Abstract
The polypeptide subunits of the photosynthetic electron transport complexes in plants and algae are encoded by two genomes. Nuclear genome-encoded subunits are synthesized in the cytoplasm by 80S ribosomes, imported across the chloroplast envelope, and assembled with the subunits that are encoded by the plastid genome. Plastid genome-encoded subunits are synthesized by 70S chloroplast ribosomes directly into membranes that are widely believed to belong to the photosynthetic thylakoid vesicles. However, in situ evidence suggested that subunits of photosystem II are synthesized in specific regions within the chloroplast and cytoplasm of Chlamydomonas. Our results provide biochemical and in situ evidence of biogenic membranes that are localized to these translation zones. A “chloroplast translation membrane” is bound by the translation machinery and appears to be privileged for the synthesis of polypeptides encoded by the plastid genome. Membrane domains of the chloroplast envelope are located adjacent to the cytoplasmic translation zone and enriched in the translocons of the outer and inner chloroplast envelope membranes protein import complexes, suggesting a coordination of protein synthesis and import. Our findings contribute to a current realization that biogenic processes are compartmentalized within organelles and bacteria.
Keywords: photosynthesis, protein trafficking, assembly, cell, mRNA
Membrane biogenesis requires the concerted synthesis and localization of component lipids and proteins. The endoplasmic reticulum (ER) organizes these processes for the biogenesis of the nuclear envelope, the endomembrane system, lysosomes, peroxisomes, and the plasma membrane. This coordination involves the localization of ribosomes and mRNAs to the rough ER for translation and the cotranslational membrane translocation of proteins destined for specific subcellular compartments (1). Other subcellular compartments and structures are also sites of localized translation. For example, localized synthesis of specific proteins occurs in eggs and polarized cells for pattern formation, in neurons for the formation and remodeling of synapses, and at the mitotic spindle, mitochondria, chloroplasts, and bacteria for biogenesis (2–4). Therefore, localized translation is a general mechanism for establishing the correct protein compositions of subcellular compartments.
Here we explore localized translation in the biogenesis of the photosynthetic thylakoid membranes in chloroplasts. Thylakoid membranes form a network of vesicles and contain the complexes of the photosynthetic electron transport system. Their biogenesis involves two distinct translation systems (5). Subunits encoded by the nuclear genome are synthesized in the cytoplasm by 80S ribosomes, imported across the chloroplast envelope, and targeted to thylakoids. Within the chloroplast, other subunits are encoded by the plastid genome and synthesized by 70S bacterial-like ribosomes. Precisely where thylakoid proteins are synthesized within the cytoplasm and chloroplast is under debate (3).
As chloroplasts enlarge and divide (e.g., in the young green tissue of vascular plants and growing populations of algae), they require protein synthesis to make new photosynthesis complexes. It is widely believed that nascent polypeptides are cotranslationally inserted into stroma-exposed thylakoid membranes because ribosomes are bound to thylakoid membranes (5). However, chloroplast ribosomes also translate the psbA mRNA to repair photochemically damaged photosystem II (PSII), making it difficult to identify ribosomes involved in the de novo biogenesis of the complex.
An alternative model proposes that a specific “translation zone” (T-zone) in the chloroplast of the green alga Chlamydomonas reinhardtii is a privileged site of protein synthesis for the de novo biogenesis of PSII and possibly other complexes (6, 7). This T-zone was defined by the colocalization of markers of the chloroplast ribosome, chloroplast mRNAs encoding PSII subunits, and the PSII translation factor RBP40 [RBP40 is also called RB38 (8, 9)], as seen by confocal microscopy (6). The T-zone is located in the outer perimeter of the pyrenoid, a spherical body in algal chloroplasts and only relevant here as a cytological landmark. The T-zone was defined only by the results of fluorescence microscopy and, therefore, its ultrastructure and biochemical nature are unknown.
We postulated that the T-zone contains a chloroplast translation membrane (CTM) as a privileged site for the synthesis of PSII subunits encoded by the chloroplast genome because chloroplasts ribosomes synthesizing PSII subunits are bound to membrane, but the chloroplast envelope and most thylakoid membranes are outside the T-zone (6, 10). This prediction provided an avenue to test the T-zone model at the biochemical level.
Here, a subcellular fractionation scheme was developed to reveal a CTM. This scheme resolves chloroplast membranes with the high density of rough ER membrane (11) because we predicted that a CTM would also have a high-density membrane as a result of its having bound ribosomes. We focused on the location of PSII subunit synthesis because the evidence for the T-zone was established for chloroplast mRNAs encoding PSII subunits (6). We also identified unique domains of chloroplast envelope that are enriched in the translocons of the outer and inner chloroplast envelope membranes (TOC-TIC) and located adjacent to cytosolic regions where previous in situ evidence supports the localized translation of the mRNA encoding a subunit of the light harvesting complex II (LHCII), which is peripherally associated with PSII (7). Taken together, the results reveal two unique biogenic membranes of the chloroplast and suggest a spatiotemporal organization of PSII-LCHII biogenesis.
Results
Subcellular Fractionation Reveals Chloroplast Translation Membranes.
To study where proteins are synthesized in the chloroplast, we used analytical subcellular fractionation to determine whether the membranes bound by the chloroplast translation machinery have the density of thylakoid membranes, the chloroplast envelope membrane, or an unknown membrane type (12). C. reinhardtii cells were broken by French Pressure Cell because other breakage methods leave unbroken cells that contaminate the membranes of interest in subsequent steps. Using isopycnic ultracentrifugation, membranes were floated from a 2.5 M sucrose cushion into a 0.5–2.2 M sucrose concentration gradient, where they banded according to density. Nonmembrane material either remained in the 2.5 M sucrose cushion or pelleted. Most previous studies of ribosome-bound chloroplast membranes used discontinuous sucrose density gradients to isolate bulk membranes in broad density ranges. Chloroplast envelope membranes are isolated in the density range of 0.4–1 M sucrose (13). Thylakoid membranes are considered to be the densest membrane of the chloroplast and, therefore, are collected as bulk dense membranes in the range of 1–2 M sucrose (14, 15). As rough ER membranes are denser than thylakoid membranes (11), it seemed plausible that previous studies inadvertently analyzed an analogous ribosome-bound CTM with thylakoids (10). Therefore, to separate membranes in this density range with high resolution, we used continuous sucrose gradients with a high maximal concentration (2.2 M). Gradient fractions were analyzed by immunoblotting to determine the density of membranes associated with markers of the chloroplast translation machinery and known chloroplast compartments (Fig. 1). Gel lanes had the same proportions of fractions so that the amount of a marker would reflect the proportion of its total cellular pool (12). In other words, samples were not normalized on the basis of mass amounts of protein because this would drastically overrepresent markers, on a per cell basis, in fractions with the least amount of protein, and vise versa.
