Abstract
The aryl hydrocarbon receptor (AHR) mediates toxic effects of dioxin and xenobiotic metabolism. AHR has an emerging role in the immune system but its physiological ligands and functional role in immunocytes remain poorly understood. Mast cells are immunocytes that are central to inflammatory responses and release a spectrum of pro-inflammatory mediators including histamine, mast cell proteases, and pro-inflammatory cytokines such as IL-6 upon stimulation. Our aim was to investigate the AHR in model mast cells and examine how both putative and known AHR ligands, e.g., kynurenine, kynurenic acid (KA), Resveratrol, indolmycin, and violacein, affect mast cell activation and signaling. We tested these ligands on calcium signaling, degranulation, and gene expression. Our data show that AHR is present in three model mast cell lines, and that various known and putative AHR ligands regulate gene expression of Cyp1a1, a gene down-stream of AHR. Furthermore, we found that calcium influxes and mast cell secretory responses were enhanced or suppressed after chronic treatment with AHR agonists or antagonists, and that AHR ligands modified RBL2H3 cell degranulation. AHR ligands can chronically change cytokine gene expression in activated mast cells, as exemplified by IL-6. The antagonist Resveratrol repressed expression of induced IL-6 gene expression. Though KA and kynurenine are both AHR agonists, these ligands behaved differently in regards to degranulation and IL-6 expression, indicating that they may function outside of AHR pathways. These data suggest considerable complexity in RBL2H3 responses to AHR ligands, with implications for our understanding of both dioxin pathology and the immunological effects of endogenous AHR ligands.
Keywords: mast cells, calcium, aryl hydrocarbon receptor, resveratrol, kynurenine
Introduction
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor (Hankinson, 1995; DiNatale et al., 2010a). Upon ligand binding, cytosolic AHR translocates to the nucleus, dimerizes with the AHR nuclear translocator, and interacts with the xenobiotic-responsive element (XRE). AHR has high affinity for 2,3,7,8-tetrachlordibenzo-p-dioxin (TCDD) and regulates detoxification/metabolism enzymes such as CYP1A1 (Poland, 1982; DiNatale et al., 2010a). However, the AHR pathway has diverse physiological roles outside of its toxicological importance, including regulation of immunity and inflammation. It is expressed in immunocytes (Kerkvliet, 2009). There are numerous immune-related genes with dioxin response elements (Takamura et al., 2011). Moreover, AHR ligands may have a wider impact on immune cell signaling through a ‘non-genomic’ pathway (Li and Matsumura, 2008; Matsumura, 2009). Here, AHR ligands can cause an increase in intracellular free calcium levels, implying the modulation of a diverse set of calcium-dependent transcriptional targets and effector functions (Matsumura, 2009).
AHR ligands are structurally diverse (DiNatale et al., 2010a). While most research on the AHR has been performed with TCDD, the endogenous ligand to AHR remains unknown. Proposed AHR ligands include Resveratrol (MacPherson and Matthews, 2010), indirubin (Adachi et al., 2004), bilirubin (Sinal and Bend, 1997), and indigo (Pocar et al., 2005). Within their structural diversity, many AHR ligands are tryptophan-derived compounds (Mezrich et al., 2010). Thus, products generated from the indoleamine-2,3-dioxygenase (IDO) pathway have been considered as potential AHR ligands. Of these, kynurenic acid (KA) (DiNatale et al., 2010a) and kynurenine (Mezrich et al., 2010) were shown to be AHR ligands. Identifying new non-toxic ligands to AHR and characterization of their effects will add insight into the physiological role of AHR, and may identify new immunomodulating compounds.
While recent evidence supports a role for the AHR in chronic immune responses, including inflammation, the AHR has not been studied in mast cells. Mast cells are central to inflammation, releasing pro-inflammatory mediators including histamine, bioactive lipids, serotonin, as well as chemokines and cytokines, including interleukin (IL)-6 (Galli and Tsai, 2008). Ligand-activated AHR coupled with inflammatory signals have recently been shown to synergistically induce IL-6 expression in human MCF-7 tumor cells (DiNatale et al., 2010a,b). This linkage between AHR and IL-6 is intriguing. Along with the documented ability in other systems for AHR ligands to induce calcium responses, it seems reasonable to propose that endogenous and exogenous AHR ligands may regulate mast cell function.
Here, we examined AHR pathways in RBL2H3 cells. We evaluated three known AHR ligands: an antagonist Resveratrol, the agonists KA and kynurenine, together with a panel of potential AHR ligands. We sought to examine how these compounds modulate mast cell secretion, calcium responses, and the production of IL-6. Our data suggest that AHR ligands affect mast cells through either the genomic or ‘non-genomic’ pathways. Moreover, calcium influxes are enhanced by AHR agonists or suppressed after chronic treatment with AHR antagonists. The AHR ligands have highly ligand-specific effects on gene expression, degranulation, and calcium influxes. These findings suggest that both exogenous and endogenous ligands of AHR may modulate mast cell function using diverse signaling pathways of considerable complexity.
Materials and Methods
Cell culture
RBL-2H3 cells are a rat basophilic leukemia cell line (American Type Culture Collection, Bethesda, MD). They have been extensively used as a model that recapitulates many features of mucosal mast cells and basophils (Passante et al., 2009). RBL-2H3 cells were maintained at 37°C in a 95% air/5% CO2 humidified atmosphere in Dulbecco’s Modified Eagle Medium (DMEM; Mediatech Inc., Herndon, VA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 2 mM L-glutamine (both Mediatech). C57.1 murine mast cells (a generous gift from Dr. Steve Galli, Stanford University) and the P815 cell line (these were used only as an expression comparison in Figure 1) were grown in RPMI (Mediatech) containing 10% FBS, 2 mM L-glutamine, 2 mM non-essential amino acids (NEAA), and 1 mM sodium pyruvate. In addition, 50 mM 2-mercaptoethanol and 5 ng IL-3/ml were added fresh to each passage for C57.1. RBL2H3 and C57.1 lines were kept below passage 40 and were regularly checked for their degranulation abilities.
