Abstract
Emerging concepts suggest that macrophage functional phenotype is regulated by transcription factors that define alternative activation states. We found that RBP-J, the major nuclear transducer of Notch signaling, augmented TLR4-induced expression of key mediators of classically activated M1 macrophages and thus innate immune responses to L. monocytogenes. Notch-RBP-J signaling controlled expression of the transcription factor IRF8 that induced downstream M1-specific genes. RBP-J promoted IRF8 protein synthesis by selectively augmenting IRAK2-dependent TLR4 signaling to the MNK1 kinase and downstream translation initiation control through eIF4E. These results define a signaling network in which Notch-RBP-J and TLR signaling are integrated at the level of IRF8 protein synthesis and identify a mechanism by which heterologous signaling pathways can regulate TLR-induced inflammatory macrophage polarization.
INTRODUCTION
Macrophages play an essential role in performing sentinel and effector functions in innate immunity and the transition to adaptive immunity. Depending on environmental cues, macrophages can assume a spectrum of activation states ranging from classically activated, M1 inflammatory macrophages, to various alternatively activated M2 macrophages that are involved in immune regulation and tissue repair1. M1 macrophages are characterized by production of inflammatory mediators, such as IL-12 and inducible nitric oxide synthase (iNOS), in response to microbial product-mediated activation of Toll-like receptors (TLRs) and cytokines such as interferon-γ (IFN-γ)1. In contrast, M2 macrophages express less inflammatory mediators and play a key role in wound healing, host defense against helminths, and resolution of inflammation1. Recent work links specific transcription factors to macrophage functional phenotypes2,3, suggesting a parallel to T cell biology, in which lineage-specific transcription factors regulate cell differentiation. Members of the interferon regulatory factor (IRF) family are transcriptional regulators of macrophage polarization, with IRF5 and IRF4 associated with M1 and M2 polarization, respectively2,3. IRF8 is induced by IFN-γ and contributes to induction of several genes, including Ifnb4, Il12b (encodes the IL-12p40 subunit)5, Il12a (encodes the IL-12p35 subunit)6 and Nos2 (encodes iNOS)7 in response to TLR stimulation, and thus plays a role in host defense against intracellular pathogens such as vaccinia virus and Leishmania major8. In the immune system, IRF8 also regulates the development of lymphoid and myeloid lineages and is indispensable for generation of plasmacytoid dendritic cell (pDC) and CD8+ DC populations9,10.
Stimulation of TLRs activates at least three major downstream signaling pathways: nuclear factor (NF)-κB, mitogen-activated protein kinases (MAPKs), and IRFs11 to induce gene transcription. However, TLR responses are also modulated by a variety of post-transcriptional mechanisms, including regulation of mRNA decay and transport, and control of translation initiation12. Translation control mechanisms often target the process of translation initiation, during which recruitment and assembly of translation initiation factors, including the main cap-binding protein eukaryotic initiation factor 4E (eIF4E), on target mRNAs activates translation13. Cytokines, chemokines, and enzymes are often targets of translational control12. Whether translational regulation controls other molecules, such as signaling intermediates and transcription factors, remains an open question.
The Notch signaling pathway regulates cell differentiation, proliferation, survival and development14. Ligation of Notch receptors by their ligands leads to cleavage of Notch by ADAM family proteases, and subsequent intramembranous cleavage by a γ-secretase to release the Notch intracellular domain (NICD). NICD translocates to the nucleus and binds to the DNA-binding protein RBP-J (also named CSL or CBF1)14. In the immune system, the most established functions for Notch signaling are in regulating lymphocyte development and function15. Recent data also suggest a role for the Notch pathway in regulating myeloid cell differentiation and function16–24. However, the mechanism of action of the Notch-RBP-J pathway in macrophage polarization is unknown.
In this study we found that the Notch-RBP-J pathway controls expression of prototypical M1 effector molecules such as IL-12 and iNOS and promotes host defense against the intracellular pathogen Listeria monocytogenes (L. monocytogenes). We identified IRF8 as a downstream target of the Notch-RBP-J pathway and showed that RBP-J regulated the translation of IRF8 by selectively modulating TLR4 signaling through IRAK2-dependent activation of the kinase MNK1 and eIF4E-controlled initiation of translation. These studies delineate a signaling network in which the Notch-RBP-J and the TLR signaling pathways are integrated at the level of IRF8 protein synthesis to regulate the induction of the M1 phenotype in macrophages.
RESULTS
RBP-J controls prototypical M1 gene expression
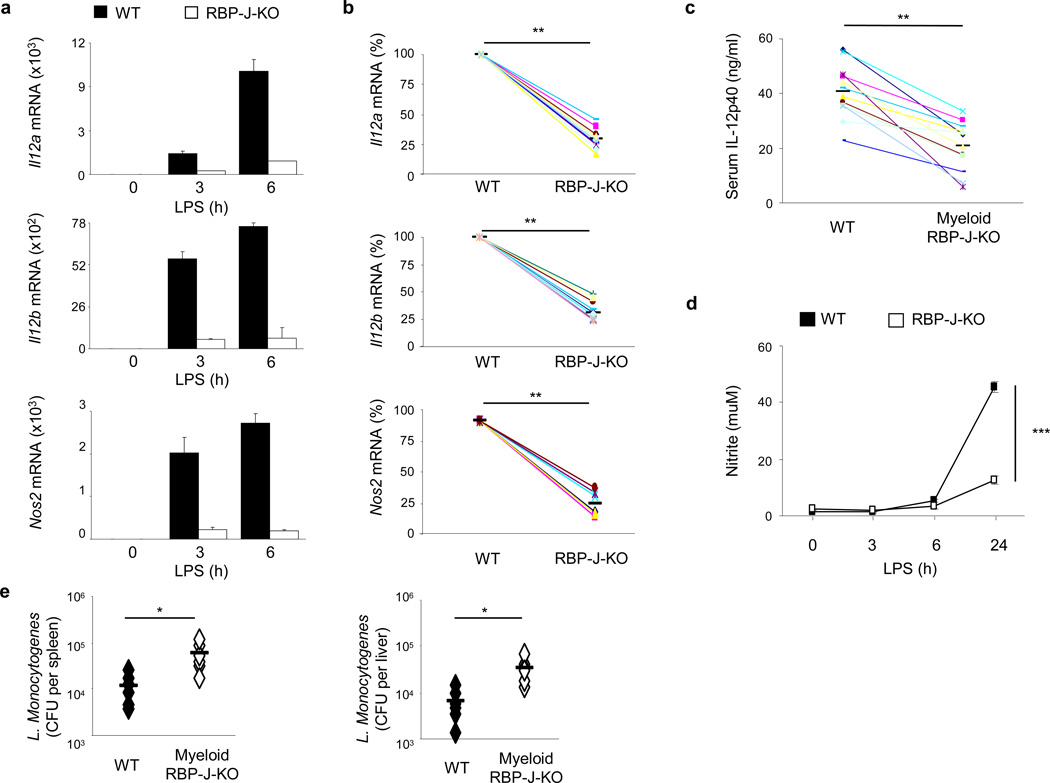
In order to investigate the role of the Notch-RBP-J pathway in macrophage activation, gene expression profiling was performed with wild-type and RBP-J-deficient bone marrow-derived macrophages (BMDMs) stimulated with the TLR4 ligand lipopolysaccharide (LPS), which induces the expression of key M1 proteins like IL-12 and iNOS1. Efficient deletion of RBP-J in BMDMs from RBP-J-deficient mice (Rbpjflox/flox, Mx1-Cre) was confirmed at the mRNA and protein expression levels (Supplementary Fig. 1a, b). Microarray analysis showed that approximately 10% of TLR4-inducible genes were partially dependent on RBP-J and that a very broad range of TLR target genes were induced to normal levels in RBP-J-deficient macrophages and thus were RBP-J-independent (Supplementary Fig. 1c and data not shown). However, a small number of LPS-induced genes (< 10) were essentially completely dependent on RBP-J (greater than 80% reduction in RBP-J-deficient cells). Among these genes, Il12a, Il12b and Nos2 dependence on RBP-J expression was confirmed by quantitative PCR (qPCR) (Fig. 1a, b).
Figure 1. RBP-J controls expression of prototypical M1 genes.
(a) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice and Rbpj+/+, Mx1-Cre (wild-type; WT) littermate controls, stimulated with LPS (1 ng/ml) for the indicated periods. Unstimulated wild-type controls were set to 1. Results are normalized relative to Gapdh mRNA. Data are shown as means + SD of triplicate determinants. (b) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice, stimulated with LPS for 3 or 6 h. Data are represented as percentage of maximal mRNA expression in wild-type cells. Cumulative data from 6–12 independent experiments is shown. (c) ELISA of IL-12p40 expression in the serum of Rbpjflox/flox, LysM-Cre (RBP-J-KO) and Rbpj+/+, LysM-Cre (WT) mice, injected intraperitoneally with 200 µg of LPS, followed by blood collection 3 h later (n = 12 per group; average values are shown as horizontal bars). (d) NO release measured, as the NO metabolite nitrite, in the supernatants of Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) BMDMs, treated with LPS. Data shown are representative of three independent experiments. (e) Bacterial CFUs in the spleens and livers of Rbpjflox/flox, LysM-Cre (RBP-J-KO) or control Rbpj+/+, LysM-Cre (WT) bone marrow chimeras infected intravenously with 3 × 103 L. monocytogenes strain 10403S and analyzed at day 3.5 post infection (n= 6 per group). * P<0.05, ** P<0.001, *** P<0.0001 (Student’s t test).