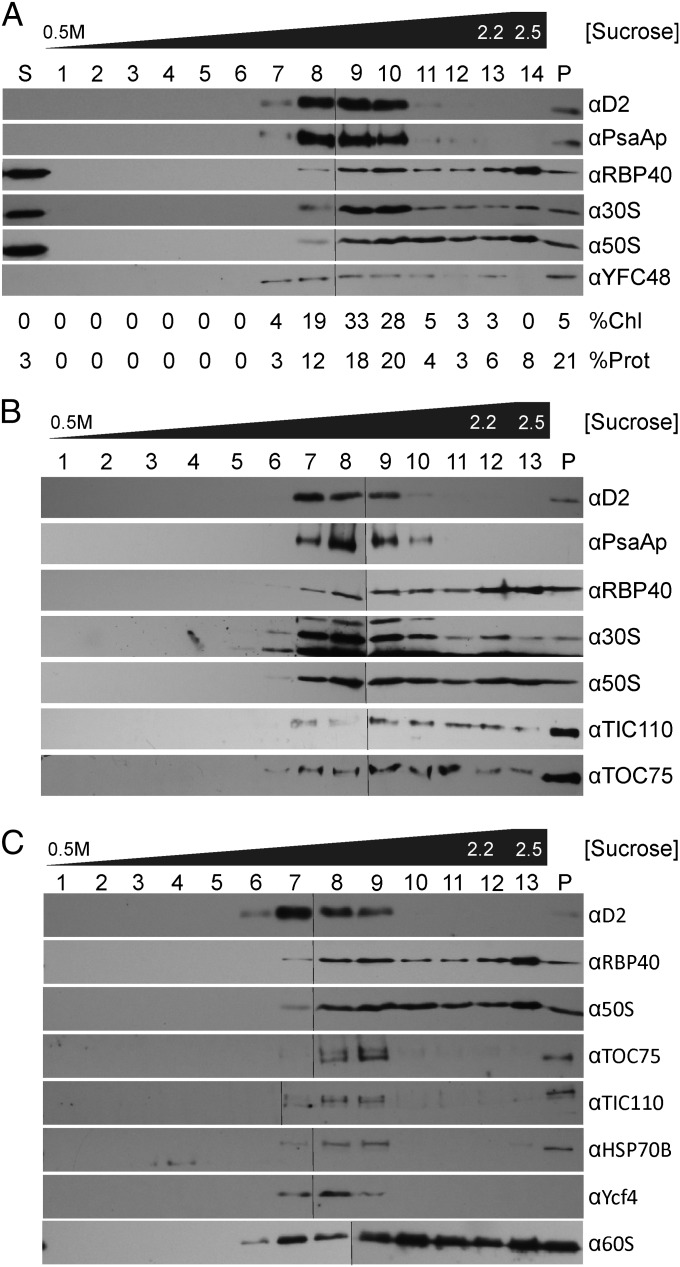
Fig. 1.
Chloroplast translation membranes were revealed by subcellular fractionation. Each panel shows the results from an independent trial of our subcellular fractionation scheme. Fractions were assayed with immunoblots for the following marker proteins: appressed (granal) thylakoid membranes (D2), stroma-exposed thylakoid membranes (PsaAp), CTM and the T-zone (RBP40, r-proteins of the 30S and 50S chloroplast ribosomal subunits), the TOC-TIC protein translocons of the outer and inner chloroplast envelope membranes (Toc75 and Tic110, respectively), chloroplast stroma (HSP70B), PSII assembly (YFCF48/HCF136), PSI assembly (YCF4p), and the cytoplasmic ribosome (60S). (A) Percentages of total chlorophyll (%Chl) and protein (%Prot) in each fraction are indicated. The supernatant of the initial high speed centrifugation is labeled “S.” Membranes of the sucrose gradient were collected as (A) fractions 1–13 or (B and C) fractions 1–12. The 2.5 M sucrose from which membranes were floated is (A) fraction 14 and (B and C) fraction 15. The pellet of the sucrose gradient (P). A thin line in each row distinguishes the images of immunoblots of two gels for which all steps were carried out in the same solutions and ECL and photographic exposures.
Our major finding was that thylakoid membranes can be resolved from denser membranes that are associated with markers of the chloroplast translation machinery and the T-zone. As seen for the three experimental trials of our subcellular fractionation scheme (Fig. 1), the fractions with thylakoid membranes could be identified by their enrichment in chlorophyll and the subunits of PSI and PSII, PsaAp, and D2, respectively (Fig. 1A, lanes 7–10; Fig. 1B, lanes 7–10; and Fig. 1C, lanes 6–9). Envelope membranes are less dense than thylakoids and should be in lanes 4–6 of Fig. 1 (see below) (13). Interestingly, denser membrane fractions had substantial proportions of the total pools of chloroplast ribosomal (r)-proteins and RBP40, and yet they had minor amounts of thylakoid membranes (Fig. 1A, lanes 11–13; Fig. 1 B and C, lanes 10–12). As predicted, these dense membranes had similar density to the canonical ribosome-bound membrane of the rough ER revealed by an r-protein of the 60S subunit of the cytoplasmic ribosome (Fig. 1C, lanes 6–12). In contrast, CTM were distinct from stroma-exposed thylakoid membranes, the accepted site of PSII subunit synthesis (Fig. 1 A and B, PsaAp). These results provided evidence of a CTM privileged for the synthesis and membrane insertion of PSII subunits and localized in the T-zone.