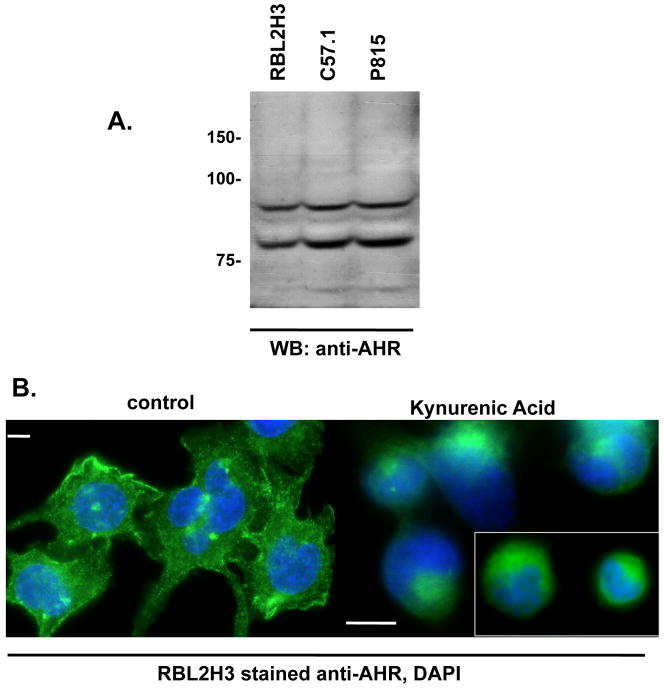
Figure 1. Model mast cells express the AHR.
(A) Lysates of RBL2H3, C57.1 and P815 were resolved by SDS-PAGE (2 × 106 cells per lane). Western blot analysis was then performed with anti-AHR (0.1 μg/ml) antibody. The predicted MW of AHR is 96 kDa. (B) Immunofluorescence analysis of sub-cellular localization of AHR in untreated (left panel) and KA-treated (1 hr, 10μM, right panel) RBL2H3 cells. Scale bars are 5 μm. Inset right panel is used to show cells from a different field.
Chemicals and Reagents
Reagents used in this study were the polyclonal rabbit anti-AHR antibody from Abcam (Cambridge, MA), anti-Grb-2 from Cell Signaling Technology (Danvers, MA), Kynurenic acid (KA), L-Kynurenine, Resveratrol, and Violacein from Sigma (St. Louis, MO), and Indolmycin from Bioaustralis (Smithfield, Australia). Di-nitrophenol-conjugated keyhole limpet hemocyanin (KLH-DNP), PMA (phorbol myristate 12,13 acetate), ionomycin and other miscellaneous chemicals were from Sigma. Fluo-4 was from Molecular Probes (Eugene, OR).
Cell Lysis and Western blot
Cells (≈ 107) were lysed on ice for 30 min by addition of 350 μl lysis buffer (50 mM HEPES [pH 7.4], 250 mM NaCl, 20 mM NaF, 10 mM iodoacetamide, 0.5% [w/v] Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 500 mg aprotinin/ml, 1.0 mg leupeptin/ml, and 2.0 mg chymostatin/ml). The lysates were then centrifuged at 17,000 × g for 20 min. For preparation of total protein, lysates were acetone-precipitated (1.4 vol acetone for 1 hr at −20°C and then centrifuged at 10,000 × g for 5 min). Protein samples were resolved by 10% SDS-PAGE under reducing conditions in a modified Laemmli buffer. All SDS-PAGE gels contained lysate from 106 cells per lane. The resolved proteins were then electrotransferred to PVDF membranes in 192 mM glycine/25 mM Tris (pH 8.8) buffer.
For Western blotting, the membranes were blocked in 5% (weight/volume) non-fat milk in phosphate-buffered saline (PBS, pH 7.4) for 1 hr at room temperature (RT). Primary antibodies were dissolved in PBS/0.05% Tween-20/0.05% NaN3 and incubated with the membranes for 16 hr at 4°C. Anti-rabbit (for AhR Westerns) or anti-mouse (for Grb2 Westerns) IgG conjugated to horseradish peroxidase (Amersham, Piscataway, NJ) were used as secondary antibodies and diluted to 0.01 μg/ml in PBS/0.05% Tween-20 and incubated with the membranes for 45 min at RT. A standard washing protocol (four washes of 5 min each in 50 ml PBS/0.1% Tween-20 at RT) was employed between primary and secondary antibodies and following the secondary antibody. Signal was visualized using enhanced chemiluminescence (Amersham) and exposure to Kodak BioMax film (Sigma).
Immunofluorescence
RBL-2H3 (seeded at 1 × 104 cells/cm2) were grown on glass coverslips and stimulated with 10 μM KA for 1 hr. After fixation in 0.4% paraformaldehyde, cells were permeabilized (0.4% Triton X-100 for 4 min) and blocked with 0.75% fish-skin gelatin before staining with 0.1μg/ml anti-AHR and Alexa-488 conjugated anti-rabbit secondary antibody (Invitrogen, Carlsbad, CA). Nuclei were visualized with DAPI (Molecular Probes).
Degranulation assessment using a β-Hexoseaminidase assay
RBL-2H3 mast cells were plated in 12-well cluster plates (CELLSTAR, Greiner Bio-one, Monroe, NC) at 105 cells/well. Where indicated, cells were pre-incubated with various ligands prior to induction of degranulation with either pharmacological or antigenic stimuli. Culture supernatants were then analyzed (see below). In an alternate protocol, degranulation responses were induced using stimuli and acute addition of AHR ligands was performed to assess the possibility of post-effects. Cells were incubated in 230 μl Tyrode’s buffer. After 30 min at 37°C, 25 μl supernatant was removed, clarified by micro-centrifugation, and transferred to a 96-well plate containing 100 μl (per well) p-NAG (1 mM p-N-acetyl glucosamine [Sigma] in 0.05 M citrate buffer [pH 4.5]) substrate solution. After 1.5 hr at 37°C, reactions were quenched by addition of 100 μl 0.2 M glycine (pH 9.0) to each well. β-Hexoseaminidase levels were read at OD = 405 nm in a Biorad Benchmark plate reader (BioRad, Hercules, CA). Results are reported as the mean (± SD) of triplicate samples.
Calcium assay
To investigate chronic effects of compounds on calcium influx, adherent RBL2H3 cells were pre-incubated for another 24 hr with AHR ligands or vehicle. In an alternate protocol, AHR ligands were added during ongoing calcium responses to assess possible post effects. Untreated or treated RBL2H3 cells were washed and incubated with 1 μM Fluo-4 for 30 min at 37°C in a standard modified Ringer’s solution (145 mM NaCl, 2.8 mM KCl, 10 mM CsCl, 10 mM CaCl2, 2 mM MgCl2, 10 mM glucose, and 10 mM HEPES·NaOH [pH 7.4]). Cells were then transferred to a 96-well plate (at 50,000 cells/well) and stimulated using pharmacologic (ionomycin) or antigenic stimuli as described above. Calcium signals were then acquired using a Flexstation 3 (Molecular Devices, Sunnydale, CA). All data were analyzed using the software SoftMax® Pro 5 (Molecular Devices) and GraphPad Prism 5 (GraphPad, La Jolla, CA).
