are unstable and, at least for miR145 and let-7d, 3′ monoadenylation enhances stability. An alignment of all the let-7 family members shows that some such as let-7f that are not stabilized by monoadenylation share 100% identity with let-7a in the last 10 nucleotides. Conversely, members that are stabilized by monoadenylation carry single or multiple base substitutions compared to let-7a in the last 10 nucleotides (let-7d, let7i, miR-98) (Figure 4C). To test whether stabilization of miRNAs by monoadenylation could be predicted based on sequence divergence from let-7a, we analyzed additional members of the let-7 family in the in vitro stability assay. We chose let-7e, which would be predicted to be stable and unaffected by monoadenylation because it is identical to let-7a in the last 10 nucleotides, and let-7b, which has two transitions (from A to G) at positions 17 and 19 and would be predicted to be unstable but be stabilized by monoadenylation. Fig. 4D and E show that these predictions were borne out: let-7e was stable in the assay irrespective of 3′ monadenylation while the unstable let-7b was stabilized by monoadenylation both in vitro and in vivo. These results suggest that, at least for let-7 family members, specific nucleotides outside of the seed region are important for monoadenylation sensitivity and stability.
DISCUSSION
The levels of mature miRNAs are the products of tightly regulated transcription, nuclear and cytoplasmic processing, and turnover (Thomson et al., 2006). Although previous observations have shown that Gld2 depletion reduces miRNA monoadenylation in various cell types (Burroughs et al. 2010; Wyman et al. 2011), the importance of this enzyme for miRNA stability has only been inferred (Burns et al., 2011; and Katoh et al., 2009). Here, we present direct evidence that Gld2-catalyzed 3′ monoadenylation is one mechanism that controls the stability of specific mature miRNA sub-populations. Although mature miRNAs are thought to be generally stable by their partnering with Argonaute (Diederichs and Haber, 2007; Pasquinelli, 2012), recent studies indicate that their turnover is a far more complex affair. For example, miRNAs in neurons turnover much faster than in other cells; in retinal neurons, the miR-183/96/182 cluster, miR-201 and miR-211 decay with particular alacrity during dark adaptation (Krol et al., 2010), perhaps indicating that turnover rates of specific miRNAs are modified by metabolic cues. Similarly, in mouse embryo fibroblasts, several members of the miR-16 family are extraordinarily unstable, and their instability allows precise and coordinated cell-cycle transitions through de-repression of specific mRNAs (Rissland et al., 2011). With this study, we show that the same species of miRNAs are not homogenous within cells, but that sub-populations have unique stabilities dependent upon their state of 3′ monoadenylation. This stabilization effect by non-templated monoadenylation also occurs in plants, where replacement of the 3′ nucleotide with an adenine results in reduced miRNA decay (Lu et al., 2009). Clearly, monoadenylation is one of multiple combinatorial determinants of miRNA stability. Indeed, similar to our results, Rissland et al. (2011) and Bail et al. (2010) found that miRNA 3′ nucleotides are important for specific miRNA stability.
Our results also raise a number of questions concerning monoadenylation: when does monoadenylation occur during miRNA biogenesis, how does the modification increase miRNA stability, and how are specific miRNAs selected for monoadenylation? Although recent reports find mono- and oligonucleotidylation of pre-miRNAs (Heo et al., 2012), the fact that we observe modifications on miRNAs deriving from both 5′ and 3′ miRNA precursor arms implies that monoadenylation occurs after Dicer cleavage. Whether this modification occurs before or after the miRNA is loaded into RISC is an unresolved question. One model posits that miRNA stability is mediated by target mRNA and Argonaute association (Pasquinelli, 2012). Because 3′ UTRs are enriched in uridylate residues (Corà et al., 2007) it is possible that the 3′ adenylate base-pairs with a uridine, thereby reinforcing the Argonaute-miRNA-target ternary complex association and reducing accessibility to 3′->5′ exonucleases. This model could also explain the partial penetrance of the 3′ monoadenylation stabilizing effect in that other target mRNAs may not have a corresponding uridylate to which the adenylate may basepair. Recent evidence suggests that extensive miRNA-substrate complementarity activates miRNA tailing and trimming (Ameres et al., 2010); however, the prevalence and physiological relevance of this phenomenon has not yet been established. Another possibility is that monoadenylated miRNAs constitute an inactive pool of molecules that can eventually be shortened and re-activated by a still unknown trimming enzyme. This model would reconcile our finding with those of Burroughs et al. (2010) in which they observe a reduction of adenylated miRNAs in Ago2 complexes (Burroughs et al. 2010).