In PSII assembly, newly synthesized subunits associate to form subcomplexes, which then associate to form the monomeric PSII complex RCC1 (reaction centre core 1) (16). In an attempt to identify membranes associated with the assembly of chloroplast-encoded subunits, fractions were immuno-probed with an antiserum against a PSII assembly factor in Synechocystis sp. PCC 6803, YCF48, the homolog of HCF136 of Arabidopsis thaliana (Fig. 1A) (17). This antiserum detected a protein of the expected molecular mass (Fig. S1). This putative YCF48/HCF136 was detected in a broad membrane density range (Fig. 1A, lanes 7–13) but not in the nonmembrane material (Fig. 1A, lanes S and 16). Notably, Fraction 7 contained YCF48/HCF136 but had little thylakoid membrane and no detectable CTM (Fig. 1A, lane 7). This result suggests that this fraction contains yet another unique biogenic membrane, one involved in PSII assembly. Although these results are preliminary, they are consistent with the general theme here: that the C. reinhardtii chloroplast may have diverse biogenic membranes.
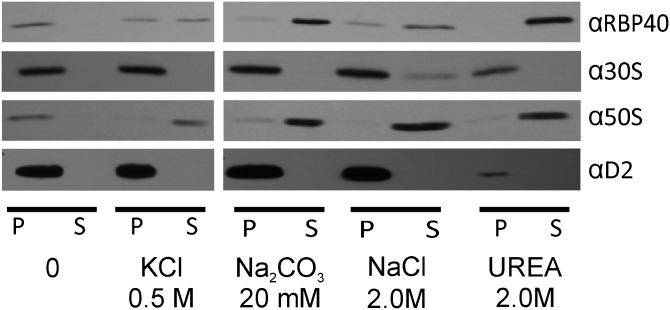
CTM should be physically bound by chloroplast ribosomes. Alternatively, a ribosome-associated membrane could be generated artifactually during cell breakage if free chloroplast ribosome subunits and RBP40 in the chloroplast stroma become trapped within vesicles that form by fragmenting membranes. Detracting from this possibility, however, is that a marker protein for the chloroplast stroma, HSP70B, was not in CTM fractions, but trace amounts were detected in the thylakoid fractions in lanes 10–12 and 7–9, respectively, of Fig. 1C. To more directly address whether chloroplast translation marker proteins are bound to the CTM, we asked whether they can be extracted by agents that remove peripheral membrane proteins. Membranes of fraction 10 in Fig. 1B were washed with one of the following: 500 mM KCl, 20 mM NaCO3, 1.0 M NaCl, or 2.0 M urea or, as a negative control, without agent. Supernatant and pellet fractions were analyzed by immunoblot to determine the degree of extraction of marker proteins for chloroplast ribosome subunits and RBP40 (8). To ensure pelleting of the membrane, we followed the low amount of thylakoid membrane in this fraction by immuno-probing for D2. The results revealed that RBP40 and the 50S subunit were extracted by each of the agents, either partially or completely (Fig. 2). Therefore, these translation components are peripherally bound to CTM. The 30S subunit r-protein was only extracted by high ionic strength (2.0 M NaCl) and only partially. Although this result alone is consistent with either of the two possibilities outlined above, the 30S ribosome subunit is probably bound to CTM because it seems improbable that it would be trapped in vesicles when most 50S subunits and RBP40 are not. Therefore, the 30S subunit is probably bound to CTM with particularly high affinity.
Fig. 2.
CTM association of ribosome subunits and RBP40. Samples of CTM (fraction 10 in Fig. 1B) were incubated with the indicated agents to extract peripheral membrane proteins. Membranes were pelleted by centrifugation and then immunoblot analyses compared the nonmembrane supernatant (S) and membrane pellet (P) fractions to reveal the degrees of extraction of RBP40 and the 30S and 50S subunits of the chloroplast ribosome. The trace amount of thylakoid membranes in this sample allowed us to ensure pelleting of membranes by immunoprobing for D2.
Blue-Native PAGE and Immunoblot Analyses Reveal Markers of PSII Biogenesis.
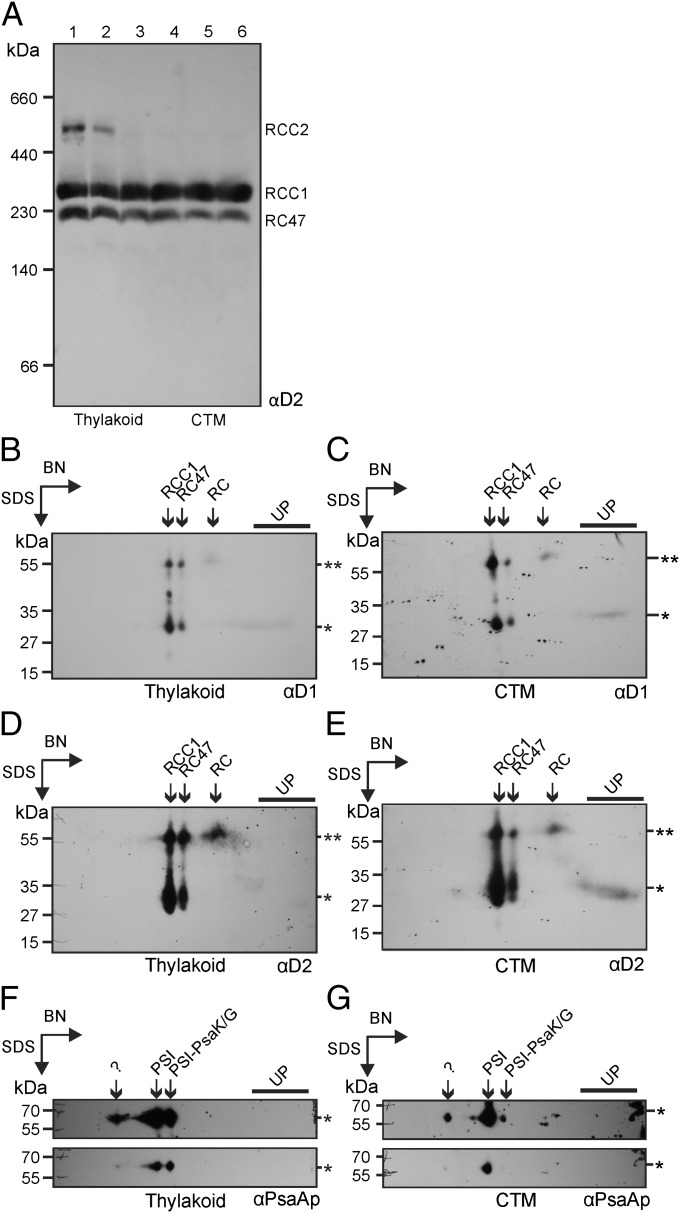
Newly synthesized PSII subunits assemble in specific combinations to form precomplexes, which then associate to form the monomeric PSII complex RCC1 (16). We reasoned that unassembled subunits could serve as a marker for a CTM, and precomplexes for specific steps in PSII assembly. Therefore, membrane fractions were compared for the assembly states of the chloroplast-encoded PSII subunits D1 and D2 using blue-native (BN) PAGE and immunoblot analysis (18). Analyses of equal amounts of membrane ensured comparable solubilisation conditions, which can affect quaternary structure artifactually (19). Because this process necessitated overrepresentation of CTM on a per cell basis, the amounts of sample were normalized to the level of the monomeric PSII complex, RCC1. In other words, we asked whether CTM are qualitatively different from thylakoid membranes in ways that support their having a role in PSII subunit synthesis and assembly. The results revealed RCC1 at constant levels across the lanes, confirming proper normalization (Fig. 3A). The higher mobility complex is RC47, the PSII monomer lacking CP43, which is generated primarily during PSII repair (20). Notably, the dimeric PSII, RCC2, was detected in the thylakoid membrane fractions but not in CTM fractions (Fig. 3A, compare lanes 1–2 with 4–6). This result suggests that thylakoid membranes are the primary location of RCC1 dimerization to form RCC2, a late step in PSII biogenesis (16).