Cell stimulation and RNA extraction
RBL-2H3 cells were plated in 6-well plates (5 × 105 cells/well) and pre-incubated for 3, 8, or 24 hr with AHR ligands as indicated. Alternately, RBL2H3 were pre-incubated for 1 hr and then subsequently stimulated with pharmacologic or antigenic stimuli as described above. RNA was then extracted using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), verified using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA), and total concentration determined using NanoVue (Healthcare Biosciences, Uppsala, Sweden).
Quantitative Real Time PCR Analysis
cDNA was synthesized from 600 ng of total RNA using the High-Capacity cDNA Transcriptase Kit (Applied Biosystems, Foster City, CA) on the DNA Engine Tetra (MJ Research, PTC-225, Peltier Thermal Cycler, Watertown, NY) according to manufacturer instructions. TaqMan Gene Expression Assays were purchased (Applied Biosystems) for: IL-6 (Assay ID Rn01410330_m1), Cyp1a1 (Assay ID Rn00487218_m1), and β-actin (Assay ID Rn00667869_m1). Amplifications were carried out in a total volume of 10 μl after optimizing the protocol containing TaqMan Fast Universal Master Mix, TaqMan Gene Expression Assay (Applied Biosystems), and purified target cDNA. Cycling parameters were initiated by 30 sec at 94°C, followed by 40 cycles with denaturation at 94°C for 3 sec, and annealing and extension at 60°C for 30 sec using StepOnePlus (Applied Biosystems). Amplifications were performed in triplicate and were normalized to the expression of the β-actin coding gene. Relative transcript levels were calculated applying the 2−ΔΔCt method described by Schmittgen and Livak (2008).
Data analysis
Results are presented as the mean ± SD. Statistical significance was determined based on one-way ANOVA followed by a Tukey’s multiple-comparison test to compare samples using GraphPad Prism v5.0 (GraphPad Software, Inc., La Jolla, CA). Adjacent to datapoints in the respective graphs, significant differences were indicated by: *p < 0.05, **p < 0.01, ***p < 0.001, and p > 0.05. The number of independent experiments is indicated in the Figure Legends. Where error bars are not presented, the data shown are representative of three independent experiments.
Results
RBL-2H3 and C.57.1 cells express the aryl hydrocarbon receptor
The aryl hydrocarbon receptor (AHR) is a 96kDa protein, which has been detected in diverse tissue and cells. Using Western blots (Figure 1A), we show that the AHR is also expressed in RBL2H3, P815, and C57 cells. Figure 1B shows positive anti-AHR immunofluorescence in resting RBL2H3 cells. Upon treatment for 1 hr with the AHR agonist KA, an increased degree of perinuclear staining of AHR is observed.
AHR ligand validation in mast cells
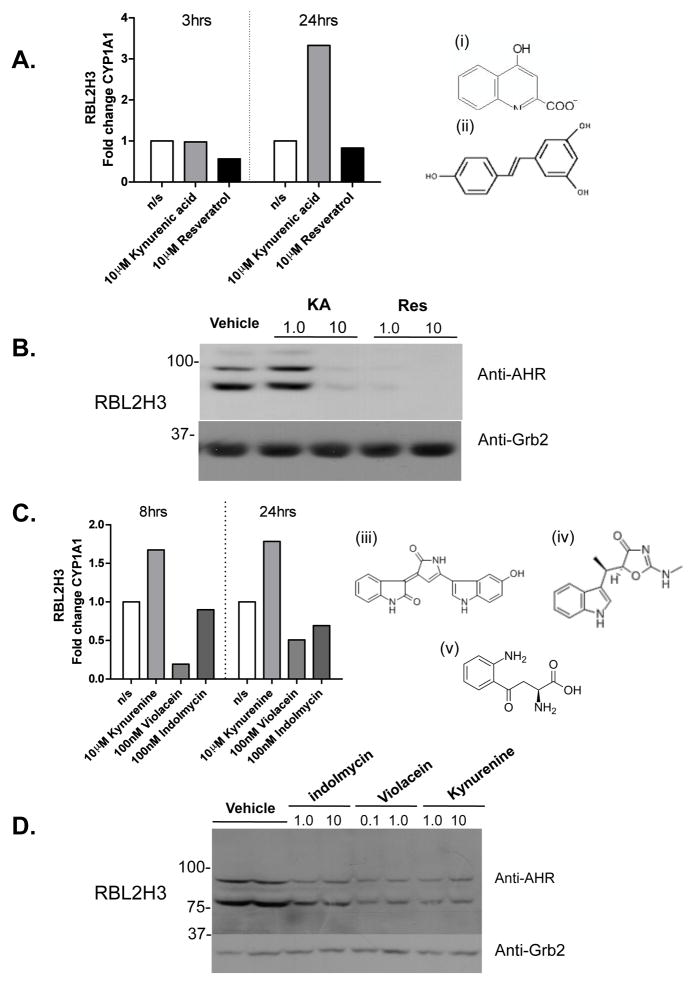
We evaluated known and putative AHR ligands for their effects in RBL2H3 cells. First, the known AHR ligands KA (an agonist) and Resveratrol (an antagonist) were tested for their ability to regulate Cyp1a1 transcription. As expected, these ligands cause up- and down-regulation of Cyp1a1 expression, respectively (Figure 2A); the structures of KA (i) and Resveratrol (ii) are illustrated in Figure 2A. Here, as in subsequent experiments, the effects are highly time-dependent. For example, Resveratrol causes a down-regulation of Cyp1a1 transcripts at 3 hr that resolves by 24 hr. In contrast, the stimulatory effect of KA is not apparent until 24 hr. In addition, we asked whether exposure to these ligands causes chronic down-regulation of AHR levels, a documented response to TCDD (Steenland et al., 2004). Figure 2B shows that AHR protein levels are down-regulated after 72 hr of exposure to both agonist KA and antagonist Resveratrol. These data suggest that the AHR shown in Figure 1A is functional in RBL-2H3 cells and can couple to transcriptional pathways similar to those seen in other immunocytes.
Figure 2. Effect of AHR ligands on CYP1A1 mRNA and AHR protein levels in RBL2H3 cells.
(A) RBL2H3 were seeded at 5 × 105 cells/well, grown for 16 hr, and then stimulated with 10 μM KA or Resveratrol; control wells received vehicle only. mRNA was then isolated from the cells after 3 or 24 hr of exposure and CYP1A1 transcript levels were measured by quantitative PCR; all signals were normalized to the relative expression of β-actin. Relative transcript levels were calculated using a 2−ΔΔCt method. Data shown represents one of three independent experiments. (i), (ii) Structures of KA and Resveratrol. (B) RBL-2H3 cells were treated for 72 hr with the indicated concentration (in μM) of KA or Resveratrol. Western blotting for either AHR or Grb2 (as loading control) was then performed. Figure shown is representative of one of at least three independent experiments. (C) Same experiment as in (A), but with cells harvested at 8 and 24 hr after exposure to the indicated concentrations of kynurenine, violacein, or indolmycin. Data are representative of one of three similar experiments. Structures for (iii) indolmycin, (iv) kynurenine, and (v) violacein. (D) Same experiment as in (B), but using the indicated concentrations (in μM) of kynurenine, violacein, or indolmycin. MW markers shown are in kDa.
