Abstract
During the last decade groundbreaking progress has been made towards the understanding of structure and function of cell’s translational machinery. Cryo-electron microscopic (cryo-EM) and X-ray crystallographic structures of cytoplasmic ribosomes from several bacterial and eukaryotic species are now available in various ligand-bound states. Significant advances have also been made in structural studies on ribosomes of the cellular organelles, such as those present in the chloroplasts and mitochondria, using cryo-EM techniques. Here we review the progress made in structure determination of the mitochondrial ribosomes, with an emphasis on the mammalian mitochondrial ribosome and one of its translation initiation factors, and discuss challenges that lie ahead in obtaining their high-resolution structures.
Certain organelles of the cell, such as mitochondria and chloroplasts, possess their own translational machineries, including ribosomes [1,2]. These ribosomes synthesize proteins with very specialized and important functions, including the proteins that are involved in oxidative phosphorylation (ATP generation) and photosynthesis. Since mitochondria and chloroplasts are thought to have evolved from endosymbiotic primitive bacteria in pre-eukaryotic host cells [3,4], structures of their ribosome are expected to be more similar to that of bacterial ribosomes [5,6] rather than to that of cytoplasmic ribosomes from eukaryotes [7••,8••]. Like all ribosomes, organellar ribosomes are made up of two unequally sized subunits, a small subunit (SSU) and a large subunit (LSU), each composed of at least one ribosomal RNA (rRNA) molecule and up to several dozens of ribosomal proteins (r-proteins). The primary functions of SSU and LSU are to facilitate the processes of mRNA decoding and peptide-bond formation, respectively.
The overall biochemical composition of the chloroplast ribosomes is very similar to that of a bacterial ribosome [9,10], whereas the composition of mitochondrial ribosomes (mitoribosome) is dramatically different (Table 1). Unlike the situation with cytoplasmic ribosomes, for which several high-resolution X-ray crystallographic structures are available [5,6,7••,8••, for recent reviews see 11,12•,13•], only a few cryo-electron microscopic (cryo-EM) structures are available for the organellar ribosomes, including two mitoribosomes, one each from mammalian tissues [14] and a protistan cell [15], and two chloroplast ribosomes [9,10]. As expected, structures of the chloroplast ribosome show greater resemblance to that of the bacterial ribosome, while a number of distinct structural features have been identified in the structures of both mitoribosomes [for a recent review see 16•]. Unfortunately there is no structure available for the mitoribosome from yeast, where the mitochondrial translation system is best understood biochemically and genetically [for a recent review see 17•]. In this article, we first summarize the information available on overall structural organization of the mammalian and protistan mitoribosomes and then describe the recent progress in structural characterization of one of the mammalian mitochondrial translation initiation factors. Since all of the polypeptides synthesized on the mammalian mitoribosome are inserted into the mitochondrial inner membrane (mtIM) [18,19], we discuss the structural relationship between the mitoribosome and mtIM in context of the recent biochemical findings.
Table 1.
Comparison of composition and physical properties of bacterial, mammalian mitochondrial, and protistan mitochondrial ribosomes*.
| Ribosome source → Properties ↓ |
Bacteria E. coli |
Mammalian mitochondria B. taurus |
Protistan mitochondria L. tarentolae |
|---|---|---|---|
| Molecular mass | 2.3 MDa | 2.7 MDa | 2.2 MDa |
|
Sedimentation coefficient |
70S | 55S | 50S |
|
RNA : protein ratio |
~2:1 | ~1:2 | ~1:3 |
| Subunits | 30S + 50S | 28S + 39S | (28S – 30S) + 40S |
|
Small subunit composition |
16S rRNA (1542 nt) + 21 proteins |
12S rRNA (950 nt) + 29 proteins (15)** |
9S rRNA (610 nt) + 56 proteins† (46) |
|
Large subunit composition |
23S rRNA and 5S rRNA (total 3024 nt) + 34 proteins |
16S rRNA (1560 nt) + 50 proteins (20) |
12S rRNA (1173 nt) + 77 proteins† (66) |
| Diameter | ~260Å | ~320 Å | ~245 Å |
Modified from [16•]
The number of mitochondrial proteins that do not have bacterial homologs are shown in brackets.