Fig. 3.
BN-PAGE revealed markers of protein synthesis for PSII de novo assembly and repair. (A) Analysis by 1D BN-PAGE compared the assembly states of D2 in thylakoids (lanes 1–2) and CTM (lanes 3–6); samples of fractions 8–13 in Fig. 1A. D2 was immunodetected in RCC1, RCC2, and RC47. Samples were normalized to the level of RCC1 to ensure comparable solubilization conditions (Results). (B–G) To reveal subcomplexes and unassembled free subunits, BN-PAGE lanes with thylakoid membranes or CTM, equivalent to lanes 1 and 6 in A, respectively, were subjected to a second dimension of SDS/PAGE before immunoblot analyses. RCC1 levels determined in A were used to normalize samples analyzed on the 2D gels. The 2D gel-immunoblots of thylakoid membranes (B, D, and F) or CTM (C, E, and G) were first immuno-probed for D1 (B and C), then for D2 (D and E), and finally for the PSI subunit PsaAp (F and G). D1 and D2 were detected in RCC1, RCC2, and RC47, in smaller assembly intermediate precomplexes [RC47 and PSII reaction center (RC)] and as unassembled subunits (UP). The expected molecular mass of each protein is indicated by an asterisk. Some D1 and D2 was shifted to higher molecular mass positions of the gels (**) because of incomplete denaturation before the second dimension of SDS/PAGE. This shift was useful because it resolved the RC (shifted*) from the free subunits (not shifted**). The same results were obtained when this shift did not occur. (F and G) PsaAp was detected in the PSI monomer (PSI), a putative PSI monomer lacking PsaK and PsaG (PSI-PsaK/G), and an unknown complex, possibly the PSI dimer (“?”).
With 1D BN-PAGE we were unable to detect free subunits and subcomplexes for use as CTM markers, possibly because of ill effects of the detergent on detection below 100 kDa (19). Therefore, BN gel lanes with either thylakoid membranes or CTM were subjected to a second dimension of denaturing SDS/PAGE and analyzed by immunoblotting. We normalized based on the relative levels of RCC1 in these samples determined by 1D BN-PAGE (Fig. 3A). (RCC2 was not detected for unknown reasons on the 2D gel immunoblot analyses.)
The results revealed that D1 is present in RCC1 and RC47, as well as in an early assembly intermediate subcomplex, the PSII reaction center, and as a free unassembled subunit. All were detected in both thylakoid and CTM samples (Fig. 3 B and C). Unassembled D1 could not serve as a marker for CTM because it is associated with both the repair and de novo biogenesis of PSII. Nevertheless, this result revealed that the assembly step in which the reaction center forms RCC1 does not occur preferentially in CTM over thylakoid membranes. Furthermore, the finding that unassembled D1 can be detected on both immunoblots serves as a positive control for the subsequent experiments.
These blots were immunoprobed for D2, a PSII subunit the synthesis of which is not induced for PSII repair. Therefore, unassembled D2 can serve as a marker for PSII biogenesis. D2 was detected in RCC1, RC47, and the reaction center (RC) in both thylakoids and CTM (Fig. 3 D and E). Most notably, however, unassembled D2 was detected only in the CTM sample (Fig. 3E). These results suggest that CTM is a privileged location of the synthesis of the plastid genome-encoded subunits for de novo assembly of PSII.
Markers of PSI Assembly Cofractionate with Thylakoid Membranes.
To determine whether CTM has a role in PSI biogenesis, we immuno-probed the 2D blots for PsaAp (Fig. 3 F and G). Although PsaAp was not detected unassembled, it was detected in the PSI monomeric complex, the PSI monomeric complex lacking PsaK and PsaG, and a larger unidentified complex of approximately 550 kDa, possibly the PSI dimer (21). Notably, the PSI monomer lacking PsaK/G was more abundant in the thylakoid membrane fraction than in the CTM fraction (Fig. 3 F and G). This result and previously reported evidence that this complex is a late intermediate in PSI assembly (21), suggest that later steps in PSI assembly occur primarily in thylakoid membranes. Earlier steps in PSI assembly may also occur in thylakoids because we detected an early PSI assembly factor, YCF4, only in thylakoid membrane fractions (Fig. 1C, compare fractions 7–9 and fractions 10–12) (22). Finally, the mRNA encoding PsaAp was not recruited to the T-zone under the same conditions that recruited two PSII subunit mRNAs (6). Taken together, these results suggest that PSI subunit synthesis and assembly occur at thylakoids, and not CTM.
Envelope Membranes with the TOC-TIC Translocons Have Higher than Expected Density.
The gradient fractions were also tested for the envelope markers, Toc75 and Tic110, subunits of the TOC and TIC protein import complexes. Instead of finding these proteins in the density range of the envelope membranes (Fig. 1, fractions 4–6) (13), they were detected with thylakoid fractions and, in certain preparations, also with denser CTM (Fig. 1 B and C). Although the basis of this unexpectedly high and variable density of envelope membranes with the TIC and TOC complexes is unknown, their occasional separation from CTM reveals these are distinct membrane types.
Chloroplast Protein Import Machinery Localizes to Unique Envelope Domains.