We took advantage of the AHR system in RBL2H3 cells to explore several marine bacteria-derived compounds that bear structural similarity to proposed endogenous AHR ligands. Recent studies revealed that the AHR binds to endogenous compounds from the tryptophan oxidation (IDO) pathway (Ball et al., 2009). This has led to the hypothesis that other tryptophan derivatives are potential ligands for the AHR. We identified two tryptophan-derived compounds (Figure 2C, violacein [iii] and indolmycin [iv]) that are synthesized in the marine bacterium Pseudoalteromonas luteoviolacea (Mansson et al., 2010). Furthermore, we included in the analysis a non-marine derived tryptophan metabolite, kynurenine (v), which has been proposed to be an AHR agonist. These putative AHR ligands were tested for their ability to regulate Cyp1a1 transcription (Figure 2C) and to cause down-regulation of AHR protein levels (Figure 2D). Our data suggest that indolmycin is without significant effect on either Cyp1a1 or AHR down-regulation. Violacein caused down-regulation of Cyp1a1, while kynurenine caused up regulation of Cyp1a1. Both of these ligands also caused AHR protein loss. These effects were observed at a later timepoint, i.e., 8 and 24 hr, compared with the 3 hr timepoint for Resveratrol. Taken together these data support the idea that kynurenine is a possible AHR agonist, and that violacein may have AHR ligand properties, possibly acting as an antagonist.
AHR ligand effects on mast cell pro-inflammatory indicators
As our data suggest that AHR is present in RBL2H3 cells and that its activity can be regulated by conventional and unconventional AHR ligands, we sought to assess the consequences of this for the mast cell-like functions of this cell line. As AHR activation is believed to play a role in controlling inflammation, we asked whether different ligands could modulate two parameters of pro-inflammatory responses in RBL2H3 cells, i.e., degranulation and IL-6 production.
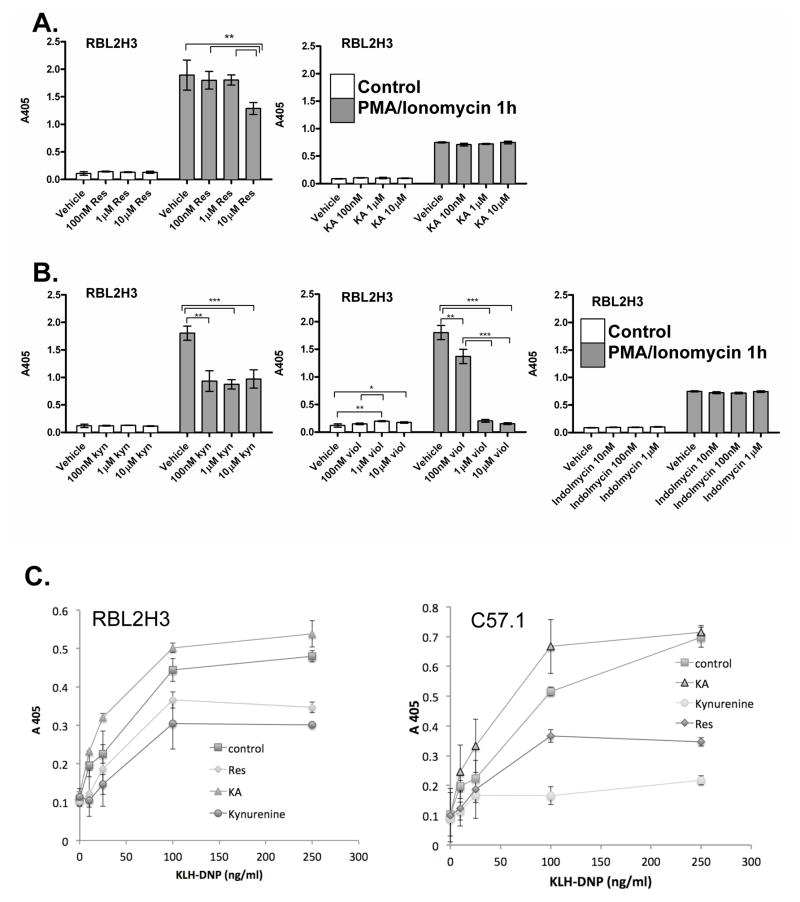
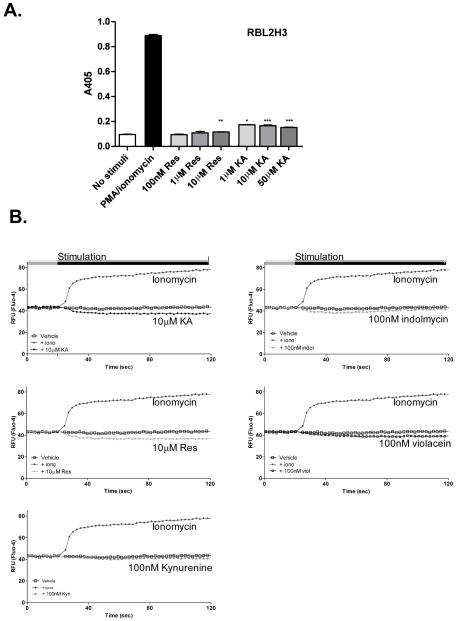
First, we asked if pre-exposure of RBL2H3 cells to AHR ligands affected degranulation and IL-6 production when these were then subsequently triggered by pharmacologic or antigenic stimulation. AHR ligands or vehicle were pre-incubated with cells for either 1 or 24 hr. The cells were then stimulated to degranulate using the pharmacological combination of PMA and ionomycin (Figures 3A and 3B). Neither of the known ligands (i.e., Resveratrol, KA) had a marked effect on degranulation with the exception of a minor inhibitory effect of Resveratrol at high doses. Indolmycin did not affect the degranulation of RBL2H3 cells and kynurenine had a minor inhibitory effect. The marked reduction of degranulation in violacein-stimulated cells reflects that violacein concentrations > 1 μM were toxic.
Figure 3. Effects of AHR ligands on degranulation in RBL2H3 mast cells.
