This study reveals a mechanism wherein brassinosteroids control the expression level of the microtubule-destabilizing protein MDP40 and regulate cortical microtubule organization to mediate hypocotyl cell elongation in Arabidopsis thaliana.
Abstract
The brassinosteroid (BR) phytohormones play crucial roles in regulating plant cell growth and morphogenesis, particularly in hypocotyl cell elongation. The microtubule cytoskeleton is also known to participate in the regulation of hypocotyl elongation. However, it is unclear if BR regulation of hypocotyl elongation involves the microtubule cytoskeleton. In this study, we demonstrate that BRs mediate hypocotyl cell elongation by influencing the orientation and stability of cortical microtubules. Further analysis identified the previously undiscovered Arabidopsis thaliana MICROTUBULE DESTABILIZING PROTEIN40 (MDP40) as a positive regulator of hypocotyl cell elongation. BRASSINAZOLE-RESISTANT1, a key transcription factor in the BR signaling pathway, directly targets and upregulates MDP40. Overexpression of MDP40 partially rescued the shorter hypocotyl phenotype in BR-deficient mutant de-etiolated-2 seedlings. Reorientation of the cortical microtubules in the cells of MDP40 RNA interference transgenic lines was less sensitive to BR. These findings demonstrate that MDP40 is a key regulator in BR regulation of cortical microtubule reorientation and mediates hypocotyl growth. This study reveals a mechanism involving BR regulation of microtubules through MDP40 to mediate hypocotyl cell elongation.
INTRODUCTION
Brassinosteroids (BRs) are crucial plant phytohormones that affect a wide range of developmental and physiological processes in plants, such as stem elongation and vascular differentiation (Clouse, 2011; Ye et al., 2011). BRs function through the BRI1 receptor-like kinase and a well-defined signal transduction pathway to activate two key transcription factors, BRASSINAZOLE-RESISTANT1 (BZR1) and BRINSENSITIVE1 (BRI1)-EMS-SUPPRESSOR1(BES1)/BZR2 (Li, 2010; Kim and Wang, 2010; Clouse, 2011; Gudesblat and Russinova, 2011). BR-deficient or -insensitive mutants generally display cell growth phenotypes, particularly affecting hypocotyl elongation. For example, BR-deficient det2 mutants and the null allele of the BR receptor mutant bri1-116 have shorter etiolated hypocotyls. The bzr1-1D mutant, which has a dominant BZR1 mutation, has longer etiolated hypocotyls (Chory et al., 1991; Li et al., 1996; Wang et al., 2001, 2002). Many upstream components, such as BR-signaling kinases (BSKs) and BRI1 suppressor 1 (BSU1), have been identified in BR signaling to regulate hypocotyl growth by altering the phosphorylated or nonphosphorylated forms of BZR1 (Tang et al., 2008a; Kim et al., 2009; Gudesblat and Russinova, 2011). However, the molecular mechanisms regarding BZR1 regulation of downstream effectors on direct participation in hypocotyl elongation are largely unknown.
Previous studies have shown that microtubules play important roles in regulating cell expansion, division, and plant cell morphogenesis. Cortical microtubules control cell growth by orientating cellulose fibrils and cellulose fibril arrays and build the mechanical properties of the cell wall (Paredez et al., 2006; Somerville, 2006; Kaloriti et al., 2007; Lloyd and Chan, 2008; Sedbrook and Kaloriti, 2008; Lloyd, 2011). The clockwise and counterclockwise rotations of cortical microtubules are dynamic features in growing hypocotyl cells as observed via long-term time-lapse imaging (Chan et al., 2007). In addition, the orientations of cortical microtubules, particularly on the inner face of the epidermis, are associated with the growth status of etiolated hypocotyls (Le et al., 2005; Li et al., 2011a; Crowell et al., 2011). For example, the parallel array of cortical microtubules is dominantly transversely oriented to the hypocotyl longitudinal growth axis in rapidly growing hypocotyl cells, while the microtubules are longitudinally oriented when cell elongation stops. Disturbing cortical microtubules with the microtubule-disrupting drug propyzamide induces a stunted hypocotyl phenotype (Le et al., 2005). Mutation or overexpression of many microtubule regulatory proteins also results in abnormal hypocotyl cell elongation by altering the stability and organization of cortical microtubules, such as SPIRAL1 (SPR1), MAP18, and MDP25 (Nakajima et al., 2004, 2006; Wang et al., 2007; Li et al., 2011a). These studies demonstrate that regulation of the organization and dynamics of cortical microtubules is crucial for hypocotyl cell growth.
Cell elongation of hypocotyls is strongly influenced by external and internal cues. Studies have detailed the mechanisms involved in hypocotyl cell elongation regulated by light, phytohormones, and transcription factors (Wang et al., 2002; Niwa et al., 2009; Luo et al., 2010; Fan et al., 2012). In addition, a recent study showed that pectin-dependent cell wall homeostasis is important for BR regulation of hypocotyl growth (Wolf et al., 2012). However, the role of microtubules in those processes is largely ambiguous. Although some hormones, such as auxin, gibberellins, and ethylene, have been reported to reorient cortical microtubules in plant cells (Shibaoka, 1994; Le et al., 2005; Li et al., 2011b; Polko et al., 2012), the molecular mechanisms regarding the effects of hormones, particularly BRs, on the regulation of microtubules in mediating hypocotyl elongation remain unknown. The identification of microtubule regulatory proteins specifically involved in BR-mediated hypocotyl cell elongation will facilitate the understanding of underlying mechanisms of BR-regulated cell growth.
Using transcript profiling and chromatin immunoprecipitation microarray (ChIP-chip) assays, many BR-regulated and BZR1 target genes have been identified in Arabidopsis thaliana (Sun et al., 2010). At1g23060 is a putative BZR1 target gene and encodes a protein that shares a ∼31% amino acid identity (calculated by DNAman version 5.22) with a potential microtubule-associated protein WAVE-DAMPENED2-LIKE7 (WDL7), suggesting that this protein has a likely role in BR-mediated cell morphogenesis by regulating microtubules (Yuen et al., 2003; Perrin et al., 2007; Sun et al., 2010).
In this study, we demonstrate that the regulation of cortical microtubule orientation and stability is essential for BR-mediated hypocotyl cell elongation. Our experiments show that BR regulates microtubules through At1g23060, which is a BZR1 target and a BR-regulated gene, to mediate hypocotyl cell elongation. The At1g23060 gene product MICROTUBULE-DESTABILIZING PROTEIN40 (MDP40), which was named based on its molecular mass of ∼40 kD and its function on microtubules, plays a role in regulating hypocotyl elongation. Our findings demonstrate that MDP40 influences BR-regulated hypocotyl cell elongation by altering the stability of cortical microtubules.
RESULTS
Regulation of Cortical Microtubules Is Essential for BR-Mediated Hypocotyl Growth
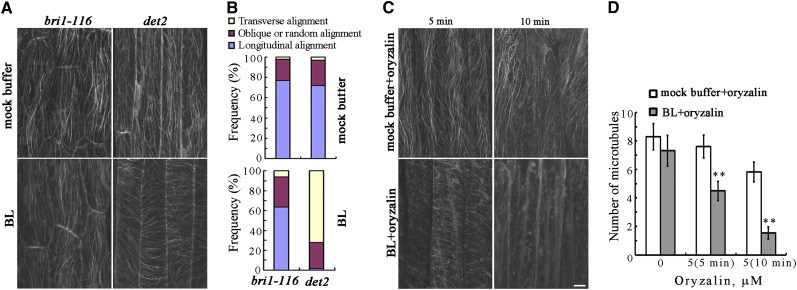
Arabidopsis lines consisting of etiolated BR-deficient det2-1 mutants and the null allele of the BR receptor mutant bri1-116 with a yellow fluorescent protein (YFP)-tubulin background were generated to test for BR regulation of cortical microtubule orientation. Treatments were performed with brassinolide (BL), which is the most active BR. Confocal observations showed that most cortical microtubules exhibited oblique and longitudinal orientations in the epidermal cells of the etiolated hypocotyls of det2-1 and bri1-116 seedlings (Figure 1A). After treatment with 1 μM BL for 60 min, the cortical microtubules were transversely reoriented to the longitudinal hypocotyl growth axis in det2-1 mutant cells, but not in the bri1-116 mutant cells (Figures 1A and 1B), demonstrating that reorientation of the cortical microtubules in etiolated hypocotyls cells is regulated by BR signaling.
Figure 1.
BRs Regulate the Orientation and Stability of Cortical Microtubules in the Epidermal Cells of Etiolated Hypocotyls.
(A) Etiolated hypocotyl epidermal cells of bri1-116 and det2-1 mutants with a YFP-tubulin background were treated with or without BL for 60 min after growth in the dark for 96 h, and cortical microtubules were observed in the hypocotyl epidermal cells.