To assess the functional and physiological relevance of RBP-J-mediated regulation of M1 genes we examined the in vivo expression of these genes in the myeloid compartment under conditions of inflammation. Upon endotoxin challenge, levels of IL-12p40 protein in serum were significantly reduced in mice with myeloid-specific deletion of Rbpj (Rbpj flox/flox, LysM-Cre) compared with wild-type littermate controls (Fig. 1c). iNOS catalyzes production of nitric oxide (NO) in macrophages. In response to LPS stimulation, RBP-J-deficient macrophages produced significantly reduced amounts of NO as measured by levels of the NO metabolite nitrite (Fig. 1d). Because Il12 and Nos2 genes mediate responses against intracellular bacteria, we assessed the role of RBP-J in vivo in host defense against L. monocytogenes, an intracellular pathogen whose successful clearance requires M1 effectors such as IL-12 and iNOS25. Compared with control animals (Rbpj+/+, LysM-Cre), mice with RBP-J deficiency in the myeloid lineage (Rbpjflox/flox, LysM-Cre), exhibited enhanced susceptibility to L. monocytogenes infection, as demonstrated by significantly increased bacterial burdens in spleens and livers of the infected animals (Fig. 1e). Taken together, these results show that RBP-J is essential for expression of genes characteristic of the core M1 response in vitro and for manifestation of key myeloid effector functions in vivo.
In addition to promoting M1 gene expression, microarray analysis showed that RBP-J suppressed expression of a group of genes characteristic of M2 macrophage phenotype, confirmed by qPCR (Supplementary Fig. 1d). RBP-J suppressed expression of the key inducer of M2 polarization JMJD33, indicating that RBP-J plays an inhibitory role in the M2 differentiation program. Although these results suggest that RBP-J might regulate the balance between M1 and M2 polarization, in this study we focused on delineating the mechanisms by which RBP-J regulates the M1 program.
RBP-J-controls M1 gene expression downstream of Notch signaling
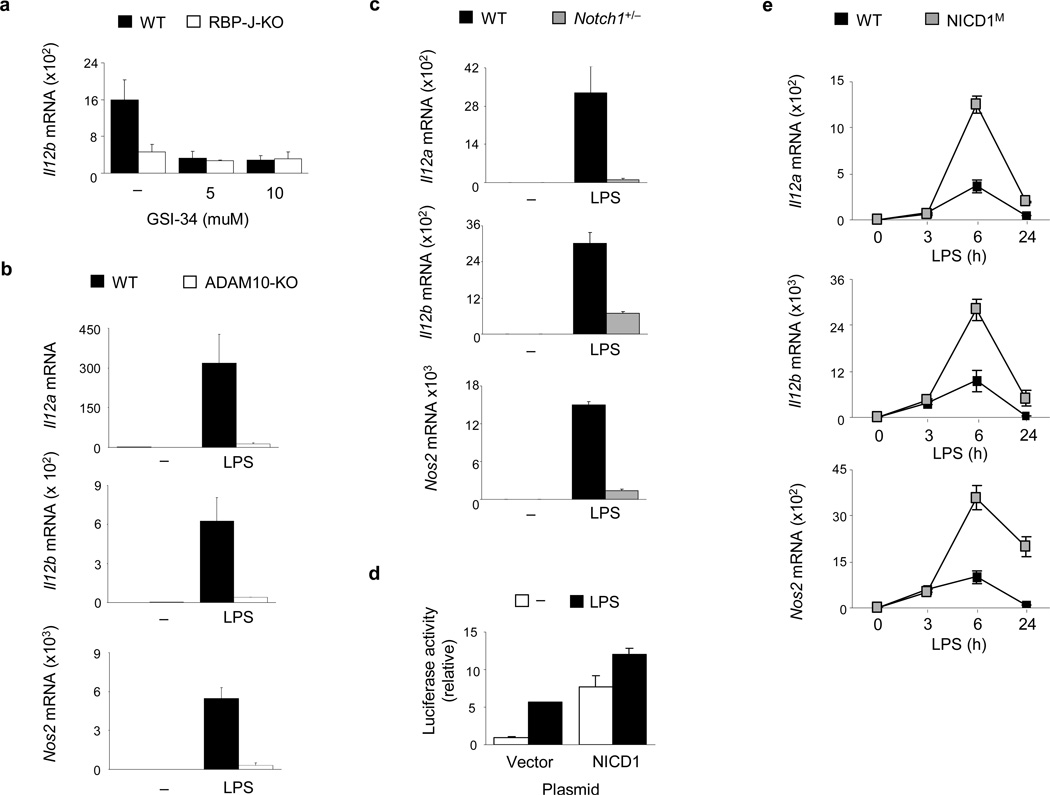
RBP-J plays a key role in signal transduction by the canonical Notch pathway. However, Notch-independent RBP-J activities have been reported14. To assess the role of canonical Notch pathway in RBP-J-mediated regulation of M1 genes, GSI-34, a chemical inhibitor of γ-secretase, was used to abolish signaling from the Notch receptors. GSI-34 treatment of wild-type mouse BMDMs did not have any detectable toxicity effects (data not shown), yet effectively suppressed LPS-induced expression of Il12b (Fig. 2a), suggesting that Il12b induction by LPS requires canonical Notch signaling. Inhibition of γ-secretase by GSI-34 had no effects on the already blunted Il12b expression in RBP-J-deficient macrophages (Fig. 2a), indicating that γ-secretase and RBP-J function in a linear pathway. Another proteolytic event required for Notch signaling activation is the cleavage of receptors by ADAM family proteases, primarily by ADAM1014. Deficiency of ADAM10 almost completely abolished the induction of RBP-J-dependent genes Il12a, Il12b and Nos2 by LPS in macrophages (Fig. 2b). In contrast, deficiency of another ADAM family protease, ADAM17, did not notably alter LPS-induced expression of RBP-J-dependent M1 genes such as Il12b (Supplementary Fig. 2a and data not shown).
Figure 2. Induction of RBP-J-dependent M1 genes requires canonical Notch signaling.
(a) Quantitative PCR of IL12b mRNA expression in BMDMs from Rbpj+/+, Mx1-Cre (wild-type; WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) paired littermates, pretreated with the γ-secretase inhibitor, GSI-34, for 48 h and then stimulated with LPS (1 ng/ml) for 3 h. (b) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from wild-type (WT) or ADAM10-deficient (ADAM10-KO) BMDMs stimulated with LPS for 3 h. (c) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from Notch1+/− mice and wild-type (WT) littermate controls, stimulated with LPS for 3 h. (d) Relative luciferase activity in lysates from RAW264.7 cells co-transfected with an Il12b reporter construct and a NICD1 expression plasmid or empty vector control, followed by stimulation for 6 h with LPS (1 µg/ml) at 36 h post-transfection. (e) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from NICD1M mice and wild-type (WT) littermate controls, stimulated with LPS for the indicated periods. Data are shown as means + SD of triplicate determinants. Results shown are representative of at least three independent experiments.
We next asked what Notch receptor(s) are responsible for M1 gene activation. Resting mouse BMDMs express predominantly Notch1 and Notch2 (data not shown). To assess the role for Notch1 in M1 gene expression, we used macrophages from Notch1 heterozygous mice, as complete Notch1 deletion leads to lethality26. Notch1 haploinsufficiency is characterized by approximately 70–80% reduction of Notch1 mRNA expression (Supplementary Fig. 2b)24. Notch1+/− macrophages exhibited profound defects in induction of RBP-J-dependent M1 genes (Fig. 2c), mimicking the effects of RBP-J deletion (Fig. 1a & 1b), γ-secretase inhibition (Fig. 2a), and ADAM10 deficiency (Fig. 2b). In contrast to Notch1, knocking down expression of Notch2 did not alter LPS-mediated induction of RBP-J-dependent genes such as Il12b (Supplementary Fig. 2c). Knockdown of Notch2 expression in Notch1+/− cells did not further reduce Il12b expression (Supplementary Fig. 2c), suggesting that Notch2, either alone or in concert with Notch1, does not significantly contribute to the induction of RBP-J-dependent M1 genes. Next, we tested Notch1 function by gain-of-function approaches. Forced expression of NICD1 activated a mouse Il12b promoter-driven reporter construct (Fig. 2d). We also generated mice constitutively expressing NICD1 in myeloid cells (referred to as NICD1M mice) by crossing LysM-Cre mice with RosaNotch mice27. BMDMs from NICD1M mice were morphologically undistinguishable from wild-type macrophages and expressed mature macrophage markers (Supplementary Fig. 3a–c). NICD1M BMDMs had increased NICD1 expression and constitutively active Notch signaling, as assessed by expression of the canonical Notch target gene Hes1 (Supplementary Fig. 3d and data not shown). Upon LPS stimulation, NICD1M macrophages showed augmented induction of M1 genes compared with control macrophages (Fig. 2e). Collectively, these results indicate that Notch signaling components, namely the Notch1-ADAM10-γ-secretase-RBP-J axis, regulate expression of M1 genes.