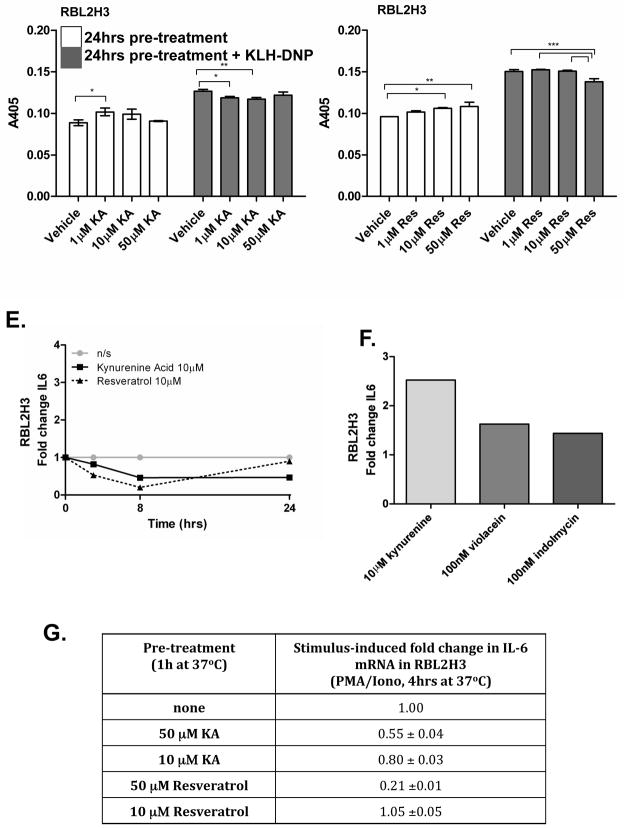
(A) RBL2H3 cells were pre-incubated with indicated concentrations of Resveratrol or KA for 24 hr. After washing, cells were stimulated with PMA and ionomycin (1 μM each) for 30 min at 37°C. Supernatants were then harvested, incubated with p-NAG substrate solution, and β-hexoseaminidase levels measured at 405 nm (B) Degranulation from RBL2H3 cells incubated for 24 hr with putative AHR ligands, followed by stimulation with PMA and ionomycin. (C) IgE anti-DNP loaded cells (RBL2H3 or C57.1 BMMC) were pre-incubated for 1 hr with 1 μM of the indicated ligand and then stimulated with KLH-DNP. (D) RBL2H3 were pre-treated for 24 hr with the indicated AHR ligand and then degranulation in response to 200 ng KLH-DNP/ml or vehicle was measured as above. Results are shown as the mean (± SD) of experiments performed in triplicate. Data are representative of one of three similar experiments. Significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) vs. vehicle treated RBL2H3 cells are indicated. (E, F, G) Induction of cytokine IL-6 gene expression after stimulation with AHR ligands.
mRNA was isolated from RBL2H3 cells treated (E) with the AHR ligands KA (10 μM) or Resveratrol (10 μM) for 3, 8, and 24 hr, (F) 10 μM kynurenine, 100 nM violacein, or 100 nM indolmycin for 24 hr, or (G) pre-incubated with the indicated amount of kynurenine, violacein, or indolmycin ligand for 1 hr before being activated (pharmacologically) using PMA/Ionomycin for 4 hr, and then IL-6 transcript levels were assessed by qPCR. The data shown in table (G) were normalized to relative expression of cells stimulated with PMA/ionomycin and depicted as mean [± SD] of amplification triplicates. Relative transcript levels were calculated applying the 2−ΔΔCt method. Figures represent one of three independent experiments.
This prompted us to carefully assess the cytotoxicity of each compound using triplicate(d) cell growth curves and trypan blue dye exclusion as the assay. These data (not shown) indicate that at a dose of 1 μM for 48 hr, the percentage of trypan blue-positive cells were 4.0 [± 0.3]% (p > 0.05; vehicle); 4.5 [± 0.12]% (p > 0.05; 10 μM resveratrol); 4.8 [± 0.2] %, (p > 0.05; 10 μM KA); 3.3 [± 0.8]% (p > 0.05; 10 μM indolmycin); 6.4 [± 0.9]%, (p > 0.05; 10 μM kynurenine); and, 33.2 [± 2.7]% (p < 0.0005; 10 μM violacein). Rates of growth were not significantly altered by any compound, except for violacein; with that agent, cells ceased to proliferate after 16 hr exposure and cultures were completely dead by the end of the 6-d exposure.
We also evaluated the effect of AHR ligands on degranulation responses that were stimulated through the high affinity receptor for IgE, FcεRI. These experiments were performed in both RBL2H3 and the C57.1 cell lines. A 1 hr pre-incubation with KA caused a slight enhancement in antigen-stimulated degranulation (Figure 3C), while both Resveratrol and kynurenine suppressed these responses. Again, this effect was sensitive to the time period of pre-incubation; Figure 3D shows that upon 24 hr pre-treatment, antigen-stimulated degranulation responses were unaffected by KA or Resveratrol.
IL-6 is a known AHR target (DiNatale et al., 2010a,b) and a pro-inflammatory cytokine secreted by activated mast cells. We used real-time RT-PCR to examine whether the compounds described above modulate expression of IL-6 over the time-course of 24 hr (Figure 3E). Both the Resveratrol and KA each transiently down-regulated IL-6 expression. This observation was unexpected, because it suggests that the agonist and antagonist compounds are having similar effects. This may be a function of time, since by 24 hr, IL-6 transcript levels in KA-treated cells had returned to baseline, while those in Resveratrol-treated cells remained suppressed. Violacein and indolmycin also up-regulated IL-6 very slightly by 24 hr (exposure), as did kynurenine (Figure 3F).
We then asked whether KA or Resveratrol could modulate the level of IL-6 expression induced by stimulation of mast cells. We found that 1 hr Resveratrol pre-treatment suppressed the achievable levels of stimulation-induced IL-6 transcription (Figure 3G) after 4 hr of incubation. Interestingly, KA also suppressed stimulus-inducible levels of IL-6 transcript; this represents a second instance wherein we noted this agonist acting in concert with the antagonist Resveratrol.
‘Non-genomic’ pathway is not regulated by AHR ligands in mast cells
AHR ligands are proposed to affect cells via two pathways. First, the genomic pathway where, over a time-course of hours, transcriptional events alter the cellular phenotype in response to AHR ligands. Second, the ‘non-genomic’ pathway refers to elevation of intracellular free calcium, which acts over a time-course of minutes. In mast cells, degranulation is critically dependent upon intracellular free calcium fluxes. Since degranulation is widely unaffected by the application of AHR ligands (Figure 4A and data not shown), we proposed that activation of the ‘non-genomic’ calcium pathway is not a major outcome of AHR ligand exposure in mast cells. Indeed, Figure 4B suggests that none of the ligands (e.g., KA, Resveratrol, kynurenine, indolmycin, or violacein) tested can mobilize intracellular free calcium when applied in isolation.