(B) The frequency of microtubule orientation patterns in the etiolated hypocotyl epidermal cells of bri1-116 and det2-1 mutants (n > 90 cells).
(C) Cortical microtubules were observed in the epidermal cells of etiolated hypocotyls in det2-1 mutants pretreated with BL or mock buffer after treatment with 5 μM oryzalin for 5 or 10 min. Bar = 10 μm.
(D) Quantification of cortical microtubules in hypocotyl epidermal cells of det2-1 mutants using ImageJ software (n > 39 cells from each sample). Vertical scale represents the number of cortical microtubules across a fixed line (∼10 μm) vertical to the orientation of the majority of cortical microtubules in the cell. The t tests compared the number of cortical microtubules in the hypocotyl epidermal cells of det2-1 mutants pretreated with BL with the number of microtubules in cells that were not pretreated with BL under the same conditions. **P < 0.01, t test. Error bars represent the se.
[See online article for color version of this figure.]
The underlying mechanism of BR reorientation of the cortical microtubules was evaluated by investigating BR regulation of cortical microtubule stability in hypocotyl cells using the microtubule-disrupting drug oryzalin. To quantify the effects of oryzalin on the stability of cortical microtubules in det2-1 mutants following treatment with BL, the density of cortical microtubules in etiolated hypocotyl epidermal cells was estimated as previously reported (Li et al., 2011a). Most of the cortical microtubules disappeared in the epidermal cells pretreated with 1 μM BL in the presence of 5 μM oryzalin for 5 min, while the microtubules in the cells that were not treated with BL were largely unaffected (Figure 1C). Increasing the duration of oryzalin treatment resulted in the disappearance of most of the cortical microtubules in the cells pretreated with BL. However, the cortical microtubules remained relatively unaffected in the cells that were not treated with BL (Figures 1C and 1D). These results demonstrate that treatment with BR increases the sensitivity of cortical microtubules to oryzalin.
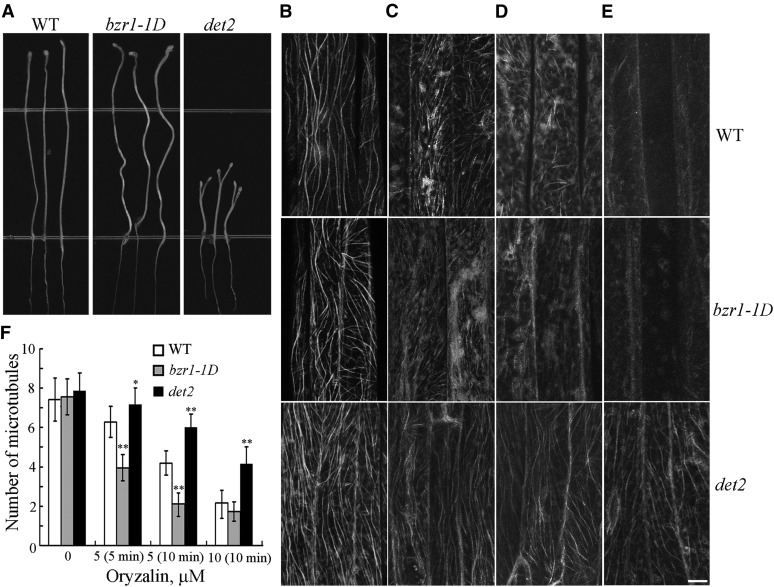
Many BR-related mutants exhibit abnormal etiolated hypocotyl elongation. BR-deficient det2-1 mutants have shorter etiolated hypocotyls, while BR perceptional bzr1-1D mutants have longer etiolated hypocotyls (Figure 2A; Wang et al., 2002). We examined the relationship between the stability of cortical microtubules and those phenotypes. The cortical microtubules in 4-d-old etiolated hypocotyl epidermal cells det2-1 and bzr1-1D mutants with a YFP-tubulin background were observed using confocal microscopy, and oryzalin was used to test the stability of the microtubules. The density of cortical microtubules in hypocotyl epidermal cells was analyzed to quantify the effects of oryzalin on the stability of cortical microtubules in the mutant seedlings. The densities of the cortical microtubules in the wild-type, det2-1, and bzr1-1D epidermal cells before treatment were not significantly different (Figures 2B and 2F). However, the density of cortical microtubules in the wild-type, det2-1, and bzr1-1D epidermal cells was significantly different following treatment (Figure 2F). The microtubules were disrupted in the bzr1-1D epidermal cells following treatment with 5 μM oryzalin for 5 min, while the microtubules in the wild-type and det2-1 cells were relatively unaffected (Figures 2C and 2F). Increasing the oryzalin concentration and duration of treatment resulted in the disappearance of most of the cortical microtubules in the wild-type and bzr1-1D cells, although the cortical microtubules remained relatively unaffected in the det2-1 mutant cells (Figures 2D to 2F). The results of this experiment showed that microtubules in bzr1-1D cells were more sensitive to the oryzalin treatment compared with the det2-1 mutant cells.
Figure 2.
Cortical Microtubules Are Hypersensitive in bzr1-1D Cells but More Resistant to Treatment with Oryzalin in det2-1 Mutant Cells.
(A) Seedlings from the wild type (WT; Columbia ecotype) and bzr1-1D and det2-1 mutants were grown on half-strength MS in the dark for 5 d.
(B) to (E) Cortical microtubules were observed in the epidermal cells of etiolated hypocotyls in the wild-type, bzr1-1D, and det2-1 mutant seedlings after treatment with 0 μM oryzalin (B), 5 μM oryzalin for 5 min (C), 5 μM oryzalin for 10 min (D), and 10 μM oryzalin for 10 min (E). Bar in (E) = 10 μm.
(F) Quantification of cortical microtubules in hypocotyl epidermal cells of the wild type and bzr1-1D and det2-1 mutants using ImageJ software (n > 42 cells from each sample). Vertical scale represents the number of cortical microtubules across a fixed line (∼10 μm) vertical to the orientation of most cortical microtubules in the cell. The t tests compared the number of cortical microtubules in the hypocotyl epidermal cells in bzr1-1D and det2-1 with the number of cortical microtubules in the wild type under the same conditions. **P < 0.01 and *P < 0.05, t test. Error bars represent the se.
MDP40 Is a BZR1 Target and BR-Upregulated Gene
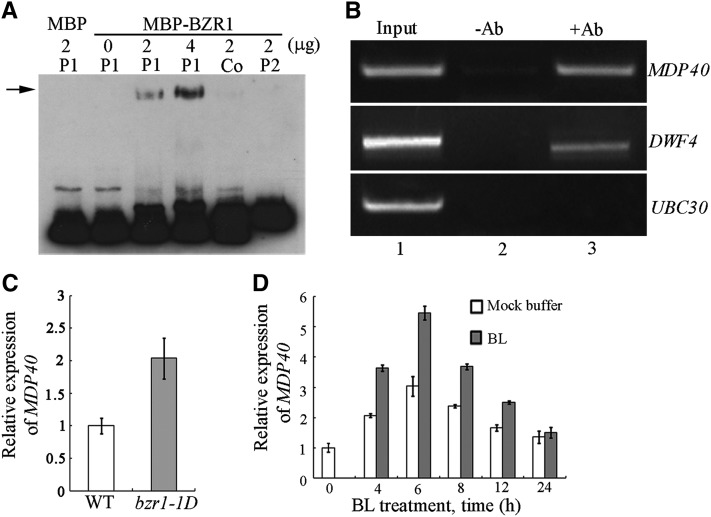
BZR1 plays a central role in BR regulation of plant growth. Many potential BR-regulated and BZR1 target genes have been identified using ChIP followed by microarray (ChIP-chip). The gene we named MDP40 herein was predicted to be a BZR1 target gene and encodes a putative microtubule-associated protein (Sun et al., 2010). We tested the direct binding of BZR1 to the promoter of MDP40 with electrophoretic mobility shift assays (EMSAs) using N-hydroxysuccinimide (NHS)-biotin–labeled DNA fragments of the MDP40 promoter and bacterially expressed BZR1 proteins fused to the maltose binding protein (MBP). The result showed that the MBP-BZR1 fusion protein bound to the –208 to –60 region (P1) but not the –502 to –175 region (P2) of the MDP40 promoter (translational start is +1). When unlabeled P1 (Co) probe was added to the system as competitor, the band was suppressed (Figure 3A), indicating that BZR1 can directly bind to the MDP40 promoter in vitro.
Figure 3.
BZR1 Directly Activates the Expression of MDP40.
(A) EMSAs with the BZR1 protein using a probe derived from the MDP40 promoter. The arrow indicates the bands caused by BZR1 binding to the MDP40 promoter P1.