The estimated number of proteins in the L. tarentolae mitoribosomal subunits has been derived from trypanosoma brucei [21], the closest known relative of Leishmania.
nt, nucleotide(s)
Structures of Mitochondrial Ribosomes
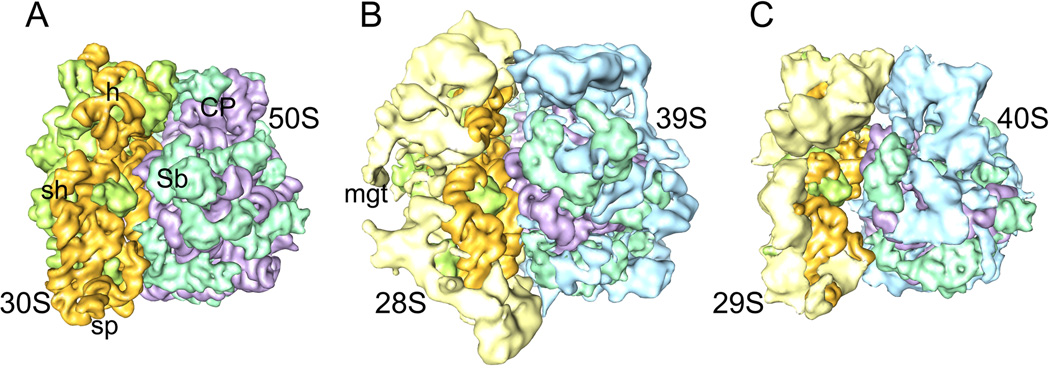
The cryo-EM structures of two mitoribosomes (Fig. 1), studied so far, have revealed several of their unique features. Both mitoribosomes are found to be highly porous structures [14,15], as compared to their bacterial counterpart despite similar molecular masses (Table 1). The porosity in structure is primarily due to the absence of several bacterial rRNA segments in those mitoribosomes [see ref 16•, for comparative secondary structures of rRNA]. Even though the proportion of r-proteins is significantly increased as compared to that in the bacterial ribosome [20,21], many of new protein masses occupy new spatial positions in the mitoribosome structures. Thus, there is limited structural compensation for missing rRNA segments by new, or enlarged homologs of bacterial, r-proteins. For example, in the mammalian (Bos taurus) mitoribosome, only ~20% of missing rRNA segments is structurally replaced by mitochondrial r-proteins (MRPs) [14], while such a compensation is much higher (>50%) in the protistan (Leishmania tarantolae) mitoribosome [15].
Fig. 1. Comparison between structures of bacterial and mitochondrial ribosomes.
RNA-protein segmented structures of ribosomes from (A) bacteria (70S, E. coli) (B) mammalian mitochondria (55S, Bos taurus), and (C) protistan mitochondria (50S, Leishmania tarentolae). In panel A, atomic structure [5] has been low-pass filtered to match the resolution of the cryo-EM map of mammalian mitoribosome shown in panel B, whereas the cryo-EM map of the protistan mitoribosome is shown at ~14 Å [15]. Mito-specific MRPs of SSU and LSU are colored yellow and blue, respectively; conserved MRPs of SSU and LSU are colored green and aquamarine, respectively; and rRNAs of SSU and LSU are colored orange and magenta, respectively. Landmarks of the small subunit: h, head; mgt, mRNA gate; sh, shoulder; sp, spur. Landmarks of the large subunit: CP, central protuberance; Sb, Stalk base or protein L11 region.
Due to significant reduction in the size of their rRNAs, both subunits of the mitoribosome possess several tunnel-like features that appear to connect the inter-ribosomal subunit space with the bulk solvent. However, the functional core of both mitoribosomes, i.e., the regions involved in mRNA decoding on the SSU [22] and peptide-bond formation on the LSU [23], are predominantly made up of conserved rRNA segments, even though there are greater occurrences of protein masses on the interface side of both their subunits as compared to that in the subunits of the cytoplasmic ribosomes. Many of these protein masses participate in the formation of protein-protein inter-subunit bridges, and paths of mRNA and tRNA interactions [14,15]. The rRNA scaffolds within the mitoribosome structures are highly shielded by MRPs, such that the solvent-facing surfaces of the mitoribosomal subunits are predominantly made up of MRPs. This situation is in sharp contrast to the pattern of distribution of rRNA and r-protein masses within the structures of bacterial or chloroplast ribosomes, where proteins are present in relatively small patches on the solvent-side surfaces (Fig. 1) [16•]. Interestingly, most (11 out of 12) bacterial 23S rRNA segments that form the tRNA-exit site (E site) [24] are absent in the mito-LSU rRNAs [14,15,25,26], implying that the E site is either absent or very weak in both these mitoribosomes and that the deacylated mitochondrial tRNA (tRNAmt) may not stay on either of these two mitoribosomes after its release from the ribosomal peptidyl site (P site) during the translation elongation cycle [27]. These unique features of the mitoribosome structures suggest that the modus operandi of these ribosomes has diverged significantly from their cytoplasmic or chloroplast counterparts.