RBP-J controls IRF8 expression and function
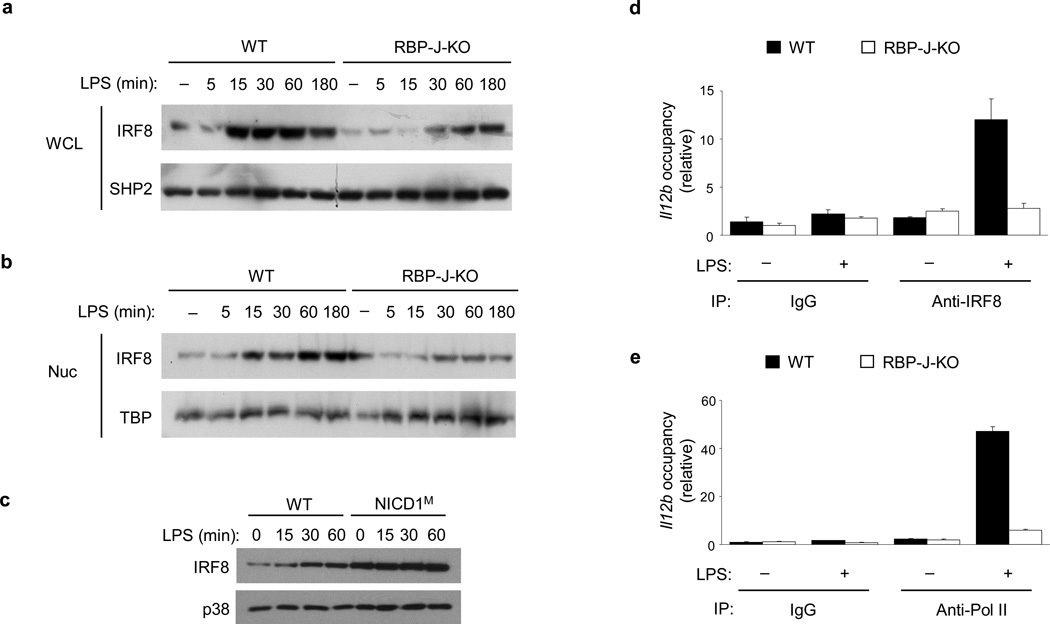
Il12a, Il12b and Nos2, are known to share common mechanisms of regulation, such as dependence on c-Rel28–30 and dependence on IRF1 and IRF86,7,31 and are all categorized as secondary response genes32 (Supplementary Fig. 4). We investigated if the Notch-RBP-J pathway regulates c-Rel, IRF1 or IRF8 expression. Neither c-Rel nor IRF1 expression was significantly altered by RBP-J deficiency (data not shown), suggesting that they are not targets of RBP-J-mediated regulation. It has been reported that IRF8 expression is regulated at the transcriptional level and that induction of IRF8 protein follows mRNA induction and occurs over the course of hours33,34. In contrast to these previous observations, LPS treatment rapidly (within 15 minutes) and robustly induced IRF8 protein expression in whole cell lysates and nuclear extracts of wild-type BMDMs (Fig. 3a, b, lanes 1–6). The specificity of IRF8 detection by immunoblotting analysis was verified using IRF8-deficient macrophages (Supplementary Fig. 5a). This rapid IRF8 protein induction was validated in various culture conditions and using several protein extraction methods (data not shown). In contrast to the robust LPS-dependent IRF8 induction in wild-type cells, decreased IRF8 expression was observed in whole cell lysates and nuclear extracts from in RBP-J-deficient macrophages (Fig. 3a, b). Expression of other IRF family members such as IRF4 and IRF5 was not affected by RBP-J deficiency (Supplementary Fig. 5b). In a gain-of-function approach, IRF8 protein expression was markedly elevated in macrophages from NICD1M mice compared with wild-type macrophages (Fig. 3c). These results suggest that Notch-RBP-J is required for the rapid induction of IRF8 protein upon TLR4 stimulation.
Figure 3. RBP-J controls IRF8 expression and function.
(a, b) Immunoblot analysis of IRF8 expression in whole cell lysates (WCL) (a) and nuclear (Nuc) extracts (b) of BMDMs from Rbpj+/+, Mx1-Cre (wild-type; WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) paired littermates, stimulated with LPS for the indicated periods. SHP2 (a) and TBP (b) served as loading controls. Data shown are representative of six independent experiments. (c) Immunoblot analysis of IRF8 expression in whole cell lysates of BMDMs from NICD1M mice and (wild-type) WT littermate controls, stimulated with LPS for the indicated periods. p38 served as a loading control. Data are representative of two independent experiments. (d, e) ChIP assays were performed with LPS-treated Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBPJ-KO) macrophages, using antibodies against IRF8 (d) and Pol II (e). Occupancy was determined by qPCR amplification over the promoter region of Il12b. Data are shown as means + SD of triplicate determinants and are representative of three independent experiments.
Recruitment of IRF8 to its target gene promoters is necessary for binding of RNA polymerase II (Pol II) and subsequent transcriptional activation4. Chromatin immunoprecipitation (ChIP) assays showed that activation of wild-type macrophages with LPS led to recruitment of IRF8 to the proximal promoter of Il12b (Fig. 3d). This effect was almost completely abolished in RBP-J-deficient macrophages (Fig. 3d). Recruitment of Pol II to the Il12b promoter was concomitantly decreased in RBP-J-deficient cells (Fig. 3e), suggesting that reduced levels of IRF8 in the absence of RBP-J were not sufficient to assemble the transcription machinery at the Il12b promoter. Overall, our data suggest that RBP-J regulates the expression and transcriptional function of IRF8 downstream of TLR signaling.
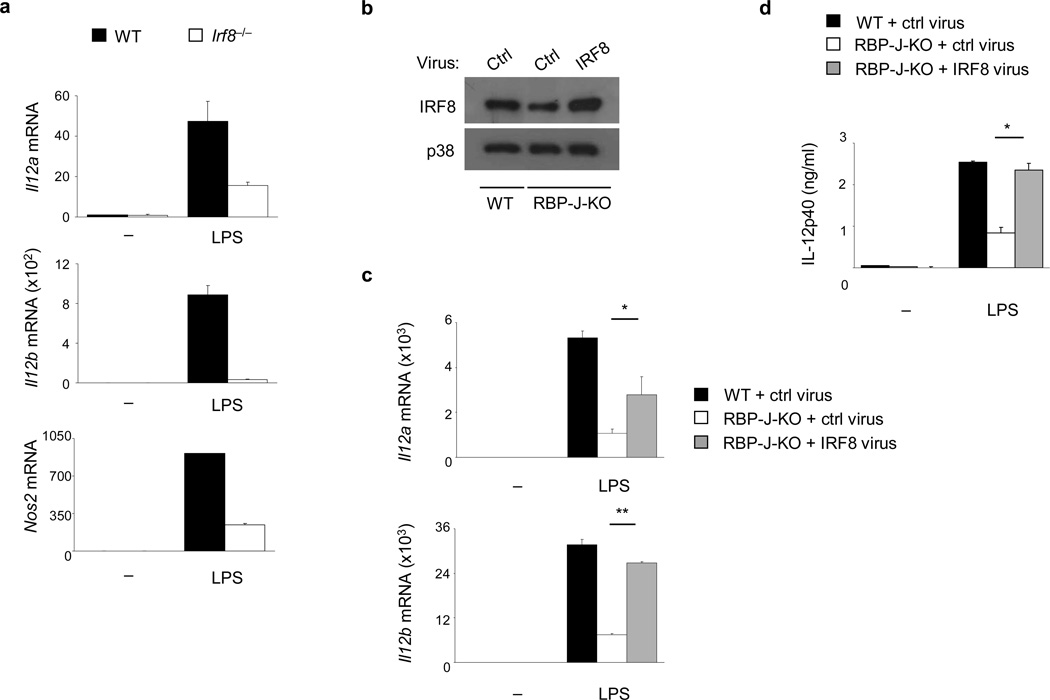
We next asked whether decreased IRF8 expression in RBP-J-deficient macrophages explained the low M1 gene expression in these cells. Induction of Il12a, Il12b and Nos2 by LPS was severely decreased in IRF8-deficient macrophages (Fig. 4a). We tested if rescue of IRF8 expression in RBP-J-deficient cells restores Il12a, Il12b and Nos2 expression. Using retroviral transduction, IRF8 expression was restored to approximately wild-type levels in RBP-J-deficient macrophages (Fig. 4b). IRF8 reconstitution nearly completely corrected the defective mRNA expression of Il12b (Fig. 4c) and IL-12p40 protein (Fig. 4d) in RBP-J-deficient cells. IRF8 reconstitution also partially restored expression of Il12a in RBP-J-deficient macrophages (Fig. 4c), while impaired Nos2 expression in RBP-J-deficient cells was not rescued by IRF8 reconstitution (Supplementary Fig. 5c), suggesting involvement of additional factors in RBP-J-regulated Nos2 expression. These results indicate that RBP-J regulates M1 gene expression, at least in part, through IRF8.
Figure 4. IRF8 mediates RBP-J-dependent activation of M1 gene expression.
(a) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from WT and Irf8−/− mice, stimulated with LPS for 3 h. Data are representative of three independent experiments. (b) Immunoblot analysis of IRF8 expression in BMDMs from Rbpj+/+, Mx1-Cre (wild-type; WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) paired littermates transduced with an IRF8-expressing retrovirus or control (ctrl) virus. p38 served as a loading control. (c) Quantitative PCR assays of mRNA expression of the indicated genes in the virally transduced BMDMs from (b), stimulated for 6 h with LPS at 48 h post-transduction. (d) ELISA analysis of IL-12p40 expression in the transduced BMDMs from (b), stimulated for 6 h with LPS at 48 h post-transduction (d). Data are shown as means + SD of triplicate determinants and are representative of two independent experiments. *P<0.05, **P<0.001 (Student’s t test).