(B) ChIP assay indicates that BZR1 is associated with the promoter of MDP40 in vivo. The names of the promoters evaluated are shown to the right of each experiment. The source of the template DNA from His-BZR1-Myc transgenic seedlings is shown as input (lane 1), DNA precipitated without addition of the antibody (-Ab) as a negative control (lane 2), and DNA precipitated with the antibody (+Ab) (lane 3). Each assay was repeated more than three times with independent biological materials.
(C) The expression level of MDP40 was determined using quantitative real-time PCR with RNA purified from the wild-type or bzr1-1D seedlings. Error bars represent ± sd (n = 3). WT, the wild type.
(D) Quantitative real-time PCR analysis of MDP40 RNA levels in 7-d-old seedlings after various treatment durations using 1 μM BL or mock buffers. EF1α was used as a reference gene. Error bars represent ± sd (n = 3).
To determine whether BZR1 binds to the MDP40 promoter in vivo, ChIP experiments were performed. The His-BZR1-Myc fusion protein was expressed using the 35S promoter and immunoprecipitated using an antibody recognizing the Myc tag. The genomic DNA fragments that coimmunoprecipitated with His-BZR1-Myc were analyzed using PCR. DWARF4 (DWF4), a known BZR1 target gene, and ubiquitin-conjugating enzyme E2 30 (UBC30), a known BZR1 nontarget gene, were used as controls (He et al., 2005; Sun et al., 2010). DNA precipitated without the anti-Myc antibody was subjected to PCR amplification, and DWF4 or MDP40 bands were not detected (Figure 3B, Lane 2). An obvious MDP40 band was amplified from the DNA that coprecipitated with the anti-Myc antibody (Figure 3B, lane 3). Similar results were obtained for DWF4, but not UBC30 (Figure 3B), demonstrating that MDP40 is a BZR1 target gene.
We analyzed BZR1 regulation of MDP40 expression. The dominant bzr1-1D mutation causes constitutive BZR1 activation due to enhanced dephosphorylation by PP2A, and BZR1 target genes are constitutively activated in the bzr1-1D mutant (Wang et al., 2002; Tang et al., 2011). If MDP40 is a BZR1-upregulated gene, a higher expression level of MDP40 is expected in bzr1-1D mutants. To test this possibility, RNA was purified from etiolated hypocotyls of the bzr1-1D mutant, and quantitative real-time PCR analysis was performed. The expression level of MDP40 was much higher in the bzr1-1D mutant than in the wild type (Figure 3C), indicating that MDP40 is a BZR1-upregulated gene.
We tested whether BRs are capable of regulating the expression of MDP40. The BR-deficient mutant det2-1 was treated with 1 μM BL. Quantitative real-time PCR showed that the expression of MDP40 was induced by BL treatment, with the peak level detected 6 h after treatment (Figure 3D). These results indicate that BZR1 directly activates the expression of MDP40.
MDP40 Colocalizes with Cortical Microtubules in Vivo
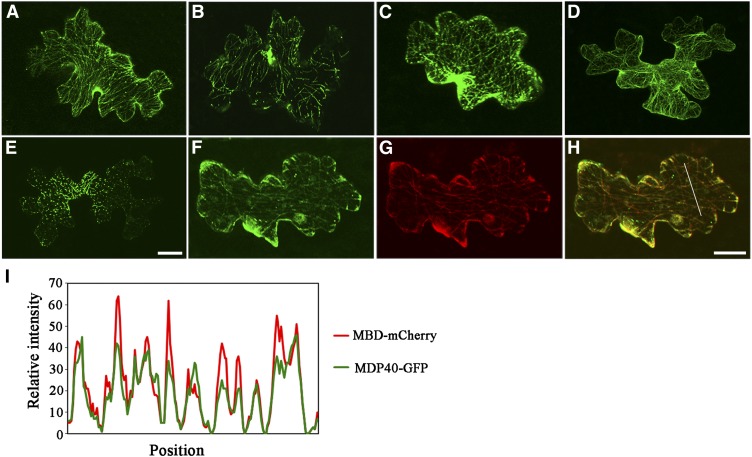
The localization of MDP40 was investigated in Arabidopsis cells. A construct encoding MDP40 with a C-terminal green fluorescent protein (GFP) tag driven by a 35S promoter was constructed and transiently introduced into cells. Confocal microscopy showed that MDP40-GFP formed filamentous structures in pavement cells (Figure 4A). The filament structures were disrupted by treatment with the microtubule-disrupting reagent oryzalin (Figure 4B) but were nearly intact in the presence of LatA, a reagent that depolymerizes actin filaments (Figure 4C). F-actin was visualized by transiently expressing fABD2-GFP in cells (Figure 4D), and the LatA treatment was shown to disrupt most of the F-actin in the pavement cells (Figure 4E). Similar phenomena were also observed in Arabidopsis cells stably expressing MDP40-GFP or fABD2-GFP (see Supplemental Figures 1A to 1E online), suggesting that this structure is related to microtubules, but not F-actin. In addition, time-lapse imaging was performed to track MDP40-GFP, and the results showed that MDP40-GFP was relatively stationary in the cells (see Supplemental Movie 1 online).
Figure 4.
MDP40 Colocalizes with Cortical Microtubules.
(A) MDP40-GFP was transiently expressed in Arabidopsis pavement cells and decorated microtubules.
(B) and (C) The filamentous pattern of MDP40-GFP was disrupted when the cells were treated with oryzalin (B) but was essentially unaffected when treated with LatA (C).
(D) F-actin was visualized by transiently expressing fABD2-GFP in pavement cells.
(E) The filamentous pattern of fABD2-GFP was disrupted when the cells were treated with LatA.
(F) to (H) Colocalization analysis of MDP40-GFP and MBD-mCherry using transient expression.
(I) Plot of a line scan showing a strong correlation between the spatial localization of MDP40-GFP and MBD-mCherry.
Bars in (E) and (H) = 20 μm.
To confirm this result, we transiently coexpressed MDP40-GFP and MBD-mCherry into Arabidopsis pavement cells. The green fluorescent signal of MDP40-GFP overlapped with the red fluorescent signal of MBD-mCherry, as shown in Figures 4F to 4H. A plot of the signal intensity analysis using ImageJ software is shown in Figure 4I. These data demonstrate that MDP40 colocalizes with cortical microtubules in vivo.
MDP40 Functions as a Positive Regulator in BR-mediated Hypocotyl Cell Elongation
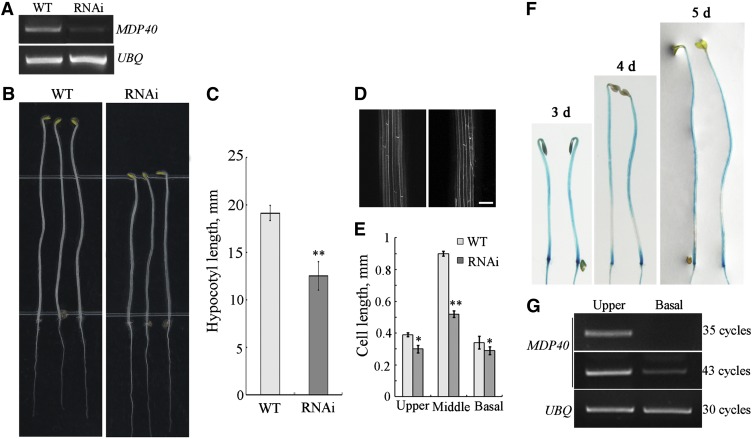
Because knockdown or knockout T-DNA insertion lines of MDP40 are unavailable, RNA interference (RNAi) lines were generated to analyze the function of MDP40 in Arabidopsis. Of the 42 MDP40 RNAi transgenic lines that were obtained, 29 showed a shorter etiolated hypocotyl. Line 6 (R6), which exhibited a typical phenotype, was selected for further analyses. The level of MDP40 transcription was considerably reduced in this RNAi line (Figure 5A). Because MDP40 shares high amino acid identity (31%, calculated by DNAman software, version 4.0) with the putative microtubule-associated protein WDL7 in the Arabidopsis genome, we detected the expression of WDL7 in the MDP40 RNAi line. RT-PCR showed that the level of WDL7 RNA in the MDP40 RNAi line was similar to the expression level in the wild type (see Supplemental Figure 2 online), demonstrating that the expression of WDL7 was not affected in the MDP40 RNAi line.
Figure 5.
MDP40 Positively Regulates Hypocotyl Cell Elongation.
(A) RT-PCR analysis of MDP40 transcripts in the wild-type (WT) Columbia ecotype (Col) seedlings and MDP40 RNAi Arabidopsis, with UBQ as a control.
(B) The MDP40 RNAi line shows shorter etiolated hypocotyls when grown on half-strength MS for 5 d.
(C) The graph shows the average hypocotyl length measured from at least 30 seedlings under dark growth. t test, **P < 0.01, t test; error bars indicate se.