Structure of the Mammalian Mitoribosome
In addition to the common features of the mitoribosome structures described in the previous section, here we highlight some of the unique features of the mammalian mitoribosome structure. Due to the absence of bacterial SSU rRNA helices 16 and 17 that are not compensated by MRPs, the mammalian mitoribosomal SSU has a narrower shoulder and body regions and a more exposed translation factor-binding region, as compared to its bacterial counterpart. However, the body of the mammalian mitoribosomal SSU is significantly elongated due to the addition of MRPs to than in its lower body region. In addition, a triangular gate-like structure, made entirely of MRPs, partially covers the mRNA entry site on the solvent side of the mitoribosomal SSU [14] (Fig. 1B). More recent 7.0 Å resolution cryo-EM maps of the mammalian mitoribosome (MR Sharma, TM Booth, ME Haque, LL Spremulli, RK Agrawal, in preparation) suggest that this gate-like feature is a highly dynamic structure, and may be involved in recruiting the generally leaderless mitochondrial mRNAs [28] to the mitoribosomal SSU.
On the mammalian mitoribosomal LSU, the central protuberance is much larger in size as compared to that in a bacterial ribosome, even though density for the 5S rRNA has not been detected within the cryo-EM map of the mitoribosomal LSU. [It should be noted that only two rRNAs, 12S and 16S, corresponding to the mitoribosomal SSU and LSU, respectively, are encoded by the mammalian mitochondrial genome.] However, a recent biochemical study suggests that the 5S rRNA is transported into the mitochondria along with MRP L18 [29]. Thus, the possibility that the 5S rRNA is present only in a very small subset of isolated mitoribosomal population, and therefore goes undetected in the averaged cryo-EM maps, cannot be ruled out. In all of the cryo-EM maps, a mito-specific MRP emerges from the inter-subunit face of the central protuberance of the LSU and contacts the P-site tRNAmt at its T-loop side [14]. This extended structure, referred to as the P-site finger, is a dynamic feature that may stabilize and regulate the binding and movements of generally short mammalian tRNAmts [30]. Another important structural aspect of the mammalian mitoribosomal LSU is its nascent polypeptide exit tunnel, which has been separately described later in this article.
Structure of the Mammalian Mitochondrial Translation Initiation Factor 2
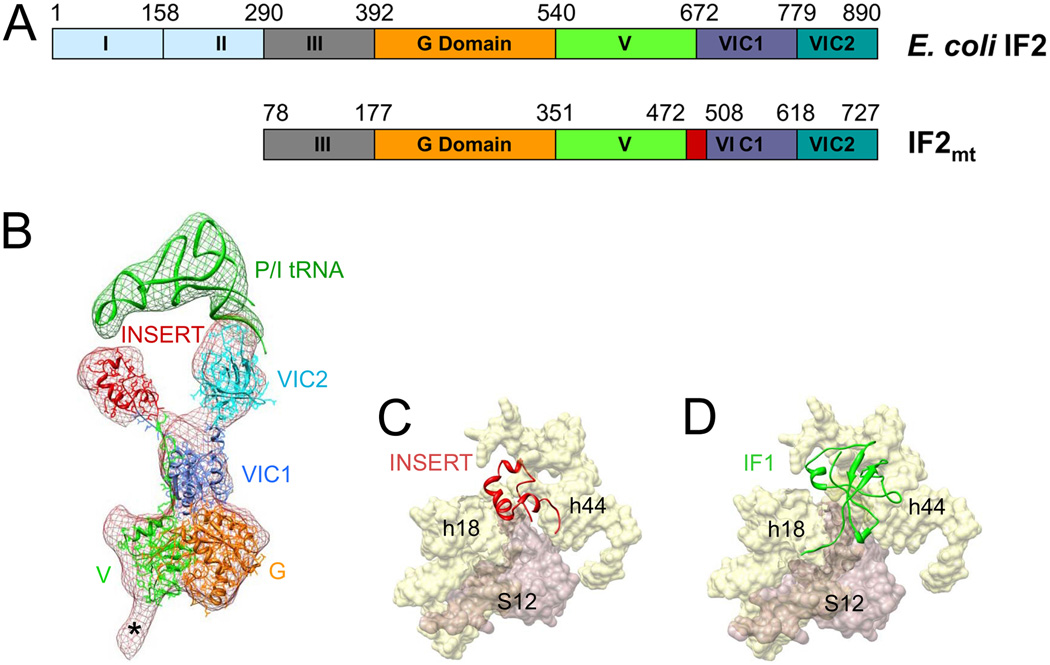
Steps in the mechanism of mammalian mitochondrial translation initiation are somewhat similar to those in a bacterial cell. However, of the three bacterial initiation factors (IF1, IF2, and IF3), homologs of only two, IF2mt and IF3mt, have been identified in the mammalian mitochondria [31]. Both these factors show significant alterations in their structural organization as compared to their bacterial counterparts [32,33•]. Interestingly, IF2mt, which stimulates the binding of fMet-tRNA to the SSU of the mitoribosome, lacks domains I and II of E. coli IF2 [34] but contains a functionally important 37 amino-acid insertion domain (Fig. 2A).