Figure 4. Acute calcium influx pathway is not regulated by AHR ligands in RBL2H3 cells.
(A) Acute degranulation of RBL2H3 cells. RBL2H3 cells were stimulated for 30 min with Resveratrol or KA (10μM), or with PMA and ionomycin as a positive control. Degranulation (β-hexoseaminidase release) was measured at 405 nm. Results are shown as the mean (± SD) and performed in triplicate. Significant differences (*p < 0.05, **p < 0.01, ***p < 0.001) vs. non-stimulated RBL2H3 cells (one-way ANOVA with Dunnett’s test comparison) are indicated. (B) Acute stimulation of calcium influx pathway in RBL2H3 cells. A baseline fluorescence signal was acquired from Fluo-4-loaded RBL2H3 for 20 sec prior to addition of the indicated stimuli. Data were collected every 2 sec over 240 sec. All stimulations were performed in triplicate and the mean was calculated and plotted.
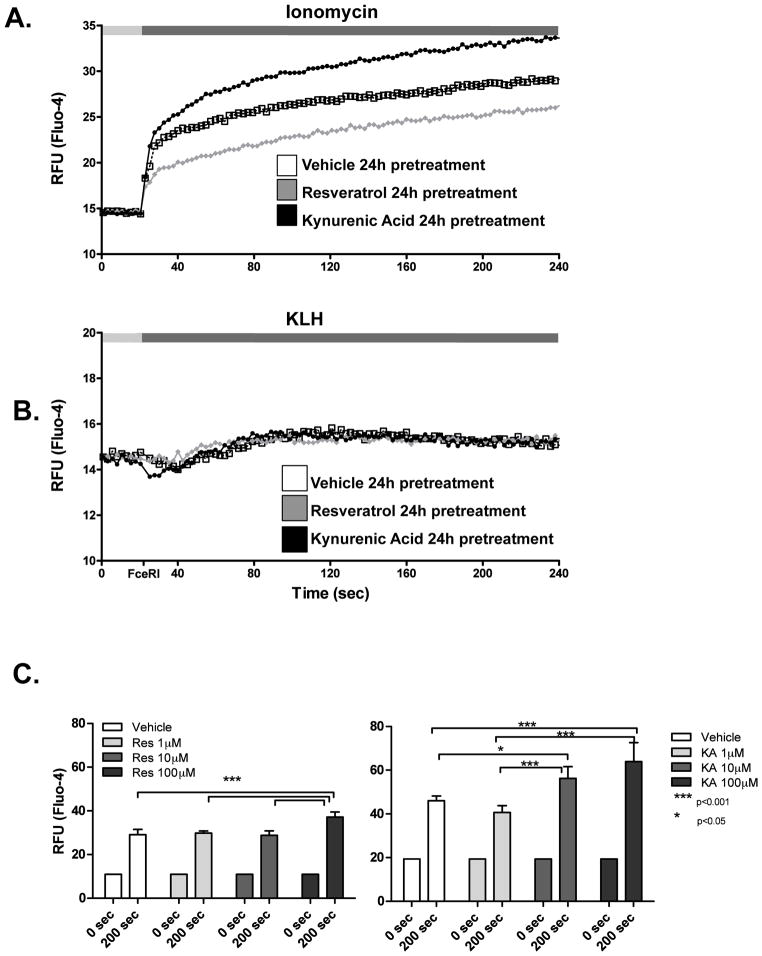
AHR ligands induce chronic events that modify calcium signals in RBL2H3 mast cells
As described above, we found that AHR ligands and candidate ligands did not exhibit an acute effect on calcium influx (Figure 4B). However, we noted that after a 24-hr pre-treatment with KA, ionomycin-induced calcium fluxes were enhanced, while pre-treatment with Resveratrol had the opposite effect (Figures 5A and 5C). These data suggest that AHR ligands do not couple to acute calcium flux, but that within the gene target set of the AHR that are regulated across a chronic time-course, there may be genes whose products play a role in generating calcium responses. We screened kynurenine, indolmycin, and violacein for their effect and found that indolmycin and kynurenine were able to up-regulate calcium influx compared with non-treated cells, while violacein did not exhibit a response (data not shown). Calcium signaling in mast cells can also be regulated through the cross-linking of the high-affinity receptor for IgE (FcεRI) by antigen, which results in calcium release and influx (Ma and Beaven, 2009). We examined this calcium flux after pre-treatment of the cells with Resveratrol and KA for 24 hr and found no effect (Figure 5B).
Figure 5. AHR ligands induce chronic events that modify calcium signals in RBL2H3 cells.
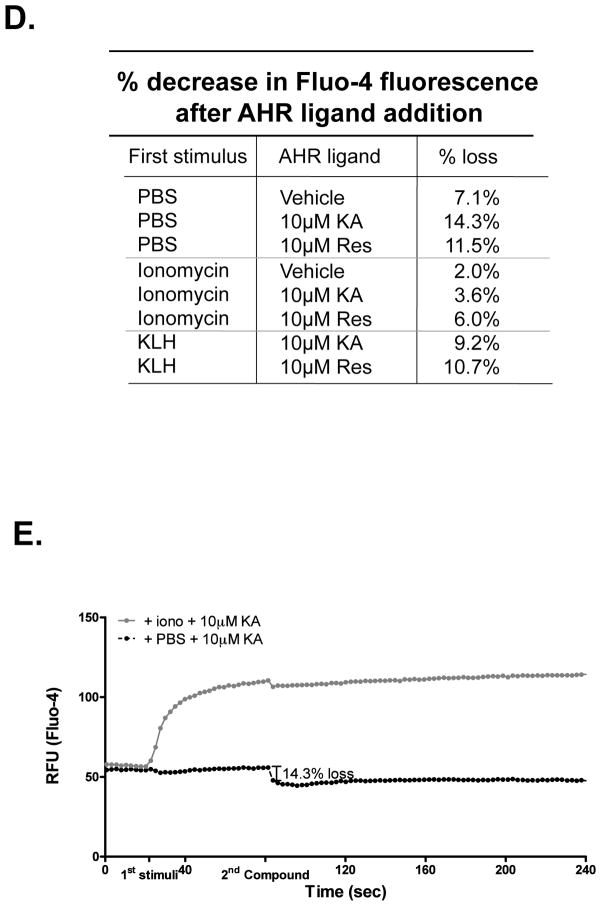
(A, B) RBL2H3 cells were pre-incubated for 24 hr with Resveratrol or KA (10 μM). Cells were incubated with Fluo-4 and a baseline fluorescence signal was acquired for 20 sec prior to addition of the indicated stimuli (Ionomycin [A], KLH-DNP [B]). Data were then collected every 2 sec over 240 sec. All stimuli were performed in triplicate and mean was calculated and plotted; data were corrected for different start points. (C) Charts summarizing calcium changes shown in (A) as a result of increasing concentrations of Resveratrol and KA. (D, E) Effect of AHR ligands on an ongoing calcium response. Cells were incubated with Fluo-4 and a baseline fluorescence signal was acquired for 20 sec before stimuli were added; a calcium response was allowed to evolve and an addition of the indicated AHR ligand occurred at 80 sec. Example trace (mean of triplicates) is shown in (E). Figures represent one of at least two similar experiments.