(D) Confocal observation showed that the profiles of etiolated hypocotyls epidermal cells are similar between the wild-type and MDP40 RNAi line. Bar = 10 μm.
(E) Size of the etiolated hypocotyl cells in the upper, middle, and basal regions of the MDP40 RNAi line. (F) and (G) MDP40 was primarily expressed in the rapidly growing region of dark-growth hypocotyls of Arabidopsis.
(F) Histochemical GUS staining of PMDP40:GUS:TMDP40 transgenic seedlings grown in the dark for 3, 4, and 5 d.
(G) RT-PCR shows that MDP40 is highly expressed in the upper region and minimally expressed in the basal region of etiolated hypocotyls. UBQ was used as a loading control. Three biological replicates showed similar results.
The hypocotyl length in 5-d-old etiolated seedlings from the MDP40 RNAi line was dramatically reduced (Figures 5B and 5C). No major differences in the cell profiles of the etiolated hypocotyls were observed between the MDP40 RNAi line and the wild type (Figure 5D). The cell numbers of an epidermal cell file of hypocotyls in the MDP40 RNAi line and the wild type were similar (∼20 to ∼22). The cell lengths in different regions of etiolated hypocotyls in the MDP40 RNAi line were much shorter than in the wild type, particularly in the middle region. Statistical analysis using paired Student’s t test indicated that this difference was significant (Figure 5E).
To confirm that the hypocotyl phenotype in the MDP40 RNAi line was linked to MDP40 expression levels, we randomly selected four additional RNAi lines for RT-PCR analysis. The results showed that the MDP40 expression levels were associated with the short etiolated hypocotyl phenotype (see Supplemental Figures 3A and 3B online). We made a second MDP40 RNAi (RNAi-1) construct using another MDP40 cDNA sequence. Nineteen MDP40 RNAi-1 lines exhibited a shorter etiolated hypocotyl phenotype, and two independent MDP40 RNAi-1 lines (lines 6 and 13) were selected for further analyses. The results showed that the transcription levels of MDP40 and the hypocotyl length in 5-d-old etiolated seedlings from the MDP40 RNAi-1 lines were dramatically reduced (see Supplemental Figures 4A to 4C online). These results confirm that the shorter etiolated hypocotyl phenotype in the MDP40 RNAi lines is dependent on the expression level of MDP40. Therefore, MDP40 plays a positive regulatory role in hypocotyl cell elongation.
The precise region of growth during hypocotyl development has been well defined. For example, the five basal hypocotyl cells stop growing from day 3 to day 4 after germination in the dark, while the cells in the upper region of the hypocotyl begin to expand (Gendreau et al., 1997). Proteins that play a positive role in hypocotyl growth are expected to be highly expressed in the upper region but not at the basal region in etiolated hypocotyls. The expression pattern of MDP40 was explored by determining the promoter activity using β-glucuronidase (GUS) as a reporter. RT-PCR and GUS staining showed that MDP40 was expressed in most of the tissues and organs of Arabidopsis, particularly in the cotyledon and hypocotyls (see Supplemental Figures 5A to 5H online), suggesting it may play a role in regulating hypocotyl growth. Detection of GUS activity in etiolated seedlings revealed that MDP40 was primarily expressed in the upper region of the hypocotyls after 3 to 5 d in the dark (Figure 5F), which is considered to be a fast growing region after 4 d (Gendreau et al., 1997). To verify this result, RNA was purified from the basal and upper regions of hypocotyls from wild-type seedlings grown in the dark for 4 d. RT-PCR showed that MDP40 was weakly expressed in the basal region, but more abundant in the upper region of etiolated hypocotyls (Figure 5G), which demonstrates that MDP40 functions as a positive regulator of hypocotyl elongation.
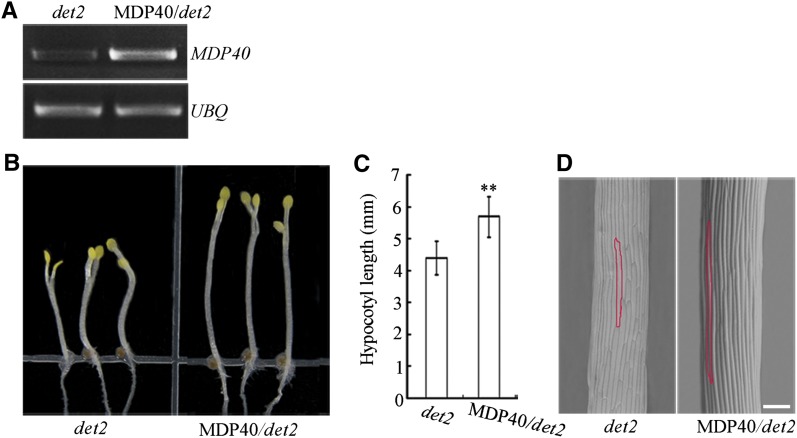
Because our results show that MDP40 is a BZR1 target and BR-upregulated gene, we hypothesized that overexpression of MDP40 could rescue the short hypocotyl phenotype induced by a deficiency of BR. Twenty-eight BR-deficient det2-1 mutant lines that overexpress MDP40 were generated, and line 8 was used for analysis. RT-PCR showed that the transcription level of MDP40 was considerably enhanced in the line that overexpressed MDP40 (Figure 6A). Overexpression of MDP40 dramatically increased the etiolated hypocotyl length of det2-1 mutants in 5-d-old etiolated seedlings (Figures 6B and 6C). Scanning electronic microscopy revealed that the cell length of etiolated hypocotyls in det2-1 mutants was increased when the expression of MDP40 was enhanced (Figure 6D), demonstrating that MDP40 is a downstream factor in the BR signaling pathway and affects hypocotyl elongation.
Figure 6.
Overexpression of MDP40 Partially Rescues Shorter Hypocotyls of the det2-1 Mutant.
(A) RT-PCR analysis of MDP40 transcripts in seedlings of det2-1 and MDP40 transgenic det2-1 mutants.
(B) The MDP40 transgenic etiolated det2-1 mutant shows longer hypocotyls grown on half-strength MS in the dark for 5 d.
(C) The graph shows the average hypocotyl length measured from at least 31 seedlings under dark growth conditions (**P < 0.01, t test). Error bars indicate the se.
(D) Scanning electron microscopy images of etiolated hypocotyl epidermal cells of det2-1 and MDP40 transgenic det2-1 mutants. Bar = 100 μm.
MDP40 Mediates Cortical Microtubule Orientation by Destabilizing Microtubules
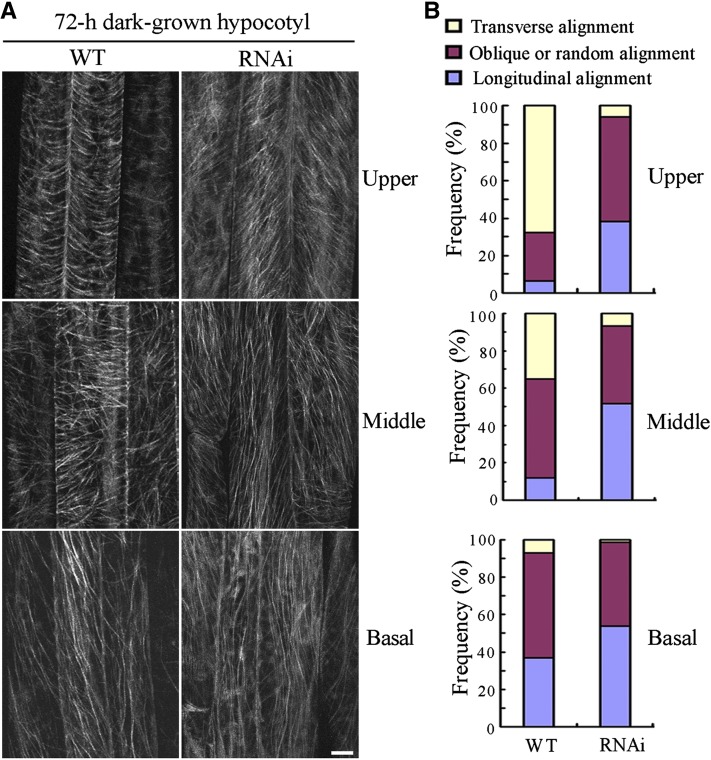
Because MDP40 colocalizes with cortical microtubules in the cell, this protein may regulate the cortical microtubule array in hypocotyl cells. Because the orientation of cortical microtubule patterns (transverse, oblique, and longitudinal) are related to the elongation rate of etiolated hypocotyl epidermal cells, etiolated hypocotyls are used to examine the correlation between cell elongation and cortical microtubule organization (Le et al., 2005; Crowell et al., 2011). To test this possibility, we observed cortical microtubules in hypocotyl epidermal cells of MDP40 RNAi transgenic Arabidopsis with a GFP-tubulin background. After 72 h of growth in the dark, parallel arrays of cortical microtubules were generally transversely oriented to the longitudinal hypocotyl growth axis in the upper and middle regions of the wild-type etiolated hypocotyls. By contrast, random, oblique, or longitudinal cortical microtubules were observed in most of the MDP40 RNAi Arabidopsis hypocotyl cells (Figures 7A and 7B), which is consistent with the significant inhibition of etiolated hypocotyl cell elongation and reduced expression of MDP40.