Fig. 2. Structure of the mammalian IF2mt as derived by cryo-EM and location of its insertion domain relative to the binding site of bacterial IF1.
(A) Domain alignment of E. coli IF2 and bovine IF2mt. Note that the mature IF2mt (i.e., after deletion of the import sequence) starts from aa residue 78. The 37-aa insertion domain in IF2mt is highlighted in red. (B) Fitting of atomic models of the IF2mt and initiator tRNA (P/I stands for P-site tRNA at initiator position) into the corresponding cryo-EM densities (meshwork) extracted from the map of the 70S•IF2mt•GMPPNP•fMet-tRNA complex [35••]. The color codes used for various domains of IF2mt are the same as in panel A. Asterisk (*) point to the region that would correspond to domain III of IF2mt but was not modeled. (C) Binding positions of the insertion domain (red), and (D) IF1 [37] (green), into a common binding pocket on the SSU of the ribosome. Landmarks of the SSU: h44 and h18, 16S rRNA helices; and S12, r-protein S12 (adopted from ref. [35••]).
Cryo-EM maps of the IF2mt•GMPPNP•fMet-tRNA in complex with both the 70S bacterial ribosome [35••] and the 55S mitoribosome (MR Sharma, AS Yassin, ME Haque, LL Spremulli, RK Agrawal, in preparation) have been determined. The overall binding positions of conserved domains of IF2mt to the ribosome are similar to those of the bacterial IF2 [36]. However, the density corresponding to the insertion domain protrudes out from the rest of the IF2mt density (Fig. 2B) to partially occupy the ribosomal aminoacyl-tRNA binding site (A-site) on the SSU, overlapping with the binding site of bacterial IF1 [37]. Thus, this structural study [35••] shows that IF2mt possesses dual functions corresponding to bacterial IF1 and IF2, corroborating a previous genetic study [38]. Despite the fact that there are significant size mismatch and no sequence homology or structural similarity between the insertion domain (37 amino acid) of IF2mt and the bacterial IF1 (71 amino acids) (Fig. 2C,D), both structures bear similar surface charge distributions, which might play an important role in recognition of the same ribosomal binding pocket [35••]. The presence of long linkers between the insertion domain and the rest of the IF2mt molecule [35••,39] suggests that the relative position of the insertion domain might vary between its native and ribosome-bound states, as also observed for the bacterial release factors [40]. An integration of a relatively small IF1-mimicking feature onto the IF2mt might have occurred during the course of evolution to serve a dual purpose: (i) one less protein had to be transported into the mitochondrial matrix, and (ii) to ensure an efficient transport of the IF1 feature to the mitochondrial protein synthesis site within highly dense mitochondrial matrix [41].