RBP-J is required for rapid synthesis of IRF8 protein
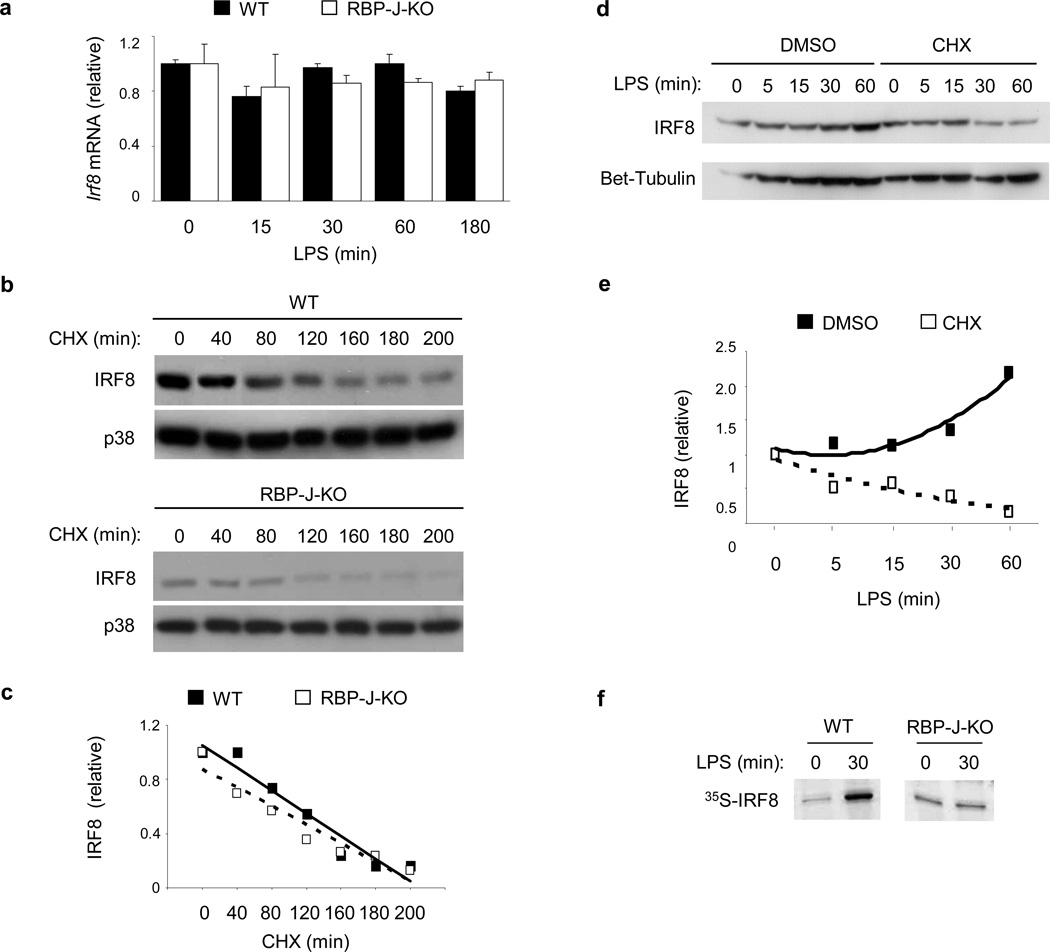
Next we investigated the mechanisms by which RBP-J regulates IRF8 protein expression. Because IRF8 expression is known to be regulated at the mRNA level by stimuli such as IFN-γ34,35, we examined if TLR4 and RBP-J induced Irf8 mRNA accumulation. LPS stimulation for up to 3 hours did not result in notable upregulation of Irf8 mRNA at any of the tested time points (0 to 180 minutes, corresponding to the observed IRF8 protein induction) in wild-type BMDMs (Fig. 5a). As a control, strong induction of Tnf mRNA was observed after LPS treatment (Supplementary Fig. 5d), and IFN-γ pretreatment resulted in induction of Irf8 mRNA in wild-type macrophages as expected (Supplementary Fig. 5e). The above results were also validated by using distinct qPCR primers, which target a different region of Irf8 mRNA33 (data not shown). These results suggest that rapid induction of IRF8 protein by TLR4 stimulation was not due to increased Irf8 mRNA expression. In addition, RBP-J deficiency did not significantly alter Irf8 mRNA levels at baseline or after LPS treatment (Fig. 5a), suggesting that rapid induction of IRF8 by TLR4 stimulation is regulated at the protein level.
Figure 5. RBP-J promotes IRF8 protein synthesis.
(a) Quantitative PCR of Irf8 mRNA in BMDMs from Rbpj+/+, Mx1-Cre (wild-type; WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) paired littermates, stimulated with LPS for the indicated periods. (b) Immunoblot analysis of IRF8 expression in whole cell lysates of BMDMs from Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) BMDMs, stimulated with LPS for 1 h and subsequently treated with the protein synthesis inhibitor, CHX (20 µg/ml) for the indicated periods. p38 served as a loading control. (c) Band intensities from (b) were quantitated using ImageJ software and normalized relative to the CHX 0 min conditions. (d) Immunoblot analysis of IRF8 expression in BMDMs from wild-type mice, pretreated with CHX (20 µg/ml) for 30 min and then activated with LPS for the indicated periods. β-tubulin served as a loading control. (e) Band intensities from (d) were quantitated using ImageJ software and normalized relative to the CHX 0 min conditions. (f) SDS-PAGE analysis of immunoprecipitated radiolabeled IRF8 in Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) BMDMs following metabolic labeling. Results shown are representative of at least three independent experiments.
IRF8 is a labile protein36, so we examined whether RBP-J regulates IRF8 protein degradation. The protein synthesis inhibitor cycloheximide (CHX) was added to LPS-stimulated wild-type or RBP-J-deficient macrophages and degradation of IRF8 protein was followed over time. Despite the expected difference in IRF8 protein levels between wild-type and RBP-J-deficient cells prior to CHX treatment, IRF8 protein decreased in a time dependent manner but independent of the RBP-J genotype after CHX treatment (Fig. 5b and Supplementary Fig. 5f). Quantification of IRF8 protein amounts by densitometry revealed that in the absence of new protein synthesis, IRF8 protein decayed at a similar rate in wild-type and RBP-J-deficient cells and that it’s half life was approximately 150 minutes in both cell types (Fig. 5c), a measurement consistent with the estimate from a previous report36. These results suggest that RBP-J does not regulate IRF8 protein degradation. However, addition of CHX to wild-type macrophages prior to LPS stimulation blocked LPS-induced upregulation of IRF8 protein (Fig. 5d, e), suggesting that induction of IRF8 by LPS was the result of new protein synthesis. Metabolic labeling assays showed that LPS stimulation upregulated the incorporation of 35S-methionine/cysteine into newly synthesized IRF8 protein in wild-type macrophages but not in RBP-J-deficient cells (Fig. 5f). These results suggest that rapid IRF8 protein synthesis induced downstream of TLR4 signaling was dependent on RBP-J.
RBP-J controls activation of the MNK1-eIF4E axis
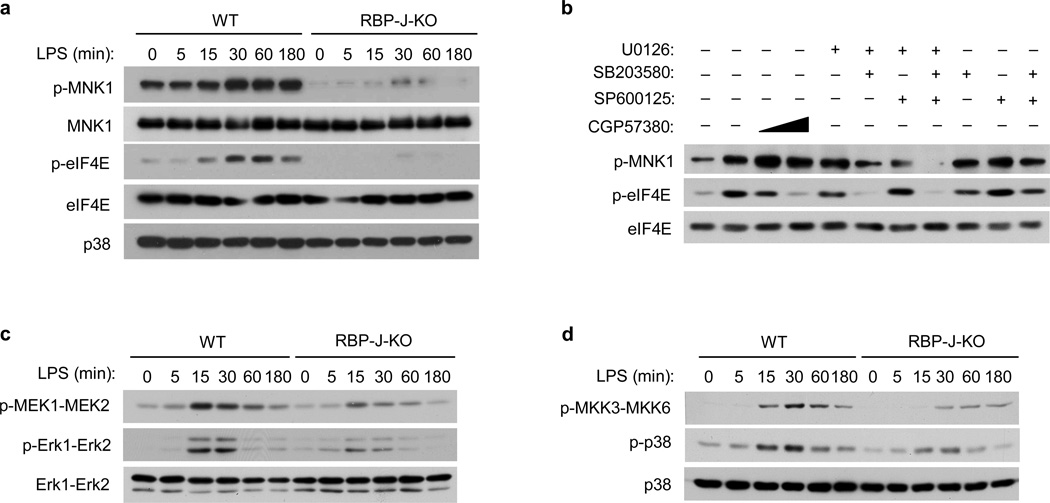
TLR stimulation induces phosphorylation and activation of MNK and subsequent MNK-mediated phosphorylation of eIF4E37,38, which is required for efficient translation of select protein transcripts39. To investigate the mechanisms by which RBP-J regulates IRF8 protein synthesis we assessed regulation of MNK-eIF4E activity by TLR4 and RBP-J. MNK-eIF4E activity is enhanced by phosphorylation of MNK1 on threonines 197 and 202, and phosphorylation of eIF4E on serine 20913. TLR4-induced phosphorylation of MNK1 and eIF4E was greatly diminished in RBP-J-deficient compared to wild-type macrophages (Fig. 6a). This was not due to decreased MNK1 or eIF4E protein expression (Fig. 6a), suggesting that activation of MNK1-eIF4E downstream of TLR4 signaling requires RBP-J.