Figure 7.
The Cortical Microtubule Array Is Significantly Altered in Etiolated Epidermal Hypocotyl Cells of MDP40 RNAi Seedlings.
(A) Cortical microtubules in etiolated hypocotyl epidermal cells of MDP40 RNAi seedlings with a GFP-tubulin background in different regions (upper hypocotyl region, middle hypocotyl region, and basal cells) after growth in the dark for 72 h were observed using confocal microscopy. WT, the wild type. Bar = 10 μm.
(B) The frequency of cortical microtubule orientation patterns in different regions of etiolated hypocotyl epidermal cells of the wild-type and MDP40 RNAi lines (n > 100 cells).
[See online article for color version of this figure.]
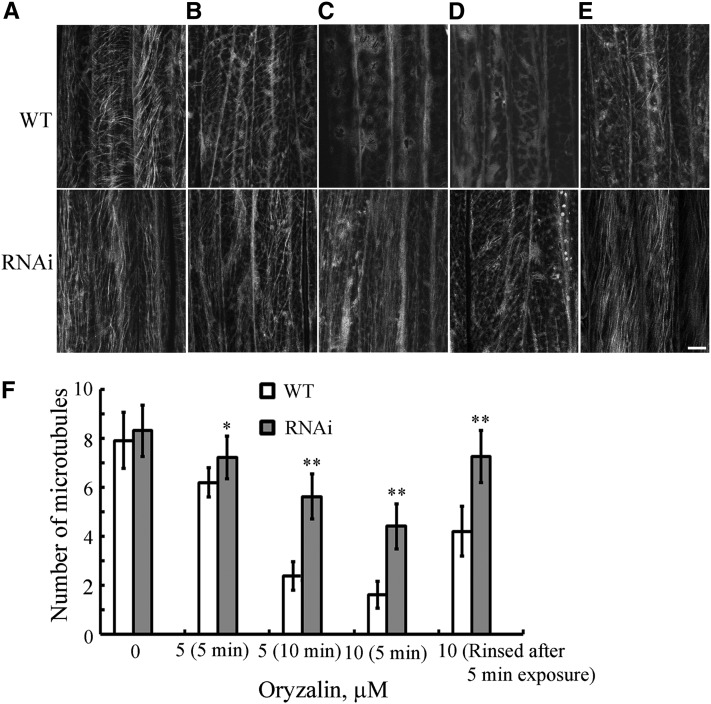
To determine the effects of MDP40 on cortical microtubule-mediated hypocotyl cell elongation, wild-type and MDP40 RNAi epidermal hypocotyl cells were treated with the microtubule-disrupting drug oryzalin. The epidermal cells in the middle region were used to compare the stability of the cortical microtubules because MDP40 is strongly expressed in the middle region of etiolated hypocotyls. The density of cortical microtubules in hypocotyl epidermal cells was measured to quantify the effects of oryzalin on the stability of cortical microtubules in the wild-type and MDP40 RNAi lines. The results revealed that the density of cortical microtubules in the epidermal cells of the wild type was similar to the density in MDP40 RNAi lines before treatment (Figures 8A and 8F). However, the densities were significantly different after the drug treatment (Figure 8F). The cortical microtubules were disrupted in the wild-type epidermal cells treated with 5 μM oryzalin for 5 min, while the microtubules in MDP40 RNAi cells were largely unaffected (Figures 8B and 8F). Increased oryzalin concentration and duration of treatment resulted in the disruption of most of the cortical microtubules in the wild-type cells. However, cortical microtubules still remained in the MDP40 RNAi cells (Figures 8C, 8D, and 8F). When the oryzalin was washed off after the treatment, most of the cortical microtubules recovered in the epidermal cells of the MDP40 RNAi cells, but not in the wild-type cells (Figures 8E and 8F). Thus, the microtubules in the MDP40 RNAi cells were less sensitive to the oryzalin treatment when the expression of MDP40 was reduced. Those results demonstrate that MDP40 functions as a microtubule destabilizer. Microtubule dynamics were further analyzed using confocal time-lapse imaging. Microtubules with clearly visible leading plus ends (identified by their growth rates) were selected for measurement in the wild-type and MDP40 RNAi Arabidopsis cells (see Supplemental Table 1 online). The results showed that the parameters of microtubule dynamics were altered in the MDP40 RNAi etiolated hypocotyl epidermal cells. The average growth rate of the leading ends of the microtubules was 9.53 ± 1.49 μm/min (mean ± sd, n = 10) in MDP40 RNAi Arabidopsis cells and 6.51 ± 1.26 μm/min (mean ± sd, n = 8) in wild-type cells. The rescue frequency of individual microtubules in MDP40 RNAi Arabidopsis cells (0.115 s−1 for rescue) was significantly higher than the frequency in wild-type cells (0.019 s−1 for rescue), suggesting that individual microtubules are more prone to growth in the absence of MDP40.
Figure 8.
Cortical Microtubules Are More Resistant to Treatment with Oryzalin in MDP40 RNAi Arabidopsis Cells.
(A) to (D) Cortical microtubules were observed in the epidermal cells in the middle region of etiolated hypocotyls in wild-type and MDP40 RNAi seedlings after treatment with 0 μM oryzalin (A), 5 μM oryzalin for 5 min (B), 5 μM oryzalin for 10 min (C), and 10 μM oryzalin for 5 min (D). WT, the wild type.
(E) After the treatment in (D), oryzalin was rinsed off, and the cortical microtubules were observed after 1 h. Bar = 10 μm.
(F) Quantification of cortical microtubules in the hypocotyl epidermal cells of the wild-type and MDP40 RNAi lines using ImageJ software (n > 46 cells from each sample). Vertical scale represents the number of cortical microtubules across a fixed line (∼10 μm) vertical to the orientation of the majority of cortical microtubules in the cell. The t tests compared the number of cortical microtubules in the hypocotyl epidermal cells of MDP40 RNAi line with the number of cortical microtubules in the wild type under the same conditions. **P < 0.01, *P < 0.05, t test. Error bars represent se.
Decreased Expression of MDP40 Affects Cortical Microtubule Reorientation in Response to BR
Because MDP40 plays a positive role in BR-mediated hypocotyl growth, we investigated the effects of MDP40 on the regulation of cortical microtubules in response to BR. The wild-type and MDP40 RNAi Arabidopsis seedlings were grown for 4 d in the dark on half-strength Murashige and Skoog (MS) medium containing brassinazole (BRZ), which specifically blocks BR biosynthesis at the C-22 hydroxylation step and decreases endogenous BR (Asami et al., 2000), and BL treatments were performed.
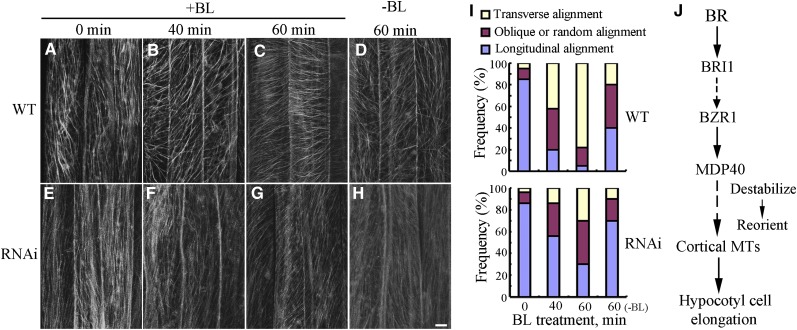
After 4 d of growth in the dark, parallel arrays of cortical microtubules were longitudinally oriented to the hypocotyl growth axis in the epidermal cells of the wild-type and MDP40 RNAi etiolated hypocotyls (Figures 9A, 9E, and 9I). After treatment with 1 μM BL for 40 min, transverse cortical microtubules were observed in the hypocotyl cells of the wild-type seedlings (Figures 9B and 9I), but not in the MDP40 RNAi Arabidopsis cells (Figures 9F and 9I). Increasing the duration of treatment induced dominantly transverse cortical microtubules in the wild-type cells, but not in the MDP40 RNAi line (Figures 9C, 9G, and 9I), indicating that cortical microtubule reorientation was hindered in MDP40 RNAi cells in response to BL treatment. Cortical microtubule arrays in the wild-type and RNAi lines did not exhibit differences after the cells were treated with mock buffer for 60 min (Figures 9D, 9H, and 9I). This demonstrates that MDP40 is necessary for BR-regulated cortical microtubule reorientation.