The Polypeptide-Exit Tunnel and Its Interaction with mtIM
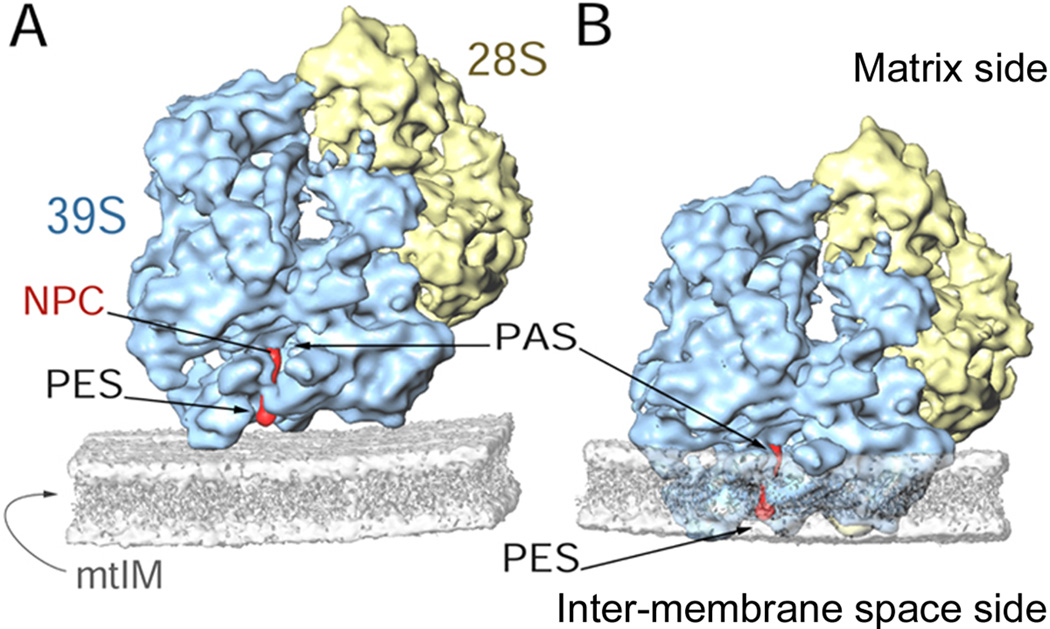
In contrast to the variety of possible destinations for the nascent polypeptide chain synthesized by the cytoplasmic and chloroplast ribosomes, all of the nascent chains synthesized by the mammalian mitoribosome are inserted into the mtIM, and it is most likely that these polypeptides are inserted co-translationally into mtIM [18,19]. The nascent chain exit tunnel in the mitoribosomes appears to be tailor made for this purpose [16•]. The exit tunnel in the mitoribosome LSU has two openings on the solvent side, one corresponding to the conventional polypeptide exit site (PES) [42], and the second site at a distance of ~25 Å from the PES. This site of premature exposure of the nascent chain is located closer to the peptidyl-transferase center and allows free access to the solvent, and is designated as the polypeptide-accessible site [PAS, 14,15]. PAS is formed because the majority of the rRNA domain I and a significant portion of domain III of LSU rRNA, which together form the inner lining of the lower portion of the exit tunnel in bacterial and archaeal ribosomes [42], are absent in the mitoribosome and are not compensated by MRPs. The solvent-side openings of both PES and PAS are predominantly encircled by mito-specific MRPs [16•], which may be involved in the adherence of the mitoribosome to the mtIM [43,44] through either PES (Fig. 3A) or PAS, such that the lower portion of the mitoribosome is embedded into the mtIM with PAS and PES exposed at opposite sides of mtIM (Fig. 3B).
Fig. 3. Hypothetical models of the mammalian mitoribosome interaction with the mtIM.
(A) The conventional polypeptide-exit site (PES) of the ribosome interacts with the matrix side of the mtIM, which is depicted as a generic lipid bilayer (semitransparent grey). (B) The lower portion of the mitoribosome is partially embedded into the mtIM, such that the conventional PES is exposed in the inter-membrane space side and the polypeptide accessible site (PAS) of the mitoribosome remains exposed on the matrix side of the mtIM. Other labels: NPC, modeled nascent polypeptide chain (red); 28S, SSU (yellow); and 39S, LSU (blue).
In yeast, Oxa1p, an integral membrane protein [45] and a mitochondrial homolog of bacterial YidC [46], interacts with the nascent chain while it is still on the ribosome. The human homolog of Oxa1p is known as Oxa1L [47]. The C-terminal tail of Oxa1L (Oxa1L-CTT) is exposed to the mammalian mitochondrial matrix and has been cross-linked to the mitoribosomal LSU [48]. Interestingly, Oxa1L-CTT does not interact with the conserved components (homologs of bacterial r-proteins L22, L23, L24, and L29) of conventional PES. Instead, it crosslinks to other homologs of bacterial r-proteins (L13, L20, and L28) and several mito-specific MRPs (MRPL48, MRPL49, and MRPL51), implying that these MRPs are situated close to the PAS, which would then be close to the matrix side of the mtIM (as depicted in Fig. 3B). A recent study showed that a mitochondrial protein ATAD3 can interact with both the mitochondrial DNA nucleoid and mitoribosome, as well as with mtIM [49•]. Another study showed an interaction between MRPL7 and the mitochondrial RNA polymerase [50•]. Both these studies suggest a coupling between mitochondrial transcription and translation machineries, and strengthen the view that mitoribosomes are either situated very close to, or partially embedded into, the mtIM. Based on these findings it is tempting to suggest that each mitoribosome could be associated with its own mitochondrial DNA (as there are more than 1000 copies of DNA in a given mitochondrion) to facilitate the synthesis and mtIM insertion of a defined set of polypeptide(s) and to ensure the high level of accuracy and efficiency needed to assemble the complexes of oxidative phosphorylation.
