Abstract
The Gene Ontology (GO) Consortium (GOC, http://www.geneontology.org) is a community-based bioinformatics resource that classifies gene product function through the use of structured, controlled vocabularies. Over the past year, the GOC has implemented several processes to increase the quantity, quality and specificity of GO annotations. First, the number of manual, literature-based annotations has grown at an increasing rate. Second, as a result of a new ‘phylogenetic annotation’ process, manually reviewed, homology-based annotations are becoming available for a broad range of species. Third, the quality of GO annotations has been improved through a streamlined process for, and automated quality checks of, GO annotations deposited by different annotation groups. Fourth, the consistency and correctness of the ontology itself has increased by using automated reasoning tools. Finally, the GO has been expanded not only to cover new areas of biology through focused interaction with experts, but also to capture greater specificity in all areas of the ontology using tools for adding new combinatorial terms. The GOC works closely with other ontology developers to support integrated use of terminologies. The GOC supports its user community through the use of e-mail lists, social media and web-based resources.
INTRODUCTION
The Gene Ontology (GO; http://www.geneontology.org) project is a bioinformatics resource that serves as a comprehensive source of functional information on gene products and descriptions of functions through the use of domain-specific ontologies (1). The project consists of a collaborative effort to create evidence-supported gene product annotations to structured, controlled vocabularies describing how and where gene products act. First defined in 1998, the GO has grown to become an integrated resource containing functional information for 347 778 species (including strains) covering plants, animals and the microbial world. GO’s usefulness to the community is also evident from the number of citations the inaugural GOC paper, Ashburner et al. (1), has received: 7637 as of October 2012 (Source: Thomson Reuters Web of Knowledge). The GOC distributes all annotations, vocabularies and tools freely via the Internet. In order to serve as an authoritative source for these functional annotations, the consortium has made several enhancements to its tools and resources and drawn policies to improve the consistency and currency of annotations.
GUIDELINES FOR GO ANNOTATIONS
The GO annotations are the core product of the GOC. There are two parts to a GO annotation: first, the association between a gene product and a descriptive GO definition; and second, the source and evidence used to make the link. The descriptive definitions, which represent an activity or process or location in the cell of a gene product, are given a name called the GO term, and a numerical identifier, the GO ID. Although these associations are viewed as being made to GO terms they are made to the descriptive definitions because sometimes names of biological concepts or terminology used in the literature can be ambiguous.
The source of the data is a specific reference (e.g. PMID: 20952387) that describes the experiment or analysis upon which the association was based and an evidence code such as Inferred from Mutant Phenotype (IMP), Inferred from Direct Assay (IDA) to reflect the type of study/analysis that supports the association.
There are two methods for making annotations: manually by curators and computationally by automated methods. Manual annotations are made by trained curators from a range of database groups such as Saccharomyces Genome Database (SGD), Mouse Genome Informatics (MGI) and UniProtKB (http://www.geneontology.org/GO.annotation.species_db.shtml).
This method involves reading relevant publications, identifying the gene product(s) of interest and translating the results from the study to a GO definition using an appropriate evidence code or by inferring a gene products role by manual examination of its sequence features. In contrast, automated methods predict functions of genes using a variety of criteria, but mostly by comparing their sequence to genes with similar sequence without any manual review.
As reports of biological data can be subject to interpretation, and the state of biological knowledge is constantly changing, to maintain consistency in curation, the GOC has come up with guidelines to help curators interpret experimental results and map them to the closest GO term definition possible (http://www.geneontology.org/GO.annotation.conventions.shtml). For example, loss of gene function can cause several phenotypes and often it can be challenging to determine whether a gene is directly involved in a process or is involved in an upstream process, perturbation of which results in the phenotypes examined. Similarly, it can sometimes be difficult to discern when a gene product is involved in a biological process as opposed to regulating the process. Another area in which the GOC has made improvements is the use of evidence codes. In order to point users to the original source of the data, during recent years, the GOC has made efforts to update literature-based annotations to use experimental evidence codes such as IMP or IDA as opposed to non-experimental codes such as Traceable Author Statement (TAS), typically made from Review articles. The GOC is also working closely with the developers of the Evidence Code Ontology (ECO, http://www .ontobee.org/browser/index.php?o=ECO) to formalize the representation of evidence used in making annotations, a result of which is that the evidence codes currently used by the GOC have been mapped to ECO identifiers. The next-generation gene association file format (GPAD format) will accommodate the ECO identifiers.