Figure 6. RBP-J augments TLR4-induced activation of the MAPK-MNK1-eIF4E pathway.
(a) Immunoblot analysis of the indicated proteins in whole cell lysates of BMDMs from Rbpj+/+, Mx1-Cre (wild-type; WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) paired littermates, stimulated with LPS for the indicated periods. p38 served as a loading control. (b) Immunoblot analysis of p-MNK1 and p-eIF4E in whole cell lysates from wild-type BMDMs, pretreated with DMSO vesicle control, a MNK inhibitor CGP57380, or MAPK inhibitors for 30 min and then stimulated with LPS for 60 min. Total eIF4E levels served as loading controls. (c & d) Immunoblot analysis of p-Erk1-Erk2 and p-MEK1-MEK2 (c) and p-p38 and p-MKK3-MKK6 (d) expression in BMDMs from Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice, stimulated with LPS for the indicated periods. Total Erk1-Erk2 (c) and total p38 (d) served as loading controls. Cumulative data from three independent experiments is shown.
Activation of MNK1 with subsequent phosphorylation of eIF4E and regulation of translation has been shown to be dependent on Erk and stress-activated MAPKs in various systems37–39. We examined the role of MAPKs in TLR4-induced activation of MNK1 using pharmacological inhibitors for MEK (U0126), p38 (SB203580) and JNK (SP600125), while an inhibitor of eIF4E phosphorylation by MNK1 (CGP57380)39 served as a positive control (Fig. 6b). While single MAPK inhibitors had modest effects, combination of MEK and p38 inhibitors effectively suppressed TLR4-induced phosphorylation of MNK1 and eIF4E (Fig. 6b), indicating that both Erk and p38 were necessary for activation of the MNK1 pathway by TLR4. RBP-J deficiency did not significantly alter TLR4-induced activation of JNK (Supplementary Fig. 6a), consistent with the idea that JNK is dispensable for MNK1 activation. In contrast, phosphorylation of Erk and MEK (which activates Erk downstream of TLR signaling) was reduced in RBP-J-deficient macrophages (Fig. 6c, Supplementary Fig. 6b). Furthermore, RBPJ deficiency led to decreased phosphorylation of p38 and its upstream kinase MKK3-MKK6 by LPS (Fig. 6d, Supplementary Fig 6b). These results indicate that regulation of TLR4-induced activation of Erk and p38 is one mechanism by which RBP-J controls activation of the MNK1-eIF4E axis. Although the dependence of Erk and p38 signaling on RBP-J was modest, MNK1 activation was dependent on RBP-J, consistent with previous work suggesting a requirement for dual activation of MNK by different MAPKs.
RBP-J targets IRAK2 upstream of MNK-eIF4E
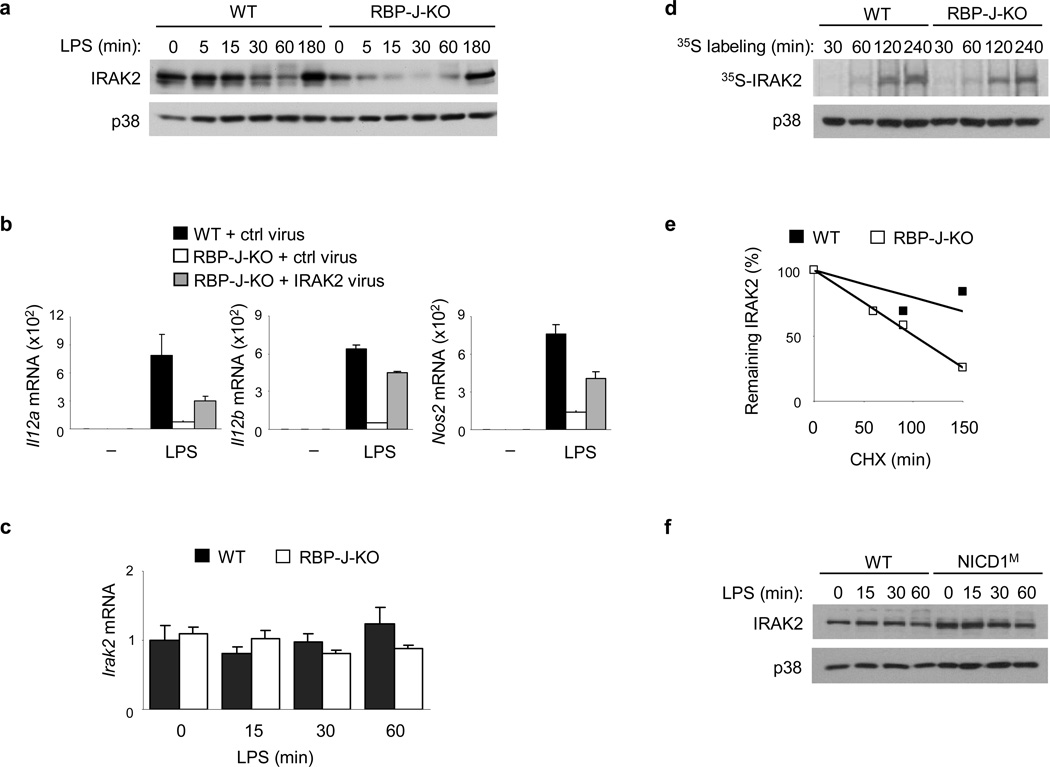
Next, we investigated potential targets of RBP-J upstream of MAPKs and MNK in the TLR4 signaling cascades. IRAK2 is a proximal TLR signaling component that plays a role in the TLR-mediated activation of MNK1 and also functions as a post-transcriptional regulator38,40. Consistent with previous reports38,41, acute LPS stimulation of wild-type macrophages did not result in upregulation of IRAK2 expression (Fig. 7a). However, IRAK2 protein expression was greatly diminished in RBP-J-deficient macrophages (Fig. 7a). This effect was specific, as expression of other IRAK family proteins such as IRAK1 was not reduced in RBP-J-deficient macrophages compared with wild-type cells (Supplementary Fig. 6c and data not shown). To determine whether decreased IRAK2 expression contributed to the reduction of M1 gene expression in RBP-J-deficient cells we restored IRAK2 expression in RBP-J-deficient macrophages by retroviral transduction. IRAK2 reconstitution partially corrected the phenotype (Fig. 7b), suggesting that the requirement of RBP-J in M1 gene induction is, at least in part, due to it’s regulation of IRAK2.
Figure 7. Notch-RBP-J signaling regulates IRAK2 protein expression.
(a) Immunoblot analysis of IRAK2 expression in BMDMs from Rbpj+/+, Mx1-Cre (wild-type; WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice, stimulated with LPS for the indicated periods. p38 served as a loading control. (b) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice, transduced with an IRAK2-expressing retrovirus, or a control (ctrl) virus, and stimulated for 6 h with LPS at 48 h post-transduction. (c) Quantitative PCR of Irak2 mRNA in BMDMs from Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice, stimulated with LPS for theindicated periods. (d) SDS-PAGE analysis of immunoprecipitated radiolabelled IRAK2 in BMDMs from Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice, following metabolic labeling for the indicated periods. Immunoblotted p38 in the whole cell lysates served as a loading control. (e) Quantitiation using ImageJ software, of IRAK2 expression in whole cell lysates of BMDMs from Rbpj+/+, Mx1-Cre (WT) and Rbpjflox/flox, Mx1-Cre (RBP-J-KO) mice, stimulated for 30 min with LPS followed by treatment with the protein synthesis inhibitor, CHX (20 µg/ml) for the indicated periods (f) Immunoblot analysis of IRAK2 expression in whole cell lyastes of BMDMs from NICD1M mice and wild-type (WT) littermate controls, stimulated with LPS for the indicated periods. Data are shown as means + SD of triplicate determinants (b, c). Data are representative of at least three independent experiments.
Next we investigated the mechanisms by which RBP-J signaling regulates IRAK2 expression. Levels of Irak2 mRNA did not significantly differ between wild-type and RBP-J-deficient cells at baseline or after LPS stimulation (Fig. 7c), indicating that RBP-J does not regulate Irak2 gene expression. We also tested if RBP-J deficiency decreases IRAK2 protein synthesis and/or increases IRAK2 protein degradation. We assessed IRAK2 protein synthesis by metabolic labeling assays and found that RBP-J deficiency resulted in attenuated synthesis of IRAK2 protein as evidenced by reduced incorporation of 35S-methionine/cysteine at multiple labeling time points (Fig. 7d). In addition, in the presence of the protein synthesis inhibitor CHX, IRAK2 degraded at a faster rate in RBP-J-deficient macrophages than in control macrophages under LPS-stimulated conditions (Fig. 7e). Therefore, both decreased synthesis and increased degradation contributed to the low IRAK2 protein levels observed in RBP-J-deficient cells. We also examined IRAK2 protein expression in NICD1M macrophages and found increased IRAK2 protein expression compared to macrophages from wild-type mice (Fig. 7f). Increased IRAK2 protein expression in NICD1M cells was not due to augmented Irak2 mRNA expression (Supplementary Fig. 6d), supporting the notion that Notch-RBP-J signaling regulates IRAK2 expression post-transcriptionally.