Because it contains no RNA-binding domains, Gld2 requires an RNA-binding partner such as CPEB or Gld3 to act on specific mRNAs (Kim and Richter, 2006; Wang et al., 2002); similarly, Gld2 could employ an as yet unidentified adaptor RNA-binding protein to recognize specific miRNAs. However, the fact that only one adenylate is added instead of a long poly(A) tail suggests that the Gld2-miRNA interaction might be an ephemeral one. Congruous with this observation is the lack of interaction between Gld2 and protein RISC components(Burroughs et al., 2010). Alternatively, monoadenylation might occur indiscriminately among miRNAs unless other molecules deny Gld2 access to certain free 3′ ends.
Interestingly, some of the miRNAs that were stabilized by Gld2 monoadenylation, such as let-7 family members and miR-145, are reduced in certain cancers (Volinia et al., 2012; Farazi et al., 2011). A gene expression meta-analysis showed that Gld2 is down-regulated in several cancers compared to healthy tissues (Rhodes et al., 2007), suggesting that the loss of this enzyme could be involved in the initiation or propagation of malignancies.
EXPERIMENTAL PROCEDURES
Monoadenylation Assays
Fibroblasts were electroporated (Amaxa, LONZA) with a Flag3X-CMV26-Gld2, inactive Gld2 (D215A), or empty expression plasmid according to the manufacturer’s instructions. Immunoprecipitation experiments were carried out following the procedure of Peritz et al. (2006) with these modifications: a) M2 anti-Flag (Sigma)-coated Dynabeads (Invitrogen) were used instead of agarose beads; b) washes with lysis buffer containing 1M urea were omitted. After the final wash, the beads were resuspended and incubated in 1X adenylation buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 25 mM MnCl2, 50 mg/mL BSA, 0.53 U/mL, 2.47 kBq/mL [α-32P]ATP (Katoh et al. 2009). After 30 min, the RNA was phenol-chloroform extracted, ethanol precipitated, and resolved by urea-PAGE. In other cases, ~5μg of GST-Gld2 was used in the same conditions as above. The radioactive signals were quantified using a Storm Phosphorimager (Amersham).
Pre-miRNA Processing Assay
Control or Gld2-depleted cells were lysed in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% NP-40, 1× COMPLETE protease inhibitors (Roche), and 10% glycerol. Fifty μg of extract was used per reaction together with in vitro T7-synthesized and α-32P-UTP labeled pre-miR-122 according to Ishizuka et al (2006). The RNA was extracted and precipitated before and after 90 min of incubation and resolved by urea-PAGE and phosphorimaging.
miRNA Stability Assays
Guide strands were T4 polynucleotide kinase (New Englad Biolabs) 5′ labeled with γ32P-ATP according to standard protocols and then precipitated and quantified. Annealing of the miRNA duplexes was carried out by incubating equimolar amounts of labeled guide with radioinert passenger strands in Tris-EDTA buffer at 95°C for 2 minutes and then used at room temperature.
For the transfection-based stability assay in Figure 2A, miRNA duplexes (~10pmol) were electroporated into nearly confluent cells; they were harvested at the times indicated and small RNAs were extracted with TRIzol (Life Technologies), precipitated and then resolved by urea-PAGE.
For the in vitro stability assays, miRNA duplexes were incubated with 110μl of master mix containing ~40pmol duplexes and 200μg of extract prepared as described previously. Twenty-five μl aliquots were removed at the times indicated, and RNA was extracted, precipitated and resolved by urea-PAGE and phosphorimaging.