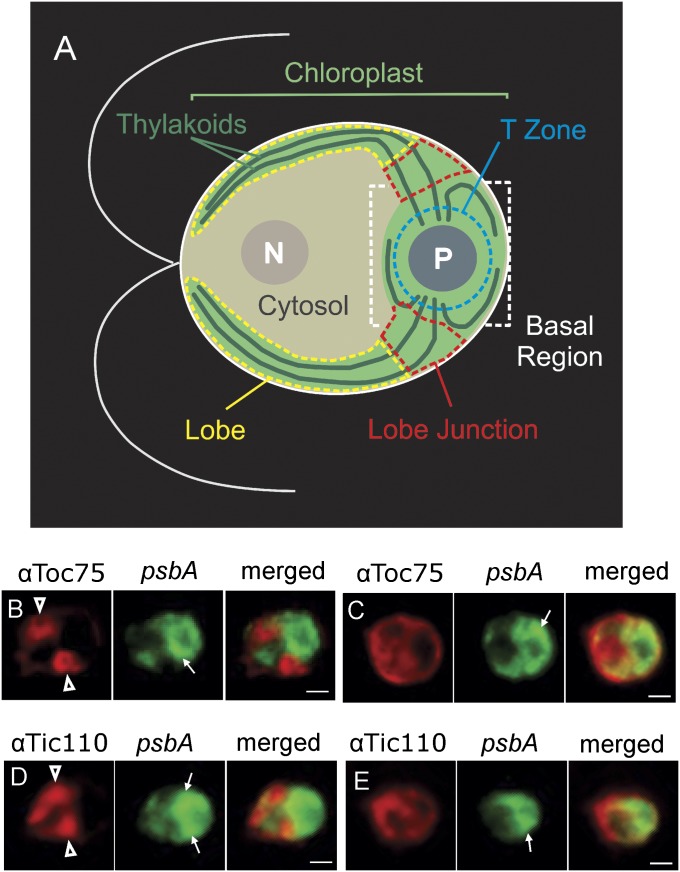
To explore the basis of the unexpectedly high density of envelope membranes with Toc75 and Tic110, the in situ distribution of these proteins was characterized by immunofluorescence (IF) staining and epifluorescence microscopy. All cells were costained for the chloroplast psbA mRNA by FISH to reveal the T-zone with strong signal localized around the pyrenoid and to stain the chloroplast with weaker diffuse signal. For a description of chloroplast anatomy, see Fig. 4A (6).
Fig. 4.
The TOC and TIC protein import complexes are localized to chloroplast envelope domains. (A) An illustration of a Chlamydomonas cell shows the nucleus (N), cytosol, and chloroplast with its lobes, lobe junctions, basal region, thylakoid lamellae, T-zone, and pyrenoid (P). The chloroplast lobes extend from the basal region to the anterior cell pole, thereby “cupping” the nuclear-cytosolic compartments. (B–E) Representative cells are oriented as in A and show the IF signal from Toc75 (B and C) or Tic110 (D and E). Costaining for the psbA mRNA by FISH (green) revealed the T-Zone (thin arrows). Cells in B and D (moderate light) show the localization of the Toc75 or Tic110 IF signal at lobe junctions, and cells in C and E (dark-adapted) do not show this localization pattern. (Scale bars, 2 μm.)
A striking pattern was observed in many cells in which the Toc75 or Tic110 IF signal localized around lobes specifically where they adjoined the basal region (Fig. 4 B and D). We named these sites “lobe junctions” (Fig. 4A). In some cases, the lobe at such a lobe junction could be seen to form a hole in the cloud of IF signal, indicating that the envelope surrounding it was enriched in the TOC-TIC protein import machinery (Fig. 4B). Examples of the cells that did not show this localization pattern are shown in Fig. 4 C and E. Of the cells examined from moderate light growth condition (ML cells), 48% showed the Toc75 IF signal localized around one or two lobe junctions (Fig. 4B) (n = 188). Similarly, of the ML cells IF-stained for Tic110, 47% showed this pattern (Fig. 4D) (n = 199). This pattern is interesting for three reasons. First, it is specific to import machinery because it was not observed for many other chloroplast proteins, the localization of which we have examined with this method (6, 7, 23). Second, this localization pattern may be physiologically relevant because the percentage of cells showing it dropped during incubation in the dark for 2 h, a condition associated with reduced rates of PSII biogenesis and chloroplast protein import in C. reinhardtii (6, 24). When ML cells were dark-adapted immediately before fixation, the percentages showing localization around lobe junctions dropped from 48 to 11% for Toc75 (n = 74) and from 47 to 15% for Tic110 (n = 52). Finally, Toc75 or Tic110 localization at lobe junction was probably present but undetected in many cells. C. reinhardtii cells are polarized and must be oriented longitudinally in the microscopy field to reveal cytological landmarks necessary to locate lobe junctions (e.g., the cytosol and pyrenoid) (Fig. 4A). Moreover, no protein marker exists for which co–IF-staining can reveal lobe junctions in a particular cell.
These results suggest that lobe junctions have special envelope domains enriched in the TOC-TIC protein import machinery. In light of these results, the unexpectedly high and variable density of envelope membranes with Toc75 and Tic110 could be explained if these import envelope domains have higher density than previously described envelope membranes, and had formed to different degrees in the various cultures used for subcellular fractionation.
Discussion
Our results provide biochemical evidence of a CTM as a privileged location of the synthesis of plastid genome-encoded PSII subunits and localized in the T-zone (6, 7). We also report biochemical and in situ evidence of a second unique chloroplast membrane compartment: domains of chloroplast envelope that are localized around lobe junctions. The possibility of a third chloroplast membrane, one privileged for PSII assembly, is suggested by the enrichment of YCF48/HCF136 in a sucrose gradient fraction with membranes that were neither of thylakoid membranes nor CTM (Fig. 1A). A previously described “low-density” membrane (LDM) of the C. reinhardtii chloroplast was suggested to have a role in the translation of chloroplast mRNAs encoding thylakoid proteins because it is physically associated with RNA-binding proteins and thylakoids (25, 26). LDM is distinct from CTM; it is less dense and not associated with chloroplast translation machinery.