Concluding Remarks
From the cryo-EM structures of mitoribosomes and their comparison with the atomic structures of the cytoplasmic ribosomes, some novel aspects of mitochondrial protein synthesis have begun to emerge, suggesting that mitoribosomes have diverged significantly to adopt different host cell types during the course of evolution. However, higher resolution structures are necessary to unravel details of the mechanisms of three very fundamental processes associated with the mammalian mitoribosome: (i) recruitment of leaderless mRNAs, (ii) decoding of atypical codons by unusual tRNAmt, and (iii) the release of nascent chains through the dramatically remodeled exit tunnels. Structural characterization of the protein components present near key functional sites, such as at the mRNA entrance, and PAS and PES of the nascent chain exit tunnel, will be critical in establishing detailed molecular mechanism of these processes. While the tracing of the rRNA scaffolds can be achieved with high confidence in current 7 Å resolution maps, identification and modeling of mito-specific MRPs, especially those with undefined secondary structure motifs, into the cryo-EM map is still challenging. In the absence of any atomic structures of MRPs or mitochondrial translational factors, molecular interpretation of structures of all mitoribosomes and their functional complexes has to be based currently on docking of homology models into the cryo-EM maps. Furthermore, mitoribosomes seem to be inherently heterogeneous in composition due to (i) dramatic reduction in size of their rRNAs and significant increase in the number of MRPs; consequently, some of the mito-specific MRPs may be less firmly attached to the rest of the mitoribosome in the absence of their direct interaction with the main rRNA scaffolds, and (ii) presence of multiple isoforms of MRPs (e.g., MRPS18) in a given mitoribosome population. Thus, the progress in determining their higher resolution structure is hindered mainly due to challenges in obtaining highly homogeneous preparations. Nevertheless, because of improved algorithms to classify particle images, cryo-EM has been very effective in providing the overall structures of the mitoribosome and associated translation factors at reasonable resolutions.
Highlights.
We reviewed structures of mitochondrial ribosomes, which are significantly different from their cytoplasmic counterparts.
Functional rRNA core of the mitochondrial ribosomes is conserved, but their surfaces were remodeled during evolution.
Cryo-EM structures suggest that the protistan and mammalian mitochondrial ribosomes must have evolved differently.
The translation initiation factor 2 of the mammalian mitochondria also possess the features of bacterial initiation factor 1.
The nascent polypeptide exit tunnel seems to be tailor made to meet the specialized needs of the mitochondrial translation.
Acknowledgements
This work was supported by a grant from the National Institutes of Health, GM61576 (to R.K.A.). Authors thank Mr. Prateek Kumar and Dr. Partha Datta for help with Figure 3, and Dr. Terence Wagenknecht for critical reading of the manuscript.
Abbreviations
- SSU
Small subunit
- LSU
Large subunit
- rRNA
ribosomal RNA
- r-protein
ribosomal protein
- mitoribosome
mitochondrial ribosome
- mtIM
mitochondrial inner membrane
- MRP
mitochondrial r-protein
- IF
translation initiation factor
- PES
polypeptide exit site
- PAS
polypeptide accessible site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Harris EH, Boynton JE, Gillham NW. Chloroplast ribosomes and protein synthesis. Microbiol Rev. 1994;58:700–754. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286:73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 3.Margulis L. Origin of Eukaryotic Cells. New Haven: Yale Univ. Press; 1970. [Google Scholar]
- 4.Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2:1018.1–1018.5. doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 6.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 7. Ben-Shem A, Garreau de Loubresse N, Mekinikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642.. •• This paper describes the first X-ray crystallographic structure of a complete eukaryotic 80S ribosome from yeast Saccharomyces cervisiae at 3 Å resolution. The structure provides an important framework for designing new biochemical experiments and analyzing large body of biochemical data available for eukaryotic translation.