ANNOTATION SUBMISSION PROCEDURES AND POLICIES
GO annotations are disseminated in a standard file format called the gene_associations file (GAF 2.0). As more genomes are being sequenced, more functional annotations are being generated by both the GOC member groups and external groups and the GOC recognizes the need to collect, integrate and disseminate all annotations in a consistent and easy-to-access format. In order to streamline this process, we have formulated and published simple guidelines which are available on the GOC website (http://www.geneontology.org/GO.Submit.Annotation.shtml). Guidelines are available for groups that want to submit annotations for a small set of gene products, for an entire genome or to correct errors in existing annotations. In addition to the annotations in the standard format, the GOC also requires a file that maps the unique database identifiers supplied in participating groups’ gene association files to UniProtKB or NCBI identifiers. These ID mapping files will allow GOC to provide a robust way to search for GO annotations using external identifiers. More information on these policies is available online (http://www.geneontology.org/IDmappingFiles.shtml).
AUTOMATED VALIDATION OF ANNOTATIONS
We have created a rule engine for checking annotations, ensuring that they conform to annotation guidelines, and do not contain logical inconsistencies, such as violating a taxon constraint (2). The engine is driven by a combination of a configurable XML (http://www.geneontology.org/GO.annotation_qc.shtml) and constraints encoded directly in the ontology.
ANNOTATION PRODUCTION STATUS
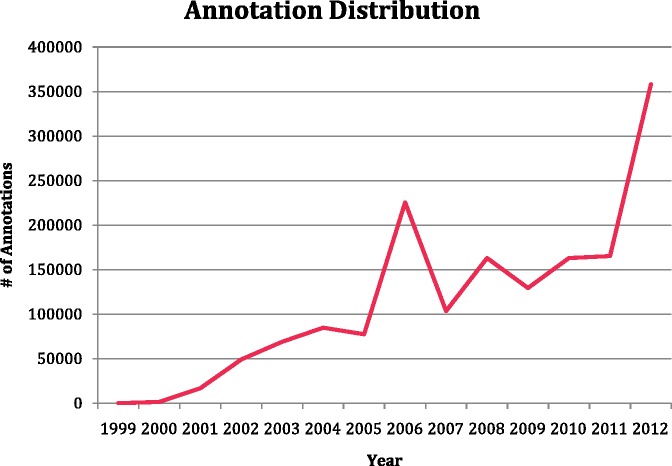
As of September 2012, there are over 96 million annotations covering 347 778 species in the GO database. Of these, 358 319 annotations were made manually (Table 1). The GOC has seen a steady increase in the number of manual annotations made by curators (Figure 1). All annotations can be queried, viewed and downloaded from AmiGO, the official web-based set of tools for searching and browsing the GO database (http://amigo.geneontology.org).
Table 1.
Status of GO as of September 2012
| Biological process terms | 23 907 |
| Molecular function terms | 9459 |
| Cellular component terms | 3050 |
| Species with annotation (includes strains) | 347 778 |
| Total annotated gene products | 96 602 850 |
| Manually annotated gene products | 358 319 |
Figure 1.
Increase in the number of manual GO annotations since 1999. Manual annotations are annotations reviewed by curators.