Any model unifying these results must explain how the PSII-LHCII supercomplex is assembled from subunits that are localized to distinct chloroplast compartments: that is, chloroplast-encoded subunits in the T-zone and nucleus-encoded subunits in lobe junctions. In addition, it must be explained how the newly assembled PSII-LHCII supercomplex is localized to thylakoid membranes throughout the chloroplast. Finally, such a model should consider that PSII biogenesis begins with the assembly of the chloroplast-encoded subunits to form RCC1, followed by RCC1 dimerization, the incorporation of nucleus-encoded PSII subunits, and the association of the oxygen evolving complex (OEC) and LHCII (16).
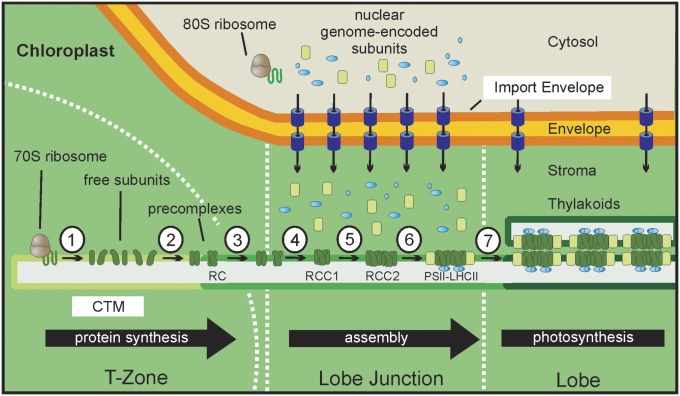
In our working model, the T-zone and lobe junctions are early and intermediate compartments in a spatiotemporal pathway of PSII-LHCII supercomplex biogenesis (Figs. 4A and 5). In the T-zone, the CTM is a platform for the synthesis of the plastid genome-encoded subunits. Chloroplast ribosome subunits and PSII subunit mRNAs are recruited to membranes in the T-zone by translation independent mechanisms (e.g., tethering by membrane bound RNA-binding proteins of LDM) (7). The particularly high affinity with which the 30S subunit is bound to the CTM (Fig. 2) may be related to its early role in the assembly of a translation-component ribosome and, consequently, a requirement to maintain its association when binding the mRNA, large subunit, and initiation factors (5). Newly assembled PSII precomplexes move by lateral diffusion within the CTM to the lobe junctions and assemble to form RCC1 (Fig. 5). The membranes involved could be CTM, thylakoid membranes, or an unknown assembly membrane (above). RCC1 dimerizes and is built upon by nucleus-encoded subunits that are imported locally by TOC-TIC import machinery of the envelope around lobe junctions. These include the subunits of the OEC and LHCII, which are peripherally associated with PSII in the PSII-LHCII supercomplex. Thus, in this model, lobe junctions are convergence points for the pathways that supply polypeptide subunits encoded by the chloroplast and nuclear genomes. Assembled PSII-LHCII supercomplexes move, again by lateral diffusion, to photosynthetic membranes of thylakoid vesicles in the lobes and at the periphery of the chloroplast basal region (Fig. 4A). At each stage, lateral diffusion of subunits and complexes could occur in a contiguous membrane because electron microscopy images have shown that thylakoid vesicles extend from the T-zone to the ends of the lobes or around the periphery of the basal domain (27). In our model, these thylakoid vesicles are laterally heterogeneous, such that their extremities in the T-zone are composed of CTM but their opposing extremities in lobes and at the periphery of the basal region are photosynthetic thylakoid membranes. This model is indirectly supported by similar findings in other organisms (see below).
Fig. 5.
A working model for the spatiotemporal organization of PSII-LHCII supercomplex biogenesis. (1) In the T-zone, plastid-encoded subunits are synthesized into CTM. (LDM might represent a mRNA-ribosome subunit recruitment membrane on the far left.) (2) Free subunits assemble to form the PSII reaction center (RC) and the other precomplexes, and then (3) move by lateral diffusion to a lobe junction (see also Fig. 4A). (4) There, precomplexes associate to form the PSII monomeric complex, RCC1. (5) RCC1 dimerizes to form RCC2. (6) Nuclear genome-encoded subunits of the OEC (blue) and LHCII (light green) are locally imported by the TOC and TIC complexes (purple) into the lobe junction and assembled upon RCC1 and RCC2. (7) The resulting PSII-LHCII supercomplex diffuses to thylakoid membrane located throughout the chloroplast.
Generality of this model is supported by the identification of a PSII biogenesis compartment in the cyanobacterium Synechocystis sp. PCC 6803 and a GFP-tagged Tic20 paralogue which was seen to be localized to specific regions of the chloroplast envelope in A. thaliana (28, 29). The effect of light on the relocalization of the TIC and TOC import machinery to lobe junctions might be relevant to the light stimulation of chloroplast protein import which has been observed in C. reinhardtii and vascular plants (24, 30). Moreover, the Rubisco holoenzyme might be assembled in the pathways described here because its small subunit is imported via the TOC and TIC pathway and the chloroplast mRNA encoding the large subunit localizes in situ in the T-zone and is translated in association with membrane (7, 31, 32). Our findings build upon growing evidence of complex cytological organizations of biogenic processes in organelles and bacteria.
Methods
Culture Conditions.
C. reinhardtii strains CC-4051 or CC-503 were cultured photoautotrophically in high-salt minimal medium with aeration at 24 °C, under a light intensity of approximately 100 µE⋅m−2⋅sec−1 until midlog phase (2–4 × 106 cells·ml−1) (33).
Analytical Subcellular Fractionation.
Cells from a 500-mL culture were pelleted by centrifugation at 4,000 × g for 5 min at 4 °C, resuspended in MKT-buffer [25 mM MgCl2, 20 mM KCl, 10 mM Tricine-Cl pH 7.5, protease inhibitor (Sigma-Aldrich)]. Cells were broken by three passes through an ice-chilled French Pressure Cell at 1,000 psi. Breakage was verified by light microscopy (400× and 1,000× magnification). The lysate was ultracentrifuged at 100,000 × g for 1 h at 4 °C. The supernatant was collected and stored at −80 °C. The pellet was resuspended in 2.5 M sucrose and overlaid with a linear sucrose gradient (0.5–2.2 M). All sucrose solutions were prepared in MKT-buffer. The gradient was ultracentrifuged at 100,000 × g for 16 h at 4 °C. Fractions (0.75 mL) were collected and the pellet was resuspended in KHEG-Buffer [60 mM KCl, 20 mM Hepes, 0.2 mM EDTA, 20% (vol/vol) glycerol]. Gradients contained only membrane and associated material based on the buoyant density of bacterial ribosomes in equilibrium CsCl gradient ultracentrifugation (1.67–1.69 g/mL) would be equivalent to 4.9 M sucrose (34).