To test the role of IRAK2 in mediating RBP-J-dependent TLR4-induced signaling events we evaluated activation of MAPKKs and MAPKs in cells in which IRAK2 expression was knocked down using RNAi. Diminished IRAK2 expression (Supplementary Fig. 6e) resulted in impaired activation of the MEK-Erk as well as MKK3-MKK6-p38 pathways upon LPS stimulation (Supplementary Fig. 6f), but did not affect JNK phosphorylation (Supplementary Fig. 6g). Overall, these results demonstrate that the Notch-RBP-J pathway controls a TLR4- activated MAPK-MNK-eIF4E signaling cascade by regulating expression of IRAK2 protein.
MNK1-eIF4E controls TLR4-induced IRF8 protein synthesis
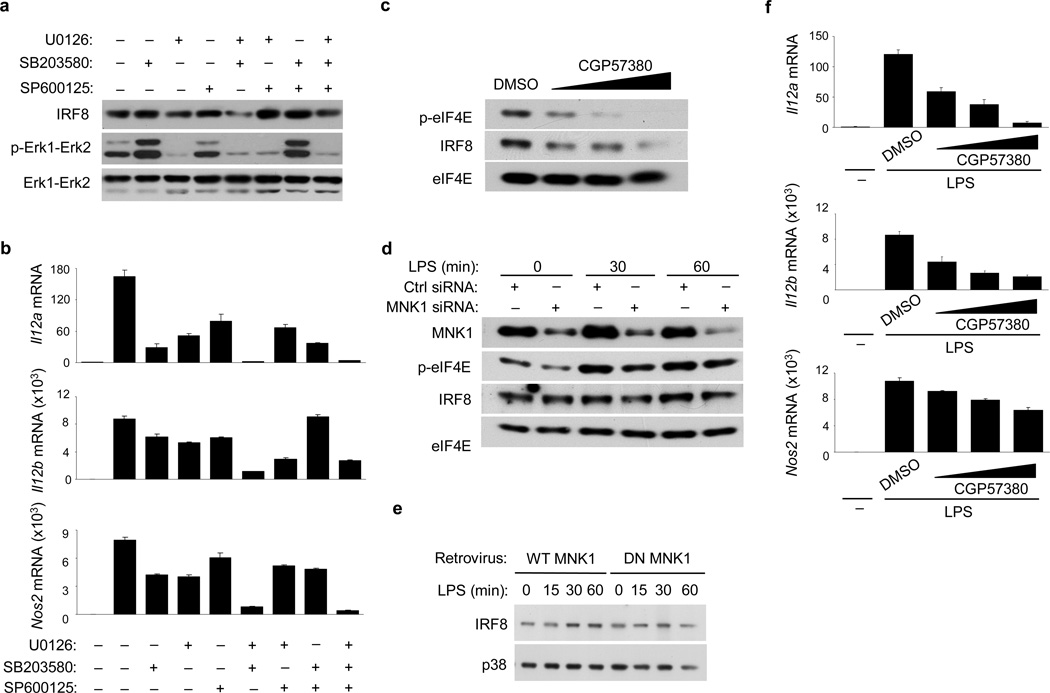
We sought to link RBP-J-mediated regulation of MAPK-MNK1-eIF4E signaling with regulation of IRF8 protein induction. In LPS-activated macrophages, IRF8 expression was strongly decreased when both Erk and p38 activation were pharmacologically inhibited using U0126 and SB203580 respectively, whereas inhibition of JNK using SP600125 did not have discernible effects on IRF8 protein amounts (Fig. 8a). Combined inhibition of Erk and p38 almost completely abolished induction of M1 genes (Fig. 8b) and IL-12 protein (Supplementary Fig. 7a) by LPS, suggesting that both Erk and p38 were necessary for IRF8 protein expression and subsequent induction of M1 genes in TLR4-stimulated macrophages. Treatment of macrophages with the MNK inhibitor CGP57380 suppressed LPS-induced eIF4E phosphorylation and IRF8 protein expression in a dose-dependent manner (Fig. 8c). RNAi knockdown of MNK1 in macrophages led to decreased eIF4E phosphorylation and resulted in attenuated induction of IRF8 by LPS (Fig. 8d, Supplementary Fig. 7b). However, it did not affect expression of Irf8 mRNA (Supplementary Fig. 7c). Transduction of macrophages with a retrovirus expressing a dominant negative mutant of MNK1 that lacks kinase activity, and thus is unable to phosphorylate eIF4E42, blunted LPS-activated IRF8 protein induction (Fig. 8e). Inhibition of MNK activity by CGP57380 suppressed TLR4-induced expression of Il12a, Il12b and Nos2 (Fig. 8f), without apparent toxicity (data not shown) and global interference with TLR responsiveness (Supplementary Fig. 7d). Altogether, these experiments support a role for MNK-eIF4E in the TLR4-induced expression of IRF8 protein and induction of IRF8 target genes. A model of the regulation of M1 macrophage polarization through Notch-TLR signaling crosstalk is shown in Supplementary Fig. 8.
Figure 8. The MAPK-MNK1-eIF4E axis promotes IRF8 synthesis and M1 gene expression.
(a) Immunoblot analysis of IRF8 and p-Erk1-Erk2 expression in BMDMs from wild-type mice, pretreated with DMSO vesicle control or MAPK inhibitors for 30 min and then stimulated with LPS for 60 min. Total Erk1-Erk2 levels served as loading controls. (b) Quantitative PCR of mRNA expression of the indicated genes in BMDMs from wild-type mice, pretreated with MAPK inhibitors and stimulated with LPS for 3 h. (c) Immunoblot analysis of p-eIF4E and IRF8 expression in wild-type BMDMs, pretreated with increasing doses of the MNK inhibitor CGP57380 or DMSO vesicle control for 30 min and then stimulated with LPS for 60 min. Total eIF4E levels served as loading controls. (d) Immunoblot for the indicated proteins in wild-type BMDMs, transfected with control (ctrl) non-targeting or MNK1-specific siRNA, and treated with LPS at 2 d post-transfection. (e) Immunoblot analysis of IRF8 expression in wild-type BMDMs, transduced with a retrovirus encoding wild-type (WT) MNK1 or a dominant negative (DN) MNK1 mutant and stimulated with LPS for the indicated periods following selection in puromycin-containing medium for 4 d. (f) Quantitative PCR of mRNA expression of the indicated genes in wild-type BMDMs, pretreated with increasing doses of CGP57380 or DMSO vesicle control for 30 min and then stimulated with LPS for 3 h. Data are representative of at least three independent experiments.
DISCUSSION
Selective transcription of functionally related subsets of genes in response to inflammatory stimuli is important for achieving appropriate immune responses11. Here we show that the Notch-RBP-J pathway selectively regulates a subset of TLR4-inducible, classical M1 genes including Il12a, Il12b and Nos2. RBP-J and TLR4 signaling converged to synergistically induce rapid expression of IRF8 protein, which in turn directly activated downstream M1 gene expression. Notch1-RBP-J signaling was required for the activity of MNK1 and eIF4E, which augmented IRF8 translation. Our findings provide a functional connection between Notch-RBP-J signaling and the IRF family of transcription factors and identify a mechanism by which RBP-J and TLR4 signaling are integrated to induce translation of a key transcription factor important in macrophage activation.
IRF8 expression is known to be transcriptionally inducible by IFN- γ34,35. Here we found that LPS alone (without IFN-γ) induced rapid protein expression of IRF8, independently of Irf8 mRNA upregulation, which activated a subset of TLR-inducible promoters such as Il12b in an RBP-J-dependent manner. The fact that MNK activation and subsequent eIF4E phosphorylation are induced by inflammatory stimuli including TLR ligands and IFNs38,39 suggests that this pathway may be important in promoting translation of a select subset of transcripts under inflammatory conditions. Furthermore, Notch-RBP-J signaling controls protein levels of IRAK2 independently of regulation of mRNA expression. Although IRAK2 is an integral component of the TLR signaling cascade and levels of IRAK2 are critical for determining TLR responsiveness43, little is known about how IRAK2 protein synthesis or degradation is regulated. The exact mechanisms by which Notch signaling regulates IRAK2 protein expression will be the subject of future investigations.
Notably, the RBP-J-dependent M1 genes identified here are all secondary response genes whose expression is dependent on new protein synthesis. The identity of factors that are responsible for induction of secondary response genes has remained elusive11. Our results indicate that IRF8 represents such a factor. However, we could not rule out the possibility that RBP-J regulates expression of TLR-inducible genes by additional mechanisms. Regulation of NF-κB activity by RBP-J has been described44. Because Il12a, Il12b and Nos2 are known c-Rel targets, we tested if NF-κB played a role in the RBP-J-mediated regulation of these genes. Acute activation of canonical NF-κB signaling, as measured by IκBα degradation and nuclear accumulation of c-Rel were however not affected by RBP-J deficiency, and expression of a number of canonical NF-κB target genes was intact in RBP-J-deficient cells (data not shown), suggesting that NF-κB is not the central point of signaling integration between RBP-J and TLR pathways in our system. Indeed, regulation of NF-κB by RBP-J would be expected to have broader effects on expression of TLR-inducible genes and could not explain the selective regulation that we observed. However, it is plausible that NF-κB may be subject to regulation by RBP-J under other conditions such as late-phase TLR responses where IRAK2 contributes to sustained NF-κB activation41 or in other cell types such as T cells and human peripheral blood mononuclear cells40,45.