Phylogenetic annotation
The GO Consortium has now established a process for creating manually reviewed GO annotations using phylogenetic inference, which is described in detail elsewhere (3). A curator views all literature-based experimental GO annotations for all genes in a family, in the context of a phylogenetic tree. The curator then integrates all of this information into a curated, consistent model of function evolution, which in turn is used to automatically predict GO annotations for unannotated family members. These annotations can be identified by their use of the new “IBA” (inferred from biological aspect of ancestor) evidence code. For users familiar with annotations using the “ISS” (inferred from sequence similarity) evidence code, these new IBA annotations can be thought of as similar in spirit, though generally based on many pieces of evidence (multiple annotations across a number of related genes, a phylogenetic tree showing all evolutionary relationships, and a multiple sequence alignment) rather than an isolated pair of related gene products and one particular annotation. As a result, these IBA annotations are in general more confident annotations, and we encourage users to take advantage of them. We have also found that this phylogenetic annotation process improves the quality of experimental GO annotations, as it has identified erroneous experimental annotations that were subsequently removed or corrected. Finally, this process creates manually reviewed GO annotations for a number of unannotated genomes. Currently IBA annotations exist for the 48 species in PANTHER version 7.2 (4) and will soon be extended to an additional 34 species (see http://www.ebi.ac.uk/reference_proteomes/ for complete list). As of October 2012, the GO database contained 25,195 annotations with the IBA evidence code, covering 3,223 genes in 47 species. This number will be rapidly increasing over the next few years.
DEVELOPMENTS IN AmiGO
To give broad access to the annotations and the ontologies, several enhancements have been implemented in AmiGO. AmiGO allows users to browse, visualize, filter, and download ontology and annotation data. Since the ontologies are growing in size, being able to view the relationships and terms in a meaningful way has become a challenge. To address this issue, several alternate displays of the ontology have been implemented in AmiGO. Ancestor and children terms of a query term can be viewed in a tabular format in a tree-like view, or in multiple graph views (SVG or PNG). Annotations can be accessed either by navigating through a GO term or by entering a gene product identifier into the search box. An Advanced Search form is also available to upload a list of gene product identifiers or GO terms and to constrain the search by a variety of filters such as species, data source or evidence code. Work is underway to provide full access to all the GO annotations for all the species through AmiGO.
IMPROVED GO TOOLS REGISTRY
The main GO website (http://www.geneontology.org/GO.tools.shtml) provides a catalog of software applications that make use of the GO, including term enrichment tools. We have improved this catalog by making use of the Neuroscience Information Framework (NIF) registry system (5). We now have 130 tools in the registry (including those developed by external groups), and provide more extensive metadata on each tool, thanks to the NIF curators.
ONTOLOGY DEVELOPMENT
To support the creation of annotations, we focused on a number of interest areas for targeted ontology development and we have improved the rigor with which we can verify and query the ontology computationally as described below.
Representing Biological Concepts in the Ontologies
The GOC continues to work with experts in the research community to expand and refine areas of the ontology. Typically this process involves face-to-face meeting with the community experts and the ontology developers to discuss how best to represent biology in the ontology. The ontology developers then make changes to the ontology (edit term strings and definitions, add or delete terms) as appropriate to reflect the current state of knowledge in that area of biology. In the last year, ontology development focused on apoptosis and cardiac conduction. A meeting with cardiac experts and the annotation group at the British Heart Foundation-University College London, (http://wiki.geneontology.org/index.php/Cardiac_conduction), resulted in the expansion of the cardiac conduction portion of the ontology to include 63 terms that represent processes and functions that contribute to the regulation of force and strength of heart contraction. We have also begun a significant top-down overhaul of the apoptosis branch of the ontology.