Quantification of Protein and Chlorophyll.
Protein concentration was determined using the bicinchoninic acid assay (35). Chlorophyll was quantified as described previously (36).
Immunoblot Analysis.
Equal proportions of the fractions were solubilized in SDS/PAGE loading buffer, denatured at 42 °C for 30 min. SDS/PAGE and immunoblot analyses were performed as described previously (37). The antisera were: αD1 (Agrisera), αS-20 (30S r-protein), αL-30 (50S r-protein) and αcyL4 (60S r-protein) (38, 39), αPsaAp (40), αHSP70B (41), and αRBP40 (42).
FISH, IF-Staining, and Fluorescence Microscopy.
FISH and IF-staining of cells of strain CC-503 were as described previously (6, 43). The psbA FISH probes were labeled with Alexa Fluor 488 and the IF-staining used Alexa Fluor 568-conjugated anti-rabbit secondary antibody (Invitrogen). Images were captured on a Leica DMI6000B microscope (Leica Microsystems) using a 40×/0.75 objective, a Hamamatsu OrcaR2 camera and Volocity acquisition software (Perkin-Elmer).
BN PAGE.
BN-PAGE was performed as described previously with the following minor modifications (18, 44). Aliquots of sucrose gradient fractions containing 6 μg of chlorophyll were concentrated by centrifugation (100,000 × g; 1 h; 4 °C) and resuspended in ACA 750 (750 mM aminocaproic acid, 50 mM Bis•Tris, and 0.5 mM EDTA, pH 7.0). Membranes were then solubilized on ice in 0.8% n-Dodecyl-β-d-Maltoside (β-DM) for 5 min. Samples were centrifuged at 17,000 × g for 30 min at 4 °C. The supernatant was added to 1/10 Vol of 5% (wt/vol) Coomassie Brilliant blue G-250, 750 mM aminocaproic acid whereupon protein complexes were then separated by electrophoresis in a 4.5–12% (wt/vol) acrylamide BN gel. To ensure that D2 signal on different 2D gels was normalized to the level of RCC1, comparable amounts of RCC1 were loaded, as determined by results from 1D BN gels, and all steps were carried out in parallel. Results of maximal ECL exposure times are shown for both.
Membrane Washing.
Aliquots of fraction 10 in Fig. 1B were diluted 25-fold in washing buffer [20 mM KCl, 10 mM Tricine and 2.0 mM EDTA pH 7.2, protease inhibitor mixture (Sigma-Aldrich)] and pelleted by centrifugation in a microfuge for 1 h at 17,000 × g at 4 °C. Pellets were resuspended in 30 µL of one of the following: washing buffer, 500 mM KCl, 20 mM NaCO3, 1.0 M NaCl, 2.0 M urea, incubated on ice for 30 min, and then subjected to the same centrifugation step. The supernatants were collected and the pellet was washed once and then resuspended in 30 µL SDS/PAGE sample buffer. SDS/PAGE and immunoblot analysis were as described previously (37).
Supplementary Material
Acknowledgments
We thank A. Piekny for assistance with microscopy, and the following colleagues for generous gifts of antisera: E. Schleiff for αTic110 and αToc75; M. Schroda for αHSP70B; Y. Takahashi for αYCF4; J. Nickelsen for αD2, αYCF48, and αRBP40; K. Redding for αPsaAp; and E. Harris for αS-20, αL-30, αcyL4. This study was funded by a Deutsche Forschungsgemeinschaft postdoctoral fellowship (to M.S.) and an operating grant from the Natural Sciences and Engineering Council of Canada (217566-03).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209860109/-/DCSupplemental.
References
- 1.Lynes EM, Simmen T. Urban planning of the endoplasmic reticulum (ER): How diverse mechanisms segregate the many functions of the ER. Biochim Biophys Acta. 2011;1813(10):1893–1905. doi: 10.1016/j.bbamcr.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vazquez-Pianzola P, Suter B. Conservation of the RNA transport machineries and their coupling to translation control across eukaryotes. Comp Funct Genomics. 2012;2012:287852. doi: 10.1155/2012/287852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weis BL, Schleiff E, Zerges W. Protein targeting to subcellular organelles via mRNA localization. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Nevo-Dinur K, Govindarajan S, Amster-Choder O. Subcellular localization of RNA and proteins in prokaryotes. Trends Genet. 2012;28(7):314–322. doi: 10.1016/j.tig.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Zerges W. Translation in chloroplasts. Biochimie. 2000;82(6–7):583–601. doi: 10.1016/s0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]
- 6.Uniacke J, Zerges W. Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell. 2007;19(11):3640–3654. doi: 10.1105/tpc.107.054882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uniacke J, Zerges W. Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc Natl Acad Sci USA. 2009;106(5):1439–1444. doi: 10.1073/pnas.0811268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarz C, Elles I, Kortmann J, Piotrowski M, Nickelsen J. Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell. 2007;19(11):3627–3639. doi: 10.1105/tpc.107.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes D, et al. Identification and characterization of a novel RNA binding protein that associates with the 5′-untranslated region of the chloroplast psbA mRNA. Biochemistry. 2004;43(26):8541–8550. doi: 10.1021/bi035909j. [DOI] [PubMed] [Google Scholar]
- 10.Mühlbauer SK, Eichacker LA. Light-dependent formation of the photosynthetic proton gradient regulates translation elongation in chloroplasts. J Biol Chem. 1998;273(33):20935–20940. doi: 10.1074/jbc.273.33.20935. [DOI] [PubMed] [Google Scholar]
- 11.Lerner RS, et al. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9(9):1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quail PH. Plant cell fractionation. Annu Rev Plant Physiol. 1979;30(1):425–484. [Google Scholar]
- 13.Salvi D, Rolland N, Joyard J, Ferro M. Purification and proteomic analysis of chloroplasts and their sub-organellar compartments. Methods Mol Biol. 2008;432:19–36. doi: 10.1007/978-1-59745-028-7_2. [DOI] [PubMed] [Google Scholar]
- 14.Chua NH, Blobel G, Siekevitz P, Palade GE. Attachment of chloroplast polysomes to thylakoid membranes in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1973;70(5):1554–1558. doi: 10.1073/pnas.70.5.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoober JK. Sites of synthesis of chloroplast membrane polypeptides in Chlamydomonas reinhardi y-1. J Biol Chem. 1970;245(17):4327–4334. [PubMed] [Google Scholar]