One potential mechanism for the effect of Resveratrol and other AHR-ligand suppressors of calcium responses is that the compound itself could act as a blocker of the calcium influx channels (primarily ICRAC) that are activated following mast cell stimulation. Figure 5D shows that post-addition of the AHR ligands to an ongoing calcium influx response that has been established by ionomycin or FcεRI causes slight but not significant diminution in the calcium signal (exemplified by the trace in Figure 5E).
Discussion
Mast cells are mediators of allergic diseases, hypersensitivity reactions and asthma (Morales et al., 2010). Recently, AHR has been shown to have an important effect in regulation and control of immune responses, showing anti-inflammatory effects and regulation of tolerance. In this study, we documented that AHR is expressed in two model mast cell lines. We studied various AHR ligands (both defined and putative) in RBL2H3 cells and described their effect on RBL2H3 cell activation and degranulation. We investigated the gene expression of Cyp1a1 and the pro-inflammatory cytokine IL-6 upon exposure to these AHR ligands and putative ligands.
In recent years, there has been intense interest in finding the endogenous ligand to the AHR, due to its importance in immunology, physiology and potential as a drug target. It has been proposed that ligands to the AHR may derive from the tryptophan degradation pathway, which we examined here. KA and kynurenine, both tryptophan derivatives from the IDO pathway, have each been shown to be AHR ligands, though with different potencies (Ball et al., 2009). Kynurenine is the first breakdown product in the IDO-dependent tryptophan degradation pathway (Ball et al., 2009), whereas KA is a secondary product (Mezrich et al., 2010). We also examined potential ligands with structural similarity to IDO-derivatives from a novel marine bacterial source. We looked at the effects of these compounds on Cyp1a1 gene expression, a gene down-stream of AHR (Hankinson, 1995). In agreement with earlier studies, we found up-regulation of Cyp1a1 after stimulation with KA and kynurenine. In mouse HepA1 cells, kynurenine was able to directly activate the AHR and up regulate gene expression of Cyp1a1 and Cyp1b1 (Mezrich et al., 2010). In contrast, another study using a lower concentration of the ligand (DiNatale et al., 2010a) found that kynurenine did not exhibit potent AHR activity in a HepG2 reporter cell line. In the RBL2H3 cell, we found that 10 μM KA increased Cyp1a1 expression, suggesting that these cells may be more sensitive than the hepatocyte lines used previously. The established AHR antagonist Resveratrol caused a time-dependent down-regulation of Cyp1a1 transcript levels, which was expected due to its function as an antagonist and also seen in the study by Beedanagari et al. (2010). In addition to these, the two tryptophan based compounds indolmycin and violacein did not show marked effect on Cyp1a1 expression.
In addition to the study of Cyp1a1, which is an established but not specific marker of AHR activity, we sought to design another assay that would enable the exploration of the compounds described above. Down-regulation of AHR protein levels has been described in response to chronic exposure to TCDD (Steenland et al., 2004). Similarly, we were able to show that both agonist and antagonist AHR ligands caused loss of AHR from RBL2H3 cells after a 72-hr exposure. Indolmycin did not have significant effects, but both violacein and kynurenine caused down-regulation. Taken together with the Cyp1a1 data presented above, our data suggest that kynurenine, and violacein display some properties consistent with action at the AHR.
Mast cells orchestrate inflammatory responses through degranulation of mediators and the production of cytokines and chemokines (Park et al., 2008). To further characterize how the AHR ligands and AHR candidate ligands affect mast cell-driven inflammatory responses, we examined their effect on degranulation. We looked at this question in two ways. First, we pre-incubated RBL2H3 cells with the AHR ligand and then stimulated degranulation with either pharmacological or antigenic stimuli. In these assays Resveratrol and kynurenine were able to suppress both pharmacologic- and antigen-stimulated secretion. In addition, KA enhances antigen-induced secretion. Second, we asked if any of these ligands were sufficient to induce degranulation; we noted some slight stimulatory effect of KA when added in isolation.
Mast cells also have a significant role in cytokine and chemokine production. Ionomycin- and FcεRI-mediated activation of mast cells increases production of IL-6 (Park et al., 2008), through the JAK/Stat signaling pathway and NF-κB. Furthermore, it was recently found that then non-ligated AHR plays a role in repressing the IL-6 promoter through regulation of NF-κB, indicating that ligand-bound AHR may be a positive regulator of IL-6 transcription (DiNatale et al., 2010b). Our data show that the AHR antagonist Resveratrol transiently down-regulates IL-6 expression in RBL2H3 cells. KA did not induce IL-6 expression on its own in RBL2H3 cells in contrast with hepatocyte data (DiNatale et al., 2010a). Kynurenine, indolmycin, and violacein induced IL-6 expression in the RBL2H3 cells. Given the lack of Cyp1a1 induction by indolmycin (see above), this may reflect a non-AHR-mediated action of this tryptophan derivative.
Motivated by previous studies, we examined the ‘non-genomic’ pathway, through which AHR is believed to function. This pathway refers to elevation of intracellular free calcium, which acts over a time-course of minutes to regulate acute responses such as mast cell degranulation. Given the large number of calcium-regulated genes in the genome, it seems premature to define the effects of AHR ligands that work through calcium signaling as ‘non-genomic’. However, we found that AHR ligands did not induce acute calcium fluxes in RBLH3 cells or significant and consistent degranulation. However, chronic treatment with AHR ligands did have an effect on the subsequent intensity of calcium influx in response to ionophore. The minor effect that these ligands have on established calcium fluxes suggest that they are not acting as channel blockers per se, and leaves the question of why there is an effect of chronic AHR ligand exposure on ionophore but not antigen-mediated calcium responses open.
Conclusions
We found that AHR is present in RBL2H3 cells and that ligands of this receptor can regulate Cyp1a1 and IL-6 expression by various pathways, as well as alter the outcome of calcium-driven mast cell activation responses. The Pseudoalteromonas luteoviolacea-derived tryptophan derivatives indolmycin and violacein display some characteristics of AHR ligands in these cells and merit further study. Moreover, the potential for longer-term exposure to AHR ligands to alter subsequent mast cell signaling may have implications for dioxin pathology, as well as for our understanding of endogenous AHR biology. The data presented here create the need for loss-of-function experiments where the effect of AHR deficiency upon critical mast cells responses, in vitro and in vivo, will be assessed. In summary, exogenous and endogenous AHR ligands may modulate mast cell function through a complex signaling pathway, implying that mast cells may play a role in both dioxin pathology and in physiological responses to endogenous AHR ligands.