Figure 9.
Orientation of Cortical Microtubules in MDP40 RNAi Arabidopsis Cells Is Insensitive to Treatment with BL.
(A) to (H) Etiolated hypocotyls of the wild type (WT; [A] to [D]) and MDP40 RNAi lines ([E] to [H]) with a GFP-tubulin background were grown on medium supplemented with 1 μM BRZ and treated in liquid medium with or without 1 μM BL. Cortical microtubules were observed in the middle region of the hypocotyl epidermal cells. (A) and (E), without BL treatment; (B) and (F), treated with BL for 40 min; (C) and (G), treated with BL for 60 min; (D) and (H), treated with a mock buffer for 60 min. Bar in (H) = 10 μm.
(I) Frequency of microtubule orientation patterns in the middle region of the etiolated hypocotyl epidermal cells of the wild-type and MDP40 RNAi lines (n > 124 cells).
(J) Model of MDP40 function on cortical microtubules in BR-mediated hypocotyl cell elongation. BR signaling from the plasma membrane receptor BRI1 to the transcription factor BZR1; BZR1 directly regulates the expression of MDP40; MDP40 alters the stability of cortical microtubules (MTs) and reorients the cortical microtubules, which results in mediation of hypocotyl cell elongation.
[See online article for color version of this figure.]
DISCUSSION
MDP40 belongs to a TPX2 (targeting protein for Xklp2) protein family and shares high amino acid identity with WDL7, which is a member of the microtubule-associated protein WVD2/WDL family in Arabidopsis (Yuen et al., 2003; Perrin et al., 2007). In this study, we demonstrated that MDP40 is a BZR1 target gene and that MDP40 participates in BR-mediated hypocotyl cell elongation by destabilizing cortical microtubules.
The Reorientation of Cortical Microtubules Is Necessary for BR-Mediated Hypocotyl Cell Growth
BRs affect a diverse array of plant growth and developmental processes, such as cell elongation and vascular differentiation (Clouse, 2011). Defective etiolated hypocotyl elongation is a typical phenotype related to many BR signaling components, including shortened etiolated hypocotyls in the receptor mutants bri1 (the null or weak alleles bri1-116 and bri1-9) and bin2 (semidominant dwarf mutant brassinosteroid insensitive2) (Jin et al., 2007; Sun et al., 2010; Su et al., 2011). Genetic and microarray studies have revealed the central role of BZR1 in BR regulation of hypocotyl elongation (Wang et al., 2002; Sun et al., 2010). BZR1 functions in rapidly growing etiolated hypocotyl cells but not in cells that are in a stationary growth phase (Wang et al., 2002). Transverse patterns of cortical microtubules, which are associated with cells in a rapid growth phase, are detected in rapidly elongating etiolated hypocotyl epidermal cells (Le et al., 2005; Li et al., 2011a; Crowell et al., 2011). The orientation of cortical microtubules is interrelated with hypocotyl cell elongation, which has been tested using microtubule-disrupting drugs and the mutant and transgenic lines of microtubule regulatory proteins (Le et al., 2005; Paredez et al., 2006; Somerville, 2006; Buschmann and Lloyd, 2008). Similarities between these mechanisms regarding the promotion of hypocotyl cell elongation suggest that BRs may mediate hypocotyl growth through cortical microtubules.
Proteomic studies indicate that the protein levels of many tubulin isoforms are altered after treatment with BR (Tang et al., 2008b). Microarray and ChIP-chip studies have identified genes encoding microtubule regulatory proteins that are responsive to BR (Sun et al., 2010). For example, the microtubule regulatory protein WAVE-DAMPENED2-LIKE3 (WDL3) is a potential BZR1 target gene. The expression levels of WDL3 are increased over 56-fold in bzr1-1D bri1-116 but are reduced by ∼0.19-fold in bri1-116, suggesting a positive role in BR-mediated plant growth (Yuen et al., 2003; Sun et al., 2010). Although the physiological contributions to the BR responses have not been evaluated through genetic and physiological analyses, the results in this study demonstrate that regulation of microtubules is crucial in BR-mediated plant growth.
This study showed that transverse patterns of cortical microtubules are induced by BRs, which is in accordance with observations that BRs promote cell growth. Decreasing expression of MDP40 resulted in significant disturbances to the reorientation of the cortical microtubules when the cells responded to BR. This demonstrates that proper reorientation of the cortical microtubules is necessary for BR responsiveness.
Cortical microtubules are also regulated by other hormones. For example, gibberellins are capable of aligning cortical microtubules transversely to the long axis of growing cells (Shibaoka, 1993; Fujino et al., 1995), and cortical microtubules reoriented longitudinally in hypocotyl cells when seedlings were grown on a medium supplemented with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (Le et al., 2005). We suggest that reorientation of cortical microtubules may be necessary for hormone-regulated cell elongation.
The Involvement of Microtubule Regulatory Proteins in BR-Regulated Hypocotyl Cell Elongation
Many microtubule regulatory proteins, such as SPR1 and MDP25, are involved in regulating hypocotyl cell elongation by altering the stability and organization of cortical microtubules (Nakajima et al., 2004, 2006; Li et al., 2011a). This study showed that the stability of cortical microtubules is decreased in hypocotyl epidermal cells following treatment with BL, which likely explains the underlying mechanism regarding cortical microtubule reorientation in response to BR. Coincidentally, cortical microtubules are more stable in the shorter hypocotyls of the det2-1 mutant but are more unstable in the longer hypocotyls of the constitutive BR signaling mutant bzr1-1D. We hypothesize that BR-regulated hypocotyl cell growth requires the destabilization of cortical microtubules.
Microtubule regulatory proteins are considered to be either microtubule stabilizers or destabilizers (Heald and Nogales, 2002). However, how the proteins coordinate to regulate hypocotyl growth is largely unknown. A previous microarray study showed that the expression of MDP40 was increased over threefold in bzr1-1D bri1-116 but was reduced in bri1-116 (Sun et al., 2010). The expression of the MDP40 gene was significantly induced by treatment with BL, and increased expression of MDP40 partially rescued the shorter etiolated hypocotyls of the BR-deficient det2 mutant, demonstrating that BZR1 directly targets MDP40, which is upregulated by BR. MDP40 plays a positive role in hypocotyl cell growth by destabilizing cortical microtubules in response to BRs. Arabidopsis SPR1 and the SPR1-like protein family also participated in the regulation of hypocotyl elongation by stabilizing cortical microtubules. Interestingly, the expression patterns of the SPR1 family members are similar to BZR1 and MDP40, which are primarily expressed in rapidly growing etiolated hypocotyl cells (Nakajima et al., 2004, 2006). Expression levels of SPR1-LIKE3, which is a homolog of SPR1, are increased by approximately twofold in the BR receptor null allele mutant bri1-116 but are reduced in bzr1-1D bri1-116 based on microarray analysis (Sun et al., 2010). Although genetic and physiological analyses are lacking, these data suggest that the SPR1 gene family may be BR downregulated and might play a negative role in BR promotion of hypocotyl growth (Nakajima et al., 2004, 2006; Sun et al., 2010). Expression of the SPR1 family members is decreased in the rapidly growing etiolated hypocotyl cells in response to BR, while expression of MDP40 is increased. Regulation of microtubule stabilizers and destabilizers results in destabilization of the cortical microtubules and promotes hypocotyl cell elongation.
Our previous study showed that the microtubule regulatory protein MDP25 is involved in the negative regulation of hypocotyl elongation by destabilizing cortical microtubules. MDP25 is dominantly expressed in the light, but not in the dark, and the expression pattern in etiolated hypocotyls of MDP25 is different than that of BZR1, which is primarily expressed in hypocotyl cells that are not in a growth phase (Li et al., 2011a). This suggests that MDP25 may function in specific cell types in BR-regulated plant cell growth in response to light in addition to playing a role in BZR1-regulated etiolated hypocotyl elongation. This evidence suggests that cell growth induced by BRs is related to the dynamics and organization of cortical microtubules. However, the molecular mechanisms regarding this regulation are complicated. For example, it is still unclear whether the destabilizing activity of MDP40 is transient or prolonged in nature for promoting hypocotyl growth and whether other microtubule regulatory proteins are also involved in maintaining a dynamic cortical microtubule array to facilitate cell elongation. To obtain a comprehensive understanding of the involvement of microtubule regulatory proteins in the regulation of BR-induced hypocotyl growth, future studies will be necessary to distinguish their diverse functions through multiple genetic and physiological assays.