- 8. Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204.. •• The paper decribes the x-ray crystallographic structure of the eukaryotic 60S LSU from Tetrahymena thermophila, in complex with eukaryotic initiation factor 6 (IF6) and cycloheximide, at 3.5 Å resolution. The study elucidates the roles of eukaryotic r-proteins, eukaryotic-specific differences in functional regions of 60S LSU, and the role of IF6 in the initiation of protein synthesis. Together with other studies [5,6,7••], this study provides an important resource for comparative structural analyses of the organellar ribosomes.
- 9.Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci USA. 2007;104:19315–19320. doi: 10.1073/pnas.0709856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manuell AL, Quispe J, Mayfield SP. Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol. 2007;5:e209. doi: 10.1371/journal.pbio.0050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 12. Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of eukaryotic ribosomes. Trends Biochem Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007.. • An excellent review describing X-ray crystallographic studies of three structures determined for the cytoplasmic 80S ribosome and its two subunits, highlighting some of the eukaryote-specific features of the cytoplasmic ribosomes.
- 13. Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313.. • Another excellent, recent review on structures of cytoplasmic ribosome from eukaryote.
- 14.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosomes reveals an expanded role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 15.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci USA. 2009;106:9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agrawal RK, Sharma MR, Yassin A, Lahiri I, Spremulli LL. Structure and function of organellar ribosomes as revealed by cryo-EM. In: Rodnina M, Wintermeyer W, Green R, editors. Ribosomes: Structure, Function, and Dynamics. New York: SpringerWien; 2011. pp. 83–96.. • A recent review on structure and function of organellar ribosomes. The article provides a side-by-side comparison of secondary structures of ribosomal rRNAs, description of overall structures of bacterial, chloroplast and mitochondrial ribosomes, and identifies some of the organelle-specific and mito-specific features of these ribosomes and their polypeptide-exit tunnels.
- 17. Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: The concept of translational activators. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.03.007. (in press) doi: 10.1016/j.bbamcr.2012.03.007.. • An excellent review on the translational control in the yeast Saccharomyces cervisiae mitochondria. The article summarizes the basic components of the yeast mitochondrial translation machinery, and describes recent progress in biochemical studies on the translational controls involving translational activators and their feed-back control loops in yeast mitochondria.
- 18.Jia L, Kaur J, Stuart RA. Mapping of the Saccharomyces cerevisiae Oxa1-mitochondrial ribosome interface and identification of MrpL40, a ribosomal protein in close proximity to Oxa1 and critical for oxidative phosphorylation complex assembly. Eukaryot Cell. 2009;8:1792–1802. doi: 10.1128/EC.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruschke S, Ott M. The polypeptide tunnel exit of the mitochondrial ribosome is tailored to meet the specific requirements of the organelle. Bioessays. 2010;32:1050–1057. doi: 10.1002/bies.201000081. [DOI] [PubMed] [Google Scholar]
- 20.Koc EC, Haque ME, Spremulli LL. Current views of the structure of the mammalian mitochondrial ribosome. Israel J Chem. 2010;50:45–59. [Google Scholar]
- 21.Zíková A, Panigrahi AK, Dalley RA, Acestor N, Anupama A, Ogata Y, Myler PJ, Stuart KD. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7:1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogle JM, Broderson DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 23.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 24.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 25.Mears JA, Cannone JJ, Stagg SM, Gutell RR, Agrawal RK, Harvey SC. Modeling a minimal ribosome based on comparative sequence analysis. J Mol Biol. 2002;321:215–234. doi: 10.1016/s0022-2836(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 26.Mears JA, Sharma MR, Gutell RR, McCook AS, Richardson PE, Caulfield TR, Agrawal RK, Harvey SC. A structural model for the large subunit of the mammalian mitochondrial ribosome. J Mol Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agrawal RK, Spahn CM, Penczek P, Grassucci RA, Nierhaus KH, Frank J. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J Cell Biol. 2000;150:447–460. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs-like members of all families, similar but different. Biochim Biophys Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smirnov A, Entelis N, Martin RP, Tarassov I. Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev. 2011;25:1289–1305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K. Unique features of animal mitochondrial translation systems: the non-universal genetic code, unusual features of the translational apparatus and their relevance to human diseases. Proc Jpn Acad Ser B. 2010;86:11–39. doi: 10.2183/pjab.86.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J Biol Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 32.Spremulli LL, Coursey A, Navratil T, Hunter SE. Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog Nucleic Acid Res Mol Biol. 2004;77:211–261. doi: 10.1016/S0079-6603(04)77006-3. [DOI] [PubMed] [Google Scholar]
- 33. Christian BE, Spremulli LL. Mechanism of protein synthesis in mammalian mitochondria. Biochim Biophys Acta. 2012;1819:1035–1054. doi: 10.1016/j.bbagrm.2011.11.009.. • An excellent, comprehensive review of the current state of mammalian mitochondrial translational studies. A large body of literature on biochemical studies has been incorporated to discuss each of the major steps of the mitochondrial translation.