The Apoptosis GO project stems from a dual need: a thorough revision of existing apoptosis-related terms in GO, and an expansion of the ontology with new terms to fully capture recent biological knowledge on apoptosis. The ontology work will be followed by a significant annotation and re-curation effort to increase the breadth (number of gene products) and depth (granularity of the GO term) of apoptosis-related GO annotations. The project was initially supported by the APO-SYS Consortium (Apoptosis Systems Biology Applied to Cancer and AIDS, http://www.apo-sys.eu/), and took advantage of direct interaction with expert researchers. At the time of writing, 75 existing apoptosis GO terms have been revisited and 60 new GO terms have been created following discussion with experts. The top-level apoptosis ontology node has been split to better represent different sub-processes of the apoptotic process (i.e. signaling phase, execution phase, changes occurring in the mitochondrion and in other sub-cellular compartments, etc.). The GO now includes specific terms to represent, e.g. apoptosis following signaling through dependence receptors, and caspase activities involved at various steps of the apoptotic process.
Use of the Web Ontology Language
The Web Ontology Language (OWL) is a standard provided by the World Wide Web Consortium (W3C) for representing ontologies and ontology-related information. OWL is now a crucial component of the GO internal infrastructure, and provides many advantages to bioinformatics users of the GO, including standardized Application Programmer Interfaces (APIs) such as the OWLAPI (6), standardized means of persistent storage and querying in RDF triplestores, and fast, powerful reasoners. In order to take advantage of the features of OWL2, we worked with members of the Open Biological Ontologies (OBO) community (7) to create a new version of the mapping between the GO ontology format (OBO) and OWL (http://oboformat.org), and a new java implementation of the obo2owl converter. The OWL2 version of the GO is available from a standard OBO library URL (http://purl.obolibrary.org/obo/go.owl). We will continue to provide the ontology in OBO format, but encourage users to consume the OWL version, and to take advantage of the increasing range of powerful tooling available for this language.
Logical Definitions
The GO is traditionally thought of as a Directed Acyclic Graph (DAG), with each term in the ontology represented as a node (1). OWL extends this model with more expressive constructs, including the ability to provide computable logical definitions for GO terms, making use of both simpler GO terms, and terms from other ontologies such as CHEBI (chemical entities of biological interest) (8). This provides additional expressive query capabilities for external users of the GO, and allows the developers of GO to make use of automated validation and classification procedures (9). Over the past year, ontology editors have continued to add computable logical definitions to GO terms. As of 12 September 2012 there are 12 597 terms in GO (35%) that have validated logical definitions. This includes 8249 terms that reference existing GO terms, and a further 4348 that reference CHEBI terms. The connections between GO and CHEBI are now complete for many part of GO, including metabolism, transport, response to stimulus and homeostasis. In addition to this fully validated set of logical definitions, we have partially validated logical definitions that make use of terms from other ontologies, such as the Cell Ontology (10) the Plant Anatomy Ontology (11) and the Uberon multi-species metazoan anatomy ontology (12). The full set is available from the GOC wiki (http://wiki.geneontology.org/index.php/Ontology_extensions). The GOC continues to work with phenotype, cell type, and anatomy ontology developers, among others, to support alignments and intersections between community biomedical ontologies.
TermGenie
We previously described our web-based term generation system TermGenie (13). We have re-implemented this system on top of the OWL API, and have created 11 standardized templates that allow instant creation of terms for GO users—this service is available from http://go.termgenie.org. This takes advantage of the logical definitions and OWL reasoning to automatically place terms in the ontology. For example, an annotator who requires a term to describe the transport of a particular molecule simply selects the ‘transport’ template, and chooses a term from CHEBI.
Automated Ontology Validation
All modifications to the ontology trigger the GO continuous integration server to launch an extensive validation procedure, including consistency checking using powerful OWL reasoners such as HermiT (14) and Elk (15). This same server is used to perform validation on the annotations, allowing ontology editors to see the impact of changes to the ontology upon annotations.
FUTURE DEVELOPMENTS
GOC will be switching to a SubVersioN (SVN) system from CVS to manage the dessimination of the ontology and annotation file formats.
GOC is working on implementing InterMine, an open source data warehouse system with a sophisticated querying interface to create GOMine. GOMine will serve as a fast and flexible data retrieval tool with custom search options and download capabilities.
Ontology development in the area of cell-cycle is in progress.
A new annotation model is being developed to increase the expressivity (richness) of GO annotations.