In this study, an in vitro microtubule binding assay was not performed because we could not obtain the MDP40 fusion protein from bacteria. Thus, how MDP40 physically binds to and regulates cortical microtubules will be a subject of future study. Characterization of MDP40 provides strong evidence for the role of microtubules as major links between BR signaling and BR-mediated hypocotyl cell elongation. We propose the following model describing the function of MDP40 in BR-induced hypocotyl cell elongation (Figure 9J): BR signaling activates the BZR1 transcription factor, BZR1 directly targets the MDP40 promoter to upregulate MDP40 expression, and MDP40 acts on cortical microtubules with microtubule destabilizing activity to maintain the dynamic features for transverse orientation of the cortical microtubules, which promotes hypocotyl cell elongation.
METHODS
Plant Materials and Growth Conditions
All plant materials used in this study were in the Columbia-0 ecotype background of Arabidopsis thaliana. Seeds were sterilized and placed on half-strength MS medium (Sigma-Aldrich) with 0.8% agar and 1% Suc. For hypocotyl measurement, plates were placed at 22°C in the light for 12 h after stratification at 4°C for 3 d and then transferred to the dark for 4 or 5 d. Mutants det2-1 (Chory et al., 1991), bzr1-1D (Wang et al., 2002), and bri1-116 (Wang et al., 2001), 35S:Tubulin5A-YFP transgenic plants (Kirik et al., 2012), and 35S:Tubulin6A-GFP transgenic plants (Wang et al., 2007) were used in this study.
Isolation of MDP40 cDNA Clones from Arabidopsis
The full-length cDNA sequence of MDP40 was amplified using RT-PCR. The primers used to amplify MDP40 were 5′-GCTCTAGAATGGCGGGAGAGGTCCAAG-3′ and 5′-GTCGACTCACAAAGCAACCTGAACCGC-3′.
Analysis of the MDP40 Promoter: GUS Activity
A DNA fragment of the MDP40 promoter containing 2000 bp upstream of the translation start site was amplified. The sequence was reconstructed into the pCAMBIA1391 vector (Invitrogen). The primers used for amplification were 5′-CAAGCTTTCTTATATTGAGAAGTAGAACAACTACTC-3′ and 5′-CGGATCCTATATAATCTAGTAAAATGTTCGACGAG-3′. The construct was transformed into Arabidopsis plants mediated by Agrobacterium tumefaciens (strain GV3101). Thirty independent transgenic lines were obtained, and the homozygous seedlings were used for histochemical localization of GUS activity in hypocotyl cells. The GUS staining procedure was performed according to Wang et al. (2007).
BL Treatment
Four-day-old etiolated det2-1 mutants or MDP40 RNAi Arabidopsis grown on half-strength MS medium with 1 μM BRZ were used for all experiments. Seedlings were treated with BL at a concentration of 1 μM for 60 min, and cortical microtubules were observed using confocal microscopy.
MDP40 Overexpression and RNAi Arabidopsis
To prepare stable MDP40 RNAi Arabidopsis lines, a MDP40 RNAi vector with 350- or 260-bp MDP40 coding sequences in the sense and antisense orientations was amplified and inserted into a pFGC5941 vector. Two pairs of primers were used for amplification of MDP40 RNAi as follows: 5′-GGATCCTTGCTTTGGAGAGAACTG-3′ and 5′-TCTAGACTTGTAGCCAACAAAGGAG-3′; 5′-CCATGGTTGCTTTGGAGAGAACTG-3′ and 5′-CTCGAGCTTGTAGCCAACAAAGGAG-3′. Another two pairs of primers were used for amplification of MDP40 RNAi-1 as follows: 5′-GGATCCCTGTGGTACGTAAAGCTC-3′ and 5′-TCTAGAGCTGTTTCTCGCGGTCTGG-3′; 5′-CCATGGCTGTGGTACGTAAAGCTC-3′ and 5′-CTCGAGGCTGTTTCTCGCGGTCTGG-3′.
To stably express MDP40 in vivo, full-length MDP40 cDNA was amplified by PCR and subcloned into the pBI221 vector (Invitrogen). GFP was amplified and inserted at the C terminus of MDP40. The cDNAs for MDP40 and GFP were amplified and reconstructed into the expression vector pCAMBIA1300 (Invitrogen), which was under the control of the 35S promoter and a nopaline synthase terminator, and transformed into the wild type (Columbia ecotype) and det2-1 mutant (Columbia ecotype background) of Arabidopsis plants with Agrobacterium (strain GV3101). The primers used to make the constructs are described in Supplemental Table 2 online.
The transgenic homozygous Arabidopsis lines from the T3 generation were used.
PCR Analysis
RT-PCR was performed to assess the MDP40 transcript levels in MDP40 RNAi and overexpressing seedlings. Total RNA was isolated using TRIzol reagent (Invitrogen). The primers used for RT-PCR were 5′-AAGCTCGTCCTTATTCTGCGACG-3′ and 5′-TCGGCCCGTTCGTTACTTCG-3′. UBQ was used as a loading control (5′-GACCATAACCCTTGAGGTTGAATC-3′ and 5′-AGAGAGAAAGAGAAGGATCGATC-3′).
The hypocotyls from Columbia ecotype seedlings after 4 d in the dark were used. Hypocotyls from the basal region (approximately eight cells) were divided into two parts under a microscope.
For quantitative real-time PCR, an ABI 7500 real-time PCR system (Applied Biosystems) was used according to the manufacturer’s instructions. The primers used for the subsequent detection of MDP40 expression were 5′-AAGTAACGAACGGGCCGAGAAGAA-3′ and 5′-CATCTCTTGCCTTTGCCTTTGCCT-3′. Three biological replicates and two to three technical replicates (for each biological replicate) were used for each treatment. The average and standard deviation were calculated from the biological replicates. EF1α was used as an internal control (5′-GACATGAGGCAGACTGTTGCA-3′ and 5′-CCGGTTGGGTCCTTCTTGT-3′).
The primers used for the detection of WDL7 transcript levels in MDP40 RNAi seedlings were 5′-TGATGAAGAAGAAGGCGGTGGTGA-3′ and 5′-ACGTTTACAGAGGAGGAGGGTGTT-3′.
EMSA
EMSA was performed according to Zhang et al. (2012). Briefly, the recombinant MBP-BZR1 was purified from Escherichia coli with amylose resin (NEB) according to the manufacturer’s instructions. The nucleotide sequences of the double-stranded oligonucleotides were MDP40 P1 (5′-GATGAAGTAACTTTGGAGCACG-3′ and 5′-GAAACGTCCGTTCGAGAACAAG-3′) and MDP40 P2 (5′-CTTCCGTAA GCCTGAAAATC-3′ and 5′-CATGGCAATTGCATGTGAC-3′). The primers were labeled using the Biotin 5′ End DNA labeling kit (Pierce). The standard reaction mixtures (20 μL) for EMSA contained 2 μg purified proteins, 2 μL biotin-labeled annealed oligonucleotides, 2 μL 10× binding buffer (100 mM Tris, 500 mM KCl, and 10 mM DTT, pH 7.5), 1 μL 50% glycerol, 1 μL 1% Nonidet P-401 μL 1 M KCl, 1 μL 100 mM MgCl2, 1 μL 200 mM EDTA, 1 μL 1 mg/mL poly (dI-dC), and 8 μL ultrapure water. The reactions were incubated at room temperature (25°C) for 20 min and loaded onto a 6% native polyacrylamide gel in TBE buffer (45 mM Tris, 45 mM boric acid, and 1 mM EDTA, pH 8.3). The gel was sandwiched and transferred to an N+ nylon membrane (Millipore) in 0.5× TBE buffer at 380 mA at 4°C for 60 min. The detection of biotin-labeled DNA by chemiluminescence was performed based on the instructions provided in the Light Shift Chemiluminescent EMSA kit (Pierce).
ChIP
ChIP was performed as previously described (Johnson et al., 2002) with 5-d-old Columbia or BZR1-Myc overexpression lines. Antibody against the Myc tag (for BZR1) was used. Equal quantities of starting plant material and ChIP reagents were used for the PCR reaction. The primers used to detect the BZR1 target MDP40 promoter were 5′-CATGTCATGTCACATGTGC-3′ and 5′-CGATCGAACGGTTGATCTTG-3′. UBC30 and DWF4 were used as controls (5′-CCATCGAACAGTTTGGC-3′ and 5′-GGAGAGAGACAGAGACTCATC-3′ for UBC30; 5′-AATAATGCATGGTGCGATTCAGAATT-3′ and 5′-GATGCTGAAATAGTTTAACAGCTATTT-3′ for DWF4). The ChIP experiments were performed independently two to four times.
Ballistics-Mediated Transient Expression in Leaf Epidermal Cells
Subcellular localization of MDP40-GFP, cortical microtubules, and F-actin was visualized using transiently expressed 35S:MDP40-GFP, 35S:MBD-mCherry, and 35S:fABD2-GFP constructs in leaf epidermal cells of Arabidopsis (Columbia ecotype), respectively. The experiments were performed according to Fu et al. (2002). We used 1 μg 35S:MDP40-GFP, 1 μg 35S:MBD-mCherry, and 0.5 μg 35S:fABD2-GFP for particle bombardment. Six to 8 h after bombardment, GFP and mCherry signal was detected using a Zeiss LSM 510 META confocal microscope.