- 34.Spencer AC, Spremulli LL. The interaction of mitochondrial translational initiation factor 2 with the small ribosomal subunit. Biochim Biophys Acta. 2005;1750:69–81. doi: 10.1016/j.bbapap.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 35. Yassin AS, Haque ME, Datta PP, Elmore K, Banavali NK, Spremulli LL, Agrawal RK. Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1. Proc Natl Acad Sci USA. 2011;108:3918–3923. doi: 10.1073/pnas.1017425108.. •• This paper describes the first structure of a mammalian mitochondrial translational factor, IF2mt, in complex with initiator tRNA and the bacterial 70S ribosome, using cryo-electron microscopy. The study provides new insights into the mechanism of translation initiation in mitochondria and the evolution of one of the mitochondrial translational initiation factors.
- 36.Simonetti A, Marzid S, Jenner L, Myasnikov A, Romby P, Yusopova G, Gualerzi CO, Klaholz BP, Yusupov M. A structural view of translation initiation in bacteria. Cell Mol Life Sci. 2009;66:423–436. doi: 10.1007/s00018-008-8416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 38.Gaur R, Grasso D, Datta PP, Krishna PD, Das G, Spencer A, Agrawal RK, Spremulli L, Varshney U. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol Cell. 2008;29:180–190. doi: 10.1016/j.molcel.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yassin AS, Agrawal RK, Banavali NK. Computational exploration of structural hypotheses for an additional sequence in a mammalian mitochondrial protein. PLoS One. 2011;6:e21871. doi: 10.1371/journal.pone.0021871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 41.Srere PA. The infrastructure of the mitochondrial matrix. Trends Biochem. 1980;5:120–121. [Google Scholar]
- 42.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 43.Liu M, Spremulli MM. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J Biol Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- 44.Vogel F, Bornhovd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrmann JM, Neupert W, Stuart RA. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Funes S, Kauff F, van der Sluis EO, Ott M, Herrmann JM. Evolution of YidC/Oxa1/Alb3 insertases: three independent gene duplications followed by functional specialization in bacteria, mitochondria and chloroplasts. Biol Chem. 2011;392:13–19. doi: 10.1515/BC.2011.013. [DOI] [PubMed] [Google Scholar]
- 47.Haque ME, Elmore KB, Tripathy A, Koc H, Koc EC, Spremulli LL. Properties of C-terminal tail of human mitochondrial inner membrane protein Oxa1L and its interactions with mammalian mitochondrial ribosomes. J Biol Chem. 2010;285:28353–28362. doi: 10.1074/jbc.M110.148262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haque ME, Spremulli LL, Fecko CJ. Identification of protein-protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J Biol Chem. 2010;285:34991–34998. doi: 10.1074/jbc.M110.163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, Walker JE, Holt IJ. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nuc Acids Res. 2012;40:6109–6121. doi: 10.1093/nar/gks266.. • This recent biochemical study is interesting because it identifies a mitochondrial protein ATAD3 that can interact with the mammalian mitochondrial ribosome, mitochondrial DNA nucleoid, and is tightly associated with the inner mitochondrial membrane, suggesting that the replication and transcription machineries could be close to the site of protein synthesis on the mitochondrial inner membrane.
- 50. Surovtseva YV, Shutt TE, Cotney J, Cimen H, Chen SY, Koc EC, Shadel GS. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc Natl Acad Sci USA. 2011;108:17921–17926. doi: 10.1073/pnas.1108852108.. • A recent biochemical finding that links the components of mitochondrial transcription and translation machineries, as it shows that one of the mitochondrial ribosomal proteins L7 associates with the mitochondrial RNA polymerase. Together with another recent study [49], this observation supports the view that both transcription and translation events could be coupled, and might be happening close to the inner mitochondrial membrane.