Notch receptors and ligands have been recently implicated in the regulation of inflammatory cytokine production18,20,24 mostly through a positive feed-forward loop in which inflammatory stimuli such as TLR ligands induce the expression of Notch receptors and/or ligands and activate canonical Notch signaling, which in turn augments TLR-induced cytokine production in a non-selective manner. In contrast, here we show that the induction of IRF8 by RBP-J and TLR signaling occurred minutes after TLR stimulation, prior to the previously reported induction of Notch receptor or ligand expression18,20,24. Furthermore, in primary macrophages, despite the fact that Notch signaling was constitutively active at baseline, it was not further activated by TLR stimulation within the experimental time frame (X.H., unpublished data), indicating a lack of acute activation of canonical Notch signaling by TLR pathways. As such, our data suggests a model where constitutive Notch signaling via RBP-J serves as a ‘tonic’ signal that is necessary but not sufficient for gene induction, and that the TLR pathway provides a ‘triggering’ signal that activates gene expression. Such a tonic signal would be delivered in vivo under conditions where Notch ligands are expressed at baseline, such as in the marginal zone of the spleen16 and in the blood circulation24. Feed-forward regulation involving the induction of Notch components would then serve as an amplification loop that is potentially important for sustaining TLR responses at later time points. Overall, our findings highlight the selective regulation of TLR-inducible gene expression by Notch signaling that modulates inflammatory macrophage phenotype.
METHODS
Cells and reagents
Murine BMDMs were obtained as described17 and maintained in DMEM supplemented with 10% FBS and 10% L929 cell supernatant as conditioned medium providing macrophage colony stimulating factor (M-CSF). Recombinant mouse IFN-γ was from Peprotech and used at 10 ng/ml. Cell culture grade LPS and CHX were purchased from Sigma-Aldrich. Pam3Cys was purchased from EMC Microcollections. LPS was used as 1 ng/ml and Pam3Cys was used at 10 ng/ml unless otherwise noted. A γ-secretase inhibitor GSI-34 was used at 10 µM as previously described46. U0126, SB203580, SP600125, and CGP57380 were from Calbiochem.
Mice
The experiments using mice were approved by Institutional Animal Care and Use Committees at the Hospital for Special Surgery, Columbia University, and Christian Albrechts Universität Kiel. C57/BL6 mice were purchased from Jackson Laboratory. Rbpjflox/flox mice were kindly provided by Tasuku Honjo. Mice with a myeloid-specific deletion of Rbpj (Rbpjflox/flox, LysM-Cre) were as described previously17 and used for in vivo experiments given the tissue specificity of gene deletion. Mice with an inducible deletion of Rbpj (Rbpjflox/flox, Mx1-Cre) were generated by crossing Rbpjflox/flox animals to animals with an Mx1 promoter driven Cre transgene on the C57/BL6 background (Jackson Laboratory). Littermates with Rbpjflox/flox, Mx1-Cre (RBPJ-KO) or Rbpj+/+, Mx1-Cre (wild-type; WT) genotypes were intraperitonially injected with 200 µg/mouse of PolyI:C three times in five days to induce deletion and mice were used for experiments two weeks later. For all in vitro experiments involving RBP-J-deficient macrophages, cells were derived from Rbpjflox/flox, Mx1-Cre animals where consistent and strong deletion of Rbpj (approximately 80%) was observed and the conditional deletion was controlled for expression of Cre and genetic background. Adam10flox/flox animals were as described47 and Adam10flox/flox, Mx1-Cre mice were generated and used in a manner similar to that of Rbpjflox/flox, Mx1-Cre animals. Adam17flox/flox, LysM-Cre animals were generated by crossing Adam17flox/flox animals48 to LysM-Cre mice on the C57/BL6 background (Jackson Laboratory). Notch1+/− mice were kindly provided by Thomas Gridley26. RosaNotch mice in which the mouse cDNA encoding constitutively active NICD1 was knocked into the ubiquitously expressed Rosa26 locus, followed by internal ribosome entry sequence-EGFP, and preceded by a STOP fragment flanked by loxP sites27 were obtained from the Jackson Laboratory. We generated mice with myeloid-specific constitutive expression of NICD1 by crossing RosaNotch mice with LysM-Cre mice (referred to as NICD1M mice). Gender and age matched LysM-Cre mice were used as controls. NICD1M macrophages were obtained by culturing bone marrow cells from NICD1M mice for 5 days in M-CSF-containing conditioned medium. At the end of the culture period, adherent cells were collected and replated for experiments. Irf8−/− mice were as described5. All experiments involving knockout mice were performed with sex-matched littermates of desired genotypes as controls. Bone marrow chimeras were generated as previously described49. Briefly, recipient C57BL/6 mice were subjected to irradiation at a dose of 1000 cGy, followed by intravenous injection of 1 × 106 donor bone marrow cells from the myeloid-specific RBP-J-deficient mice with the genotype Rbpjflox/flox, LysM-Cre or the WT littermate controls with the genotype Rbpj+/+, LysM-Cre. Chimeric mice were used for experiments 6 weeks after the initial bone marrow transfer.
mRNA isolation and quantitative PCR (qPCR)
RNA was extracted from whole cell lysates with an RNeasy Mini kit (Qiagen) and was reverse transcribed with a First Strand cDNA synthesis kit (Fermentas). qPCR was performed in triplicate wells with an iCycler IQ thermal cycler and detection system (Biorad) using gene-specific primers. Threshold cycle numbers were normalized to triplicate samples amplified with primers specific for glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
Enzyme-linked immunosorbent assay (ELISA)
Cytokine secretion was quantified by ELISA kits from BD Pharmaingen and nitric oxide production was measured using Greiss reagent (Sigma) according to manufacturers’ instructions.
Listeria monocytogenes infection
Mice were infected intravenously with 3 × 103 L. monocytogenes strain 10403S, as described previously49. At day 3.5 post infection, spleens and livers were harvested and dissociated in PBS containing 0.05% Triton X-100, and bacterial colony-forming units (CFUs) were determined by plating on brain-heart infusion agar plates.
Immunoblot analysis
Whole cell lysates were prepared by direct lysis in SDS loading buffer. For immunoblot analysis, lysates were separated by 10% SDS-PAGE and transferred to a PVDF membrane for probing with antibody. Polyclonal antibodies against IRF8, p38, c-Rel, TBP and SHP2 were from Santa Cruz Biotechnology. Antibodies against IRAK2 and β-tubulin were from Abcam. Anibodies against p-MEK1-MEK2 (Ser217/221), p-MKK3-MKK6 (Ser189/207), p-JNK (Thr183/Tyr185), p-Erk (Thr202/Tyr204), p-p38 (Thr180/Tyr182), p-MNK1 (Thr197/202), p-eIF4E (Ser209), MNK1, eIF4E, IκBα, IRF4, IRF5, Erk1-Erk2, and NICD1 were from Cell signaling. An antibody against NICD2 was from University of Iowa Developmental Studies Hybridoma Bank. Anti-RBP-J rabbit serum was a generous gift from Elliott Kieff and Jon C. Aster50.
Transient transfection and luciferase assay
Murine Il12b promoter reporter plasmid containing sequences from positions −356 to +55 was kindly provided by Stephen T. Smale. RAW264.7 cells were cotransfected in duplicates with the Il12b reporter plasmid and an expression vector encoding NICD1, a gift from Raphael Kopan using Lipofectamine LTX reagent (Invitrogen). pRL-TK plasmid encoding Renilla luciferase (Promega) was used as an internal control. Dual-Luciferase Reporter Assay System (Promega) was utilized to detect the luciferase activity of cell lysates 36 hours after transfection.
RNA interference (RNAi)
Prevalidated specific short interfering RNAs (siRNA) targeting murine Notch2, IRAK2, MNK1, and non-targeting control siRNA were purchased from Dharmacon. siRNAs were transfected into mouse BMDMs using the TransIT TKO transfection reagent (Mirus Bio) according to the manufacturers’ instructions.
Chromatin immunoprecipitation (ChIP)
This assay was performed as previously described17 with slight modifications. In brief, 8–10 × 106 cells were crosslinked with 0.75 % formaldehyde. After being lysed in 8 ml of lysis buffer, the pellets were re-suspended in cold RIPA buffer and sonicated on ice at power setting 5 in 20 s bursts of 6 cycles. Lysates were then cleared by centrifugation, and incubated with 2 µg of goat anti-IRF8 (C-19, Santa Cruz Biotechnology) or of monoclonal anti-RNA polymerase II antibody (Millipore) overnight with rotation. Same amounts of normal goat IgG or normal mouse IgG were used as controls, respectively. Antibody incubation was followed by incubation with 45 µl of 33% Protein A/G agarose slurry (Santa Cruz Biotechnology) for 1.5 hours at 4°C. Then the agarose slurry bond complexes were digested with proteinase K and phenol: chloroform was used to purify DNA for qPCR. The unrelated 28S rRNA was used for qPCR normalization. The value of IgG control in untreated cells was set to 1. Primers used for qPCR were as follows: Il12b locus forward: 5’-CACACTGGACCAAAGGGACT-3’; Il12b locus reverse: 5’-CTTTGCTTCCCTAGCACCT-3’; 28S rRNA forward: 5’-GATCCTTCGATGTCGGCTCTTCCTATC-3’; 28S rRNA reverse: 5’-AGGGTAAAACTAACCTGTCTCACG-3’.