The filament structures of MDP40-GFP and fABD2-GFP in the leaf epidermal cells were treated with 5 μM oryzalin and 200 nM LatA for 10 min, respectively.
Measurement of Individual Microtubule Dynamics
To analyze the dynamics of individual microtubules, cells from the middle part of the hypocotyls of 4-d-old seedlings from the wild-type and MDP40 RNAi lines with GFP-tubulin backgrounds were used. Time series images 200 s in length (with 5-s intervals) were obtained under a spinning disc confocal microscope. The measurements were performed using ImageJ tools as described by DeBolt et al. (2007). Microtubules with clearly visible leading plus ends and at least 10 times of phase transitions were selected for the measurements in the wild type (n = 36 microtubules from eight seedlings) and MDP40 RNAi Arabidopsis cells (n = 40 microtubules from 10 seedlings). The rescue and catastrophe event frequencies were measured and analyzed according to Kirik et al. (2012). All of the data were processed in Excel software (Microsoft Office 2003).
Quantification of Cortical Microtubules in the Cell
ImageJ software (http://rsb.info.nih.gov/ij/) was used to quantify the density of cortical microtubules in the cell. A vertical line that oriented to the majority of the cortical microtubules with a fixed length (∼10 μm) was drawn, and the density of cortical microtubules across the line was measured. Four repeated measures were performed for each cell, and at least 36 cells from each treatment were used. The values were recorded and the significance was analyzed using the paired Student’s t test.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under accession numbers At1g23060 (MDP40) and At1g70950 (WDL7).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MDP40 Decorates Cortical Microtubules in MDP40-GFP Transgenic Arabidopsis.
Supplemental Figure 2. The Expression of WDL7 Is Unaffected in MDP40 RNAi Lines.
Supplemental Figure 3. The Expression of MDP40 Is Associated with the Hypocotyl Phenotype in MDP40 RNAi Lines.
Supplemental Figure 4. Decreased MDP40 Expression Inhibits Hypocotyl Growth in MDP40 RNAi-1 Arabidopsis.
Supplemental Figure 5. MDP40 Expression in Arabidopsis Tissues and Organs.
Supplemental Table 1. Microtubule Dynamic Parameters in Wild-Type and MDP40 RNAi Lines.
Supplemental Table 2. Primers Used to Make MDP40-GFP Transgenic Arabidopsis Constructs.
Supplemental Movie 1. MDP40-GFP Is Relatively Stationary in the Cell.
Supplementary Material
Acknowledgments
We thank Zhiyong Wang (Stanford University, Stanford, CA) and Bo Liu (University of California, Davis, CA) for generously providing the BR-related Arabidopsis mutant seeds and the Arabidopsis expressing YFP- and GFP-tubulin seeds. We also thank Kang Chong (Chinese Academy of Sciences) and Ying Fu (China Agricultural University) for kindly providing the MBP-BZR1 and MBD-mCherry constructs. This research was supported by grants from the National Basic Research Program of China (2012CB114200 to M.Y.), the Natural Science Foundation of China (31222007 and 31070258 to T.M. and 30830058 to M.Y.), and the Chinese Universities Scientific Fund (2011JS108 to T.M.).
AUTHOR CONTRIBUTIONS
T.M. designed the project. X.W. and J.Z. performed specific experiments and analyzed the data. T.M. wrote the article. M.Y. and T.M. revised and edited the article.
Glossary
- BR
brassinosteroid
- ChIP
chromatin immunoprecipitation
- BL
brassinolide
- YFP
yellow fluorescent protein
- EMSA
electrophoretic mobility shift assay
- GFP
green fluorescent protein
- RNAi
RNA interference
- GUS
β-glucuronidase
- MS
Murashige and Skoog
- BRZ
brassinazole
References
- Asami T., Min Y.K., Nagata N., Yamagishi K., Takatsuto S., Fujioka S., Murofushi N., Yamaguchi I., Yoshida S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H., Lloyd C.W. (2008). Arabidopsis mutants and the network of microtubule-associated functions. Mol. Plant 1: 888–898 [DOI] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chory J., Nagpal P., Peto C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D. (2011). Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E.F., Timpano H., Desprez T., Franssen-Verheijen T., Emons A.M., Höfte H., Vernhettes S. (2011). Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. Plant Cell 23: 2592–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S., Gutierrez R., Ehrhardt D.W., Melo C.V., Ross L., Cutler S.R., Somerville C., Bonetta D. (2007). Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc. Natl. Acad. Sci. USA 104: 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.Y., Sun Y., Cao D.M., Bai M.Y., Luo X.M., Yang H.J., Wei C.Q., Zhu S.W., Sun Y., Chong K., Wang Z.Y. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 5: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Li H., Yang Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K., Koda Y., Kikuta Y. (1995). Reorientation of cortical microtubules in the sub-apical region during tuberization in single-node stem segments of potato in culture. Plant Cell Physiol. 36: 891–895 [Google Scholar]
- Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Höfte H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat G.E., Russinova E. (2011). Plants grow on brassinosteroids. Curr. Opin. Plant Biol. 14: 530–537 [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Nogales E. (2002). Microtubule dynamics. J. Cell Sci. 115: 3–4 [DOI] [PubMed] [Google Scholar]
- Jin H., Yan Z., Nam K.H., Li J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L., Cao X., Jacobsen S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Kaloriti D., Galva C., Parupalli C., Khalifa N., Galvin M., Sedbrook J.C. (2007). Microtubule associated proteins in plants and the processes they manage. J. Integr. Plant Biol. 49: 1164–1173 [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kirik A., Ehrhardt D.W., Kirik V. (2012). TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., De Cnodder T., Van Der Straeten D., Verbelen J.P. (2005). Cell elongation and microtubule behaviour in the Arabidopsis hypocotyl: responses to ethylene and auxin. J. Plant Growth Regul. 24: 166–178 [Google Scholar]
- Li J. (2010). Regulation of the nuclear activities of brassinosteroid signaling. Curr. Opin. Plant Biol. 13: 540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., et al. (2011b). Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23: 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J., Wang X., Qin T., Zhang Y., Liu X., Sun J., Zhou Y., Zhu L., Zhang Z., Yuan M., Mao T. (2011a). MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 23: 4411–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C. (2011). Dynamic microtubules and the texture of plant cell walls. Int. Rev. Cell Mol. Biol. 287: 287–329 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2008). The parallel lives of microtubules and cellulose microfibrils. Curr. Opin. Plant Biol. 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Luo X.M., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19: 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Furutani I., Tachimoto H., Matsubara H., Hashimoto T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Kawamura T., Hashimoto T. (2006). Role of the SPIRAL1 gene family in anisotropic growth of Arabidopsis thaliana. Plant Cell Physiol. 47: 513–522 [DOI] [PubMed] [Google Scholar]
- Niwa Y., Yamashino T., Mizuno T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Perrin R.M., Wang Y., Yuen C.Y., Will J., Masson P.H. (2007). WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 49: 961–971 [DOI] [PubMed] [Google Scholar]
- Polko J.K., van Zanten M., van Rooij J.A., Marée A.F., Voesenek L.A., Peeters A.J., Pierik R. (2012). Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol. 193: 339–348 [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Kaloriti D. (2008). Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 13: 303–310 [DOI] [PubMed] [Google Scholar]
- Shibaoka H. (1993). Regulation by gibberellins of the orientation of cortical microtubules in plant cells. Aust. J. Plant Physiol. 20: 461–470 [Google Scholar]
- Shibaoka H. (1994). Plant hormone-induced changes in the orientation of cortical microtubules—Alterations in the cross-linking between microtubules and the plasma-membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45: 527–544 [Google Scholar]
- Somerville C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2011). Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Deng Z., Oses-Prieto J.A., Suzuki N., Zhu S., Zhang X., Burlingame A.L., Wang Z.Y. (2008b). Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics 7: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008a). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhu L., Liu B.Q., Wang C., Jin L.F., Zhao Q., Yuan M. (2007). Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wolf S., Mravec J., Greiner S., Mouille G., Höfte H. (2012). Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr. Biol. 22: 1732–1737 [DOI] [PubMed] [Google Scholar]
- Ye H., Li L., Yin Y. (2011). Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J. Integr. Plant Biol. 53: 455–468 [DOI] [PubMed] [Google Scholar]
- Yuen C.Y., Pearlman R.S., Silo-Suh L., Hilson P., Carroll K.L., Masson P.H. (2003). WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis. Plant Physiol. 131: 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu Y., Guo S., Zhu J., Huan Q., Liu H., Wang L., Luo G., Wang X., Chong K. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.