- 16.Komenda J, Sobotka R, Nixon PJ. Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol. 2012;15(3):245–251. doi: 10.1016/j.pbi.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Komenda J, et al. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem. 2008;283(33):22390–22399. doi: 10.1074/jbc.M801917200. [DOI] [PubMed] [Google Scholar]
- 18.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199(2):223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 19.Reisinger V, Eichacker LA. How to analyze protein complexes by 2D blue native SDS-PAGE. Proteomics. 2007;7(Suppl 1):6–16. doi: 10.1002/pmic.200700205. [DOI] [PubMed] [Google Scholar]
- 20.Aro EM, et al. Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J Exp Bot. 2005;56(411):347–356. doi: 10.1093/jxb/eri041. [DOI] [PubMed] [Google Scholar]
- 21.Ozawa S, Onishi T, Takahashi Y. Identification and characterization of an assembly intermediate subcomplex of photosystem I in the green alga Chlamydomonas reinhardtii. J Biol Chem. 2010;285(26):20072–20079. doi: 10.1074/jbc.M109.098954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 1997;16(20):6095–6104. doi: 10.1093/emboj/16.20.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uniacke J, Zerges W. Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 2008;182(4):641–646. doi: 10.1083/jcb.200805125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Q, Schild C, Schumann P, Boschetti A. Varying competence for protein import into chloroplasts during the cell cycle in Chlamydomonas. Eur J Biochem. 2001;268(8):2315–2321. doi: 10.1046/j.1432-1327.2001.02111.x. [DOI] [PubMed] [Google Scholar]
- 25.Zerges W, Rochaix JD. Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 1998;140(1):101–110. doi: 10.1083/jcb.140.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zerges W, Wang S, Rochaix JD. Light activates binding of membrane proteins to chloroplast RNAs in Chlamydomonas reinhardtii. Plant Mol Biol. 2002;50(3):573–585. doi: 10.1023/a:1020246007858. [DOI] [PubMed] [Google Scholar]
- 27.Ohad I, Siekevitz P, Palade GE. Biogenesis of chloroplast membranes. I. Plastid dedifferentiation in a dark-grown algal mutant (Chlamydomonas reinhardi) J Cell Biol. 1967;35(3):521–552. doi: 10.1083/jcb.35.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machettira AB, et al. The localization of Tic20 proteins in Arabidopsis thaliana is not restricted to the inner envelope membrane of chloroplasts. Plant Mol Biol. 2011;77(4–5):381–390. doi: 10.1007/s11103-011-9818-5. [DOI] [PubMed] [Google Scholar]
- 29.Nickelsen J, et al. Biogenesis of the cyanobacterial thylakoid membrane system—An update. FEMS Microbiol Lett. 2011;315(1):1–5. doi: 10.1111/j.1574-6968.2010.02096.x. [DOI] [PubMed] [Google Scholar]
- 30.Hirohashi T, Hase T, Nakai M. Maize non-photosynthetic ferredoxin precursor is mis-sorted to the intermembrane space of chloroplasts in the presence of light. Plant Physiol. 2001;125(4):2154–2163. doi: 10.1104/pp.125.4.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breidenbach E, Jenni E, Boschetti A. Synthesis of two proteins in chloroplasts and mRNA distribution between thylakoids and stroma during the cell cycle of Chlamydomonas reinhardii. Eur J Biochem. 1988;177(1):225–232. doi: 10.1111/j.1432-1033.1988.tb14366.x. [DOI] [PubMed] [Google Scholar]
- 32.Mühlbauer SK, Eichacker LA. The stromal protein large subunit of ribulose-1,5-bisphosphate carboxylase is translated by membrane-bound ribosomes. Eur J Biochem. 1999;261(3):784–788. doi: 10.1046/j.1432-1327.1999.00337.x. [DOI] [PubMed] [Google Scholar]
- 33.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas Reinhardi. Proc Natl Acad Sci USA. 1960;46(1):83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenwick ML. The density of ribosomes bearing messenger RNA in phage-infected and normal bacteria. J Cell Sci. 1971;8(3):649–658. doi: 10.1242/jcs.8.3.649. [DOI] [PubMed] [Google Scholar]
- 35.Smith PK, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 36.Porra RJ. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res. 2002;73(1-3):149–156. doi: 10.1023/A:1020470224740. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. Vol 3, pp A8.52–A8.55. [Google Scholar]
- 38.Randolph-Anderson BL, Gillham NW, Boynton JE. Electrophoretic and immunological comparisons of chloroplast and prokaryotic ribosomal proteins reveal that certain families of large subunit proteins are evolutionarily conserved. J Mol Evol. 1989;29(1):68–88. doi: 10.1007/BF02106183. [DOI] [PubMed] [Google Scholar]
- 39.Fleming GH, Boynton JE, Gillham NW. Cytoplasmic ribosomal proteins from Chlamydomonas reinhardtii: Characterization and immunological comparisons. Mol Gen Genet. 1987;206(2):226–237. doi: 10.1007/BF00333578. [DOI] [PubMed] [Google Scholar]
- 40.Redding K, et al. Photosystem I is indispensable for photoautotrophic growth, CO2 fixation, and H2 photoproduction in Chlamydomonas reinhardtii. J Biol Chem. 1999;274(15):10466–10473. doi: 10.1074/jbc.274.15.10466. [DOI] [PubMed] [Google Scholar]
- 41.Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman F-A. The chloroplastic GrpE homolog of Chlamydomonas: Two isoforms generated by differential splicing. Plant Cell. 2001;13(12):2823–2839. doi: 10.1105/tpc.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz C, Elles I, Kortmann J, Piotrowski M, Nickelsen J. Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell. 2007;19(11):3627–3639. doi: 10.1105/tpc.107.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uniacke J, Colón-Ramos D, Zerges W. FISH and immunofluorescence staining in Chlamydomonas. Methods Mol Biol. 2011;714:15–29. doi: 10.1007/978-1-61779-005-8_2. [DOI] [PubMed] [Google Scholar]
- 44.Schottkowski M, et al. Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem. 2009;284(3):1813–1819. doi: 10.1074/jbc.M806116200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.