Luciferase Assay
The luciferase constructs described in Burns et al. (2011) together with miR-122, miR-122A, or no miRNA were electroporated into fibroblasts according to manufacturer’s protocol (Amaxa, LONZA). The cells were harvested at the times indicated and extracts were prepared according to Dual Luciferase Assay (Promega) instructions. Luminescence was measured with an Infinite Reader (Tecan). Firefly activity was normalized to Renilla activity to control for electroporation efficiency.
Small RNA Cloning and Bioinformatic Analysis
Small RNA libraries were prepared as described in Gu et al. (2009). Human genome GRC37.65 and miRBase release 18 were used as genomic and miRNA annotation databases, respectively. Custom PERL scripts were used to “de-barcode” the sample and remove the 5′ and 3′ linkers. Bowtie (Langmead et al., 2009) 0.12.7 with parameters -v 3 -a --best --strata -m 200 was used to map deep-sequence reads to the genome. Total count linear scaling was used as a normalization method for differential miRNA analysis. Matches were assigned to miRNA loci plus 4 nucleotides upstream and downstream to allow for non-templated nucleotide addition analysis; only loci with at least 500 matched reads were considered. The mutation rates for each miRNA nucleotide position plus and minus 4 nucleotides was computed. Divergent nucleotides at position +1 over the wild-type were calculated with custom PERL scripts available upon request. Illumina sequencing is available on the GEO, accession number GSE41786.
Supplementary Material
HIGHLIGHTS.
Gld2 monoadenylates specific miRNAs.
Monoadenylation stabilizes miRNA sub-populations and prolongs their activity.
Sensitivity to monoadenylation and stability depend on nucleotides in the miRNA 3′ end.
ACKNOWLEDGMENTS
We thank members of the Richter, Mello, and Victor Ambros laboratories for discussions. Author contributions: A.D’A and J.D.R conceived the experiments; A.D’A performed the majority of the experiments; W.G. and A.D’A prepared the small RNA libraries and analyzed the deep-sequencing data; T.U. cloned, expressed, and purified GST-Gld2; A.D’A., W.G., C.C.M. and J.D.R. analyzed and interpreted the data. A.D’A prepared the figures, and A.D’A. and J.D.R. wrote the manuscript. This work was supported by NIH grant GM46779 and AG30323.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional experimental procedures are provided in the supplementary material.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung J-H, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 Are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Bartel D. MicroRNA genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, De Conti L, Stuani C, Romano M, Baralle M, Baralle F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- Burns DM, D’Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJL, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. A comprehensive survey of 3“ animal miRNA modification events and a possible role for 3” adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corà D, Di Cunto F, Caselle M, Provero P. Identification of candidate regulatory sequences in mammalian 3′ UTRs by statistical analysis of oligonucleotide distributions. BMC Bioinformatics. 2007;8:174. doi: 10.1186/1471-2105-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Horlings HM, Hoeve, Ten JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Molecular Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Hung J-H, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′; Exoribonuclease Nibbler Shapes the 3′; Ends of MicroRNAs Bound to Drosophila Argonaute1. Curr. Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon M-J, Park J-E, Kwon SC, Chang H, Kim VN. Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II let-7 MicroRNAs. Cell. 2012 doi: 10.1016/j.cell.2012.09.022. In press: http://dx.doi.org/10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S-I, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & Development. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Molecular Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. Characterizing Light-Regulated Retinal MicroRNAs Reveals Rapid Turnover as a Common Property of Neuronal MicroRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Current Biology. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks G-J, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease nibbler controls 3 prime end processing of microRNAs in Drosophila. Curr. Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. Adenylation of plant miRNAs. Nucleic Acids Research. 2009;37:1878–1885. doi: 10.1093/nar/gkp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. Non-coding RNA: MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genetics. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiríksdóttir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Hong S-J, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Molecular Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Sana ME, Wise TF, Palatini J, Huebner K, Croce CM. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.