Acknowledgments
This work was funded by the NSF EPSCOR EPS-0903833 (to H.T.) and The Hawaii Community Foundation (Victoria S. and Bradley L. Geist Foundation) award 45408 to H.T. The study was supported by the Programme Commission on Health, Food and Welfare under the Danish Council for Strategic Research and the present work was carried out as part of the Galathea 3 expedition under the auspices of the Danish Expedition Foundation.
Footnotes
Declarations of Interest
The authors report no declaration of interest. The authors alone are responsible for the content of this manuscript.
References
- Adachi J, Mori Y, Matsui S, Matsuda T. Comparison of gene expression patterns between 2,3,7,8-tetrachlorodibenzo-p-dioxin and a natural aryl hydrocarbon receptor ligand, indirubin. Toxicol Sci. 2004;80:161–169. doi: 10.1093/toxsci/kfh129. [DOI] [PubMed] [Google Scholar]
- Ball HJ, Yuasa HJ, Austin CJD, Weiser S, Hunt NH. Indoleamine 2,3-dioxygenase-2: A new enzyme in the kynurenine pathway. Int J Biochem Cell Biol. 2009;41:467–471. doi: 10.1016/j.biocel.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Baolin L, Inami Y, Tanaka H, Inagaki N, Iinuma M, Nagai H. Resveratrol inhibits the release of mediators from bone marrow-derived mouse mast cells in vitro. Planta Med. 2004;70:305–309. doi: 10.1055/s-2004-818940. [DOI] [PubMed] [Google Scholar]
- Beedanagari SR, Bebenek I, Bui P, Hankinson O. Resveratrol inhibits dioxin-induced expression of human CYP1A1 and CYP1B1 by inhibiting recruitment of the aryl hydrocarbon receptor complex and RNA polymerase II to the regulatory regions of the corresponding genes. Toxicol Sci. 2010;116:693–693. doi: 10.1093/toxsci/kfp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, Omiecinski CJ, Perdew GH. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci. 2010a;115:89–97. doi: 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNatale BC, Schroeder JC, Francey LJ, Kusnadi A, Perdew GH. Mechanistic insights into the events that lead to synergistic induction of interleukin-6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J Biol Chem. 2010b;285:24388–24397. doi: 10.1074/jbc.M110.118570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli SJ, Tsai M. Mast cells: Versatile regulators of inflammation, tissue remodeling, host defense, and homeostasis. J Dermatol Sci. 2008;49:7–19. doi: 10.1016/j.jdermsci.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Ann Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Kerkvliet NI. AHR-mediated immunomodulation: Role of altered gene transcription. Biochem Pharmacol. 2009;77:746–760. doi: 10.1016/j.bcp.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Matsumura F. Significance of the non-genomic, inflammatory pathway in mediating the toxic action of TCDD to induce rapid and long-term cellular responses in 3T3-L1 adipocytes. Biochemistry. 2008;47:13997–14008. doi: 10.1021/bi801913w. [DOI] [PubMed] [Google Scholar]
- Ma HT, Beaven MA. Regulation of Ca2+ signaling with particular focus on mast cells. Crit Rev Immunol. 2009;29:155–186. doi: 10.1615/critrevimmunol.v29.i2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson L, Matthews J. Inhibition of aryl hydrocarbon receptor-dependent transcription by Resveratrol or kempferol is independent of estrogen receptor-α expression in human breast cancer cells. Cancer Lett. 2010;299:119–129. doi: 10.1016/j.canlet.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson M, Phipps RK, Gram L, Munro MHG, Larsen TO, Nielsen KF. Explorative solid-phase extraction (E-SPE) for accelerated microbial natural product discovery, de-replication, and purification. J Natural Prod. 2010;73:1126–1132. doi: 10.1021/np100151y. [DOI] [PubMed] [Google Scholar]
- Matsumura F. The significance of the non-genomic pathway in mediating inflammatory signaling of the dioxin-activated Ah receptor to cause toxic effects. Biochem Pharmacol. 2009;77:608–626. doi: 10.1016/j.bcp.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Mezrich JD, Fechner JH, Zhang XJ, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T-cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales JK, Falanga YT, Depcrynski A, Fernando J, Ryan JJ. Mast cell homeostasis and the JAK-STAT pathway. Genes Immun. 2010;11:599–608. doi: 10.1038/gene.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Lee S, Son HY, Park SB, Kim MS, Choi EJ, Singh T, Ha JH, Lee MG, Kim JE, Hyun M, Kwon T, Kim Y, Kim SH. Flavonoids inhibit histamine release and expression of pro-inflammatory cytokines in mast cells. Arch Pharmacol Res. 2008;31:1303–1311. doi: 10.1007/s12272-001-2110-5. [DOI] [PubMed] [Google Scholar]
- Passante E, Ehrhardt C, Sheridan H, Frankish N. RBL-2H3 cells are an imprecise model for mast cell mediator release. Inflamm Res. 2009;58:611–618. doi: 10.1007/s00011-009-0028-4. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocar P, Fischer B, Klonisch T, Hombach-Klonisch S. Molecular interactions of the aryl hydrocarbon receptor and its biological and toxicological relevance for reproduction. Reproduction. 2005;129:379–389. doi: 10.1530/rep.1.00294. [DOI] [PubMed] [Google Scholar]
- Poland A. Induction of the drug-metabolizing enzymes. IARC Sci Publ. 1982;39:351–364. [PubMed] [Google Scholar]
- Sinal CJ, Bend JR. Aryl hydrocarbon receptor-dependent induction of CYP1A1 by bilirubin in mouse hepatoma hepa 1c1c7 cells. Mol Pharmacol. 1997;52:590–599. doi: 10.1124/mol.52.4.590. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protocol. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Steenland K, Bertazzi P, Baccarelli A, Kogevinas M. Dioxin revisited: Developments since the 1997 IARC classification of dioxin as a human carcinogen. Environ Health Perspect. 2004;112:1265–1268. doi: 10.1289/ehp.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura T, Harama D, Fukumoto S, Nakamura Y, Shimokawa N, Ishimaru K, Ikegami S, Makino S, Kitamura M, Nakao A. Lactobacillus bulgaricus OLL1181 activates the aryl hydrocarbon receptor pathway and inhibits colitis. Immunol Cell Biol. 2011 Feb 15; doi: 10.1038/icb.2010.165. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]