Metabolic labeling
8–10 × 106 of BMDMs cultured in complete medium were starved for 30 minutes in methonine and cysteine-free DMEM supplemented with 5% dialysed FBS. Then the cells were labeled with 35S-Methionine/cysteine Labeling Mix (Perkin Elmer) at a final concentration of 100 µCi/ml. At the end of the labeling period, cells were washed twice with cold PBS and lysed in 500 µl of lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM EGTA, 0.2 mM EDTA, 1% Nonidet P-40, 1 mM dithiothreitol) supplemented with protease inhibitors (Roche). Immunoprecipitation of IRF8 and IRAK2 was performed using a goat polyclonal anti-IRF8 antibody (C-19, Santa Cruz Biotechnology) and a rabbit polyclonal anti-IRAK2 antibody (Abcam) respectively. Immunoprecipitates were resuspended in 20 µl of SDS-PAGE loading buffer for subsequent electrophoresis on a 7.5% SDS-PAGE. The gel was dried and placed on a film for autoradiography overnight at −80°C.
Retroviral transduction
Plat-E cells seeded at a density of 2 × 106 per well into 6-well plates were cultured overnight and then transfected using Fugene HD (Roche) with 3 µg of the retroviral vector. After 48 hours, the viral supernatant was collected and centrifuged at 1500 rpm. 1.5 ml of viral supernatant was used to transduce 4 × 105 BMDMs in the presence of 8 µg/ml of polybrene (Sigma). BMDMs were used for assays 48 hours after viral transduction. For pMx-Puro vector-based transduction, virally-transduced macrophages were selected in the puromycin-containing medium for 4 days prior to being replated for assays. pMX-Irf8-IRES-EGFP retroviral vector and pMX-IRES-EGFP control vector were kindly provided by Masamichi Takami. A retroviral construct encoding IRAK2 and an empty-vector control construct were generous gifts from Shizuo Akira41. WT and DN (Thr197Ala-Thr202Ala mutant) MNK1 expression constructs were kindly provided by Jonathan A. Cooper42 and cDNA fragments encoding WT and DN MNK1 were subcloned into the pMx-Puro retroviral vector.
Flow cytometry
BMDMs were harvested after 5 days of culture in M-CSF-containing conditioned medium and stained with an APC-conjugated anti-CD11b antibody (BD Pharmingen) and a PE-conjugated anti-F4/80 antibody (eBioscience). Cells were washed three times and analyzed on a FACScan flow cytometer (BD) using CellQuest software (BD).
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Honjo for the Rbpjflox/flox mice, T. Gridley for Notch1+/− mice, R. Kopan for the NICD1 expression plasmids, S. Smale for the Il12b promoter reporter constructs, M. Takami for the IRF8 expression construct, S. Akira for the IRAK2 retroviral constructs, J. A. Cooper for the MNK1 expression plasmids, E. Kieff and J. C. Aster for anti-RBP-J rabbit serum, and E. G. Pamer for helpful discussion regarding the L. monocytogenes infection experiments. We also thank K. Au for technical assistance. This work was supported by grants from the American College of Rheumatology (X.H.) and the National Institutes of Health (L.B.I. & X.H.).
Footnotes
AUTHOR CONTRIBUTIONS
H.X., J.Z., J.F., A.Y.C., and C.S. performed experiments and analyzed data. S.S. performed experiments, analyzed data, and prepared the manuscript. B.Z. generated NICD1M mice, provided the IRF8-expressing retroviral vector, and assisted with the experiments using IRF8−/− mice. H.O. and J.K. provided Notch1+/− mice. S.W. and P.S. provided ADAM10-deficient mice. Y.L. provided GSI-34. K.O. provided IRF8−/− mice. C.P.B provided ADAM17flox/flox mice and advice for experiments. L.B.I. provided advice for experiments and contributed to manuscript preparation. X.H. designed research, supervised experiments, and prepared the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krausgruber T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 3.Satoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 4.Tailor P, et al. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007;27:228–239. doi: 10.1016/j.immuni.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtschke T, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Guan X, Tamura T, Ozato K, Ma X. Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. J. Biol. Chem. 2004;279:55609–55617. doi: 10.1074/jbc.M406565200. [DOI] [PubMed] [Google Scholar]
- 7.Xiong H, et al. Complex formation of the interferon (IFN) consensus sequence-binding protein with IRF-1 is essential for murine macrophage IFN-gamma-induced iNOS gene expression. J. Biol. Chem. 2003;278:2271–2277. doi: 10.1074/jbc.M209583200. [DOI] [PubMed] [Google Scholar]
- 8.Giese NA, et al. Interferon (IFN) consensus sequence-binding protein, a transcription factor of the IFN regulatory factor family, regulates immune responses in vivo through control of interleukin 12 expression. J. Exp. Med. 1997;186:1535–1546. doi: 10.1084/jem.186.9.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Morse HC., 3rd IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol. Res. 2009;43:109–117. doi: 10.1007/s12026-008-8055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J. Interferon. Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 11.Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 13.Mazumder B, Li X, Barik S. Translation control: a multifaceted regulator of inflammatory response. J. Immunol. 2010;184:3311–3319. doi: 10.4049/jimmunol.0903778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 16.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monsalve E, et al. Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J. Immunol. 2006;176:5362–5373. doi: 10.4049/jimmunol.176.9.5362. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Cheng P, Youn JI, Cotter MJ, Gabrilovich DI. Notch and wingless signaling cooperate in regulation of dendritic cell differentiation. Immunity. 2009;30:845–859. doi: 10.1016/j.immuni.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foldi J, et al. Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J. Immunol. 2010;185:5023–5031. doi: 10.4049/jimmunol.1001544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang YC, et al. Notch signaling determines the M1 versus M2 polarization of macrophages in antitumor immune responses. Cancer Res. 2010;70:4840–4849. doi: 10.1158/0008-5472.CAN-10-0269. [DOI] [PubMed] [Google Scholar]
- 22.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB. TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J. Exp. Med. 2012;209:319–334. doi: 10.1084/jem.20111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Outtz HH, Wu JK, Wang X, Kitajewski J. Notch1 deficiency results in decreased inflammation during wound healing and regulates vascular endothelial growth factor receptor-1 and inflammatory cytokine expression in macrophages. J. Immunol. 2010;185:4363–4373. doi: 10.4049/jimmunol.1000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 27.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plevy SE, Gemberling JH, Hsu S, Dorner AJ, Smale ST. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grumont R, et al. c-Rel regulates interleukin 12 p70 expression in CD8(+) dendritic cells by specifically inducing p35 gene transcription. J. Exp. Med. 2001;194:1021–1032. doi: 10.1084/jem.194.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie QW, Kashiwabara Y, Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J. Biol. Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 31.Masumi A, Tamaoki S, Wang IM, Ozato K, Komuro K. IRF-8/ICSBP and IRF-1 cooperatively stimulate mouse IL-12 promoter activity in macrophages. FEBS Lett. 2002;531:348–353. doi: 10.1016/s0014-5793(02)03556-1. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Activation of IL-27 p28 gene transcription by interferon regulatory factor 8 in cooperation with interferon regulatory factor 1. J. Biol. Chem. 2010;285:21269–21281. doi: 10.1074/jbc.M110.100818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kantakamalakul W, et al. Regulation of IFN consensus sequence binding protein expression in murine macrophages. J. Immunol. 1999;162:7417–7425. [PubMed] [Google Scholar]
- 35.Zhao J, et al. IRF-8/interferon (IFN) consensus sequence-binding protein is involved in Toll-like receptor (TLR) signaling and contributes to the cross-talk between TLR and IFN-gamma signaling pathways. J. Biol. Chem. 2006;281:10073–10080. doi: 10.1074/jbc.M507788200. [DOI] [PubMed] [Google Scholar]
- 36.Xiong H, et al. Ubiquitin-dependent degradation of interferon regulatory factor-8 mediated by Cbl down-regulates interleukin-12 expression. J. Biol. Chem. 2005;280:23531–23539. doi: 10.1074/jbc.M414296200. [DOI] [PubMed] [Google Scholar]
- 37.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan Y, et al. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control. J. Biol. Chem. 2009;284:10367–10375. doi: 10.1074/jbc.M807822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joshi S, et al. Essential role for Mnk kinases in type II interferon (IFNgamma) signaling and its suppressive effects on normal hematopoiesis. J. Biol. Chem. 2011;286:6017–6026. doi: 10.1074/jbc.M110.197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flannery SM, Keating SE, Szymak J, Bowie AG. Human interleukin-1 receptor-associated kinase-2 is essential for Toll-like receptor-mediated transcriptional and post-transcriptional regulation of tumor necrosis factor alpha. J. Biol. Chem. 2011;286:23688–23697. doi: 10.1074/jbc.M111.248351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawagoe T, et al. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat. Immunol. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 42.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. Embo J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conner JR, Smirnova, Poltorak A. A mutation in Irak2c identifies IRAK-2 as a central component of the TLR regulatory network of wild-derived mice. J. Exp. Med. 2009;206:1615–1631. doi: 10.1084/jem.20090490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab. Invest. 2008;88:11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- 45.Shin HM, et al. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. Embo J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shelton CC, Tian Y, Frattini MG, Li YM. An exo-cell assay for examining real-time gamma-secretase activity and inhibition. Mol. Neurodegener. 2009;4:22. doi: 10.1186/1750-1326-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorissen E, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horiuchi K, et al. Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J. Immunol. 2007;179:2686–2689. doi: 10.4049/jimmunol.179.5.2686. [DOI] [PubMed] [Google Scholar]
- 49.Shi C, et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34:590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. U S A. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.