ACCESSING DATA AT THE GO CONSORTIUM
GO Consortium—http://www.geneontology.org
AmiGO, the primary web application that provides access to the annotations and ontologies—http://amigo.geneontology.org
Annotations, ontologies and other relevant files can be downloaded from the main GO website—http://www.geneontology.org/GO.downloads.shtml
Documentation on the Gene Association file format 2.0 can be found at—http://www.geneontology.org/GO.format.gaf-2_0.shtml
Documentation on several ongoing projects can be found on the consortium Wiki—http://wiki.geneontology.org/
Contact GOC at: go-helpdesk@ebi.ac.uk
FUNDING
National Human Genome Research Institute (NHGRI) [U41HG002273 to the Gene Ontology Consortium, 1U41HG006104-03 to the European Bioinformatics Institute]; British Heart Foundation [SP/07/007/23671]. Funding for open access charge: NHGRI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The GO acknowledges the annotation effort from the annotators at: Gramene, Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, USA; The J. Craig Venter Institute, Rockville, MD, USA; PAMGO, Wells College, Aurora, NY, USA and PAMGO, Virginia Bioinformatics Institute, VA, USA; AspGD, Stanford, CA, USA; CGD, Stanford, CA, USA; Sanger GeneDB, Hinxton, UK; InterPro EBI, Hinxton, UK; IntAct, EBI, Hinxton, UK; seudoCAP, British Columbia, Canada; SGN, Ithaca, NY, USA.
Appendix
J.A. Blake, M. Dolan, H. Drabkin, D.P. Hill, Li Ni, D. Sitnikov (MGI, The Jackson Laboratory, Bar Harbor, ME, USA); S. Bridges, S. Burgess, T. Buza, F. McCarthy, D. Peddinti, L. Pillai (AgBase, Mississippi State University, MS, USA); S. Carbon, H. Dietze, A. Ireland, S.E. Lewis, C.J. Mungall (BBOP, LBNL, Berkeley, CA, USA); P. Gaudet, R.L. Chisholm, P. Fey, W.A. Kibbe, S. Basu (dictyBase, Northwestern University, Chicago, IL, USA); D.A. Siegele, B.K. McIntosh, D.P. Renfro, A.E. Zweifel, J.C. Hu (EcoliWiki, Departments of Biology and Biochemistry and Biophysics, Texas A&M Univ., College Station, TX, USA); N.H. Brown, S. Tweedie (FlyBase, Gurdon Institute and Department of Genetics, University of Cambridge, Cambridge, UK); Y. Alam-Faruque, R. Apweiler, A. Auchinchloss, K. Axelsen, B. Bely, M-C. Blatter, C. Bonilla, L. Bougueleret, E. Boutet, L. Breuza, A. Bridge, W.M. Chan, G. Chavali, E. Coudert, E. Dimmer, A. Estreicher, L. Famiglietti, M. Feuermann, A. Gos, N. Gruaz-Gumowski, R. Hieta, U. Hinz, C. Hulo, R. Huntley, J. James, F. Jungo, G. Keller, K. Laiho, D. Legge, P. Lemercier, D. Lieberherr, M. Magrane, M.J. Martin, P. Masson, P. Mutowo-Muellenet, C. O’Donovan, I. Pedruzzi, K. Pichler, D. Poggioli, P. Porras Millán, S. Poux, C. Rivoire, B. Roechert, T. Sawford, M. Schneider, A. Stutz, S. Sundaram, M. Tognolli, I. Xenarios (UniProtKB: EBI, Hinxton, UK and SIB, Swiss Institute of Bioinformatics, Geneva, Switzerland and PIR, Georgetown, USA); R. Foulger, J. Lomax, P. Roncaglia (GO-EBI, Hinxton, UK); V.K. Khodiyar, R.C. Lovering, P.J. Talmud (Institute of Cardiovascular Science, University College London, London, UK); M. Chibucos, M. Gwinn Giglio (Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD, USA); H-Y. Chang, S. Hunter, C. McAnulla, A. Mitchell, A. Sangrador (InterPro, EBI, Hinxton, UK); R. Stephan, (MTBBASE, Berlin); M.A. Harris, S.G. Oliver, K. Rutherford, V. Wood (PomBase, University of Cambridge, Cambridge, UK); J. Bahler, A. Lock (PomBase, University College, London, UK); P.J. Kersey, M.D. McDowall, D.M. Staines (PomBase, EBI, Hinxton, UK); M. Dwinell, M. Shimoyama, S. Laulederkind, T. Hayman, S.-J. Wang, V. Petri, T. Lowry (RGD, Medical College of Wisconsin, Milwaukee, WI, USA); P. D’Eustachio, L. Matthews (Reactome, Department of Biochemistry, NYU School of Medicine, New York, NY, USA); R. Balakrishnan, G. Binkley, J.M. Cherry, M.C. Costanzo, S.S. Dwight, S.R. Engel, D.G. Fisk, B.C. Hitz, E.L. Hong, K. Karra, S.R. Miyasato, R.S. Nash, J. Park, M.S. Skrzypek, S. Weng, E.D. Wong (SGD, Department of Genetics, Stanford University, Stanford, CA, USA); T.Z. Berardini, D. Li, E. Huala (TAIR, Department of Plant Biology, Carnegie Institution for Science, Stanford, CA, USA); H. Mi, P. D. Thomas (USC, Los Angeles, CA); J. Chan, R. Kishore, P. Sternberg, K. Van Auken (WormBase, California Institute of Technology, Pasadena, CA, USA); D. Howe, M. Westerfield (ZFIN, University of Oregon, Eugene, OR, USA).
REFERENCES
- 1.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deegan née Clark JI, Dimmer EC, Mungall CJ. Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development. BMC Bioinform. 2010;11:530. doi: 10.1186/1471-2105-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudet P, Livestone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;5:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis SE, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology. Consortium. 2010:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta A, Bug W, Marenco L, Qian X, Condit C, Rangarajan A, Muller HM, Miller PL, Sanders B, Grethe JS, et al. Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF) Neuroinformatics. 2008;6:205–217. doi: 10.1007/s12021-008-9033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horridge M, Bechhofer S. The OWL API: a Java API for OWN ontologies. Semantic Web. 2009;2:11–21. [Google Scholar]
- 7.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotech. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degtyarenko K, Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mungall CJ, Bada M, Berardini TZ, Deegan J, Ireland A, Harris MA, Hill DP, Lomax J. Cross-product extensions of the Gene Ontology. J. Biomed. Inform. 2011;44:80–86. doi: 10.1016/j.jbi.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meehan T, Masci AM, Abdulla A, Cowell L, Blake J, Mungall CJ, Diehl A. Logical development of the cell ontology. BMC Bioinform. 2011;12:6. doi: 10.1186/1471-2105-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls RL, Athreya B, Cooper L, Elser J, Gandolfo MA, Jaiswal P, Mungall CJ, Preece J, Rensing S, Smith B, et al. Ontologies as integrative tools for plant science. Am. J. Bot. 2012;99:1263–1275. doi: 10.3732/ajb.1200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gene Ontology Consortium. The Gene Ontology: enhancements for 2011. Nucleic Acids Res. 2012;40:D559–D564. doi: 10.1093/nar/gkr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shearer R, Motik B, Horrocks I. Proceedings of the 5th International Workshop on OWL: Experiences and Directions. 2008. HermiT: a highly-efficient OWL reasoner. [Google Scholar]
- 15.Yevgeny K, Markus K, František S. ELK Reasoner: architecture and evaluation. In: Horrocks I, Yatskevich M, Jimenez-Ruiz E, editors. Proceedings of the 1st International Workshop on OWL Reasoner Evaluation (ORE-2012) 2012. CEUR Workshop Proceedings 2012. [Google Scholar]