Abstract
Using a yeast two-hybrid screen, we identified human nucleosome assembly protein 1 (hNAP-1) as a protein interacting with the activation domain of the transcriptional activator encoded by papillomaviruses (PVs), the E2 protein. We show that the interaction between E2 and hNAP-1 is direct and not merely mediated by the transcriptional coactivator p300, which is bound by both proteins. Coexpression of hNAP-1 strongly enhances activation by E2, indicating a functional interaction as well. E2 binds to at least two separate domains within hNAP-1, one within the C terminus and an internal domain. The binding of E2 to hNAP-1 is necessary for cooperativity between the factors. Moreover, the N-terminal 91 amino acids are crucial for the transcriptional activity of hNAP-1, since deletion mutants lacking this N-terminal portion fail to cooperate with E2. We provide evidence that hNAP-1, E2, and p300 can form a ternary complex efficient in the activation of transcription. We also show that p53 directly interacts with hNAP-1, indicating that transcriptional activators in addition to PV E2 interact with hNAP-1. These results suggest that the binding of sequence-specific DNA binding proteins to hNAP-1 may be an important step contributing to the activation of transcription.
The transcriptional activation of genes repressed by nucleosomes requires the presence of activators that bind sequence specifically. These increase the efficiency of assembly of the transcriptionally competent preinitiation complex (PIC) and counteract the repressive effects of chromatin through the recruitment of chromatin remodeling complexes and histone acetyltransferases (HATs) (2, 15, 30, 31, 40, 52, 53, 64). Chromatin remodeling is accomplished by large, ATP-dependent chromatin remodeling complexes that alter chromatin structure by transiently disrupting histone-DNA interactions (reviewed in references 6, 30, and 63). Posttranslational modifications of chromatin, such as acetylation by transcriptional coactivators, also contribute to gene regulation (22). HATs are thought to catalyze the addition of acetyl groups to the N-terminal tails of core histones, a process which usually correlates with the activation of transcription (55). The transcriptional coactivator p300 and its homologue CREB binding protein (CBP) possess HAT activity, and both are implicated in the regulation of transcription by a large number of sequence-specific activator proteins (reviewed in references 11, 19, and 21). p300 and CBP are associated with other HATs, such as p/CAF, ACTR, and SRF, in a multiprotein complex. Functional studies have shown that the coactivator function of p300 and CBP requires their acetyltransferase activity (32, 33, 41). Moreover, additional functions of these cofactors, such as the interaction with components of the PIC, are necessary for their stimulatory activity (3, 45, 51).
The assembly of nucleosomes is tightly linked to DNA replication. The naked daughter strands of newly replicated DNA are rapidly assembled into chromatin by a multistep process. Chromatin assembly factor 1 and replication-coupling assembly factor/anti-silencing function 1 protein (ASF1) act as histone chaperones to deposit histones H3 and H4. Nucleosome assembly protein (NAP) is a histone chaperone responsible for the incorporation of two histone H2A-H2B dimers to complete the nucleosome (reviewed in reference 62). NAP-1 may act as a nucleocytoplasmic shuttling protein that delivers H2A-H2B dimers from the cytoplasm to the chromatin assembly machinery in the nucleus (47). In addition to its function in chromatin assembly, NAP-1 also may play a role in cell cycle progression. Yeast genetic experiments have shown that NAP-1 has a role in cell cycle progression during G1 phase and mitosis. NAP-1 binds to cyclin B (29) and a kinase, Gin4p (1).
Furthermore, histone chaperones seem to facilitate transcriptional activation through their chromatin-modifying activity. Recent data suggest that HAT complexes as well as ATP-dependent chromatin remodeling complexes cooperate with histone chaperones in altering chromatin structure during the activation of transcription. ASF1 was found to functionally interact with the Brahma (SWI/SNF) ATP-dependent chromatin remodeling complex, involved in the activation of transcription (48). A functional interaction between p300/CBP and NAPs also has been reported (4, 27, 58). It has been demonstrated that the acetylation of histones by p300 facilitates the transfer of histones H2A and H2B to NAP-1 in vitro. Thus, the structure of the histones may be altered by histone acetylation facilitating the loss of H2A-H2B dimers that have been remodeled by the action of ATP-dependent chromatin remodeling complexes (27). This model is supported by the observation that NAPs may augment activation by factors which use p300 as a coactivator (58). Furthermore, NAP-1 has been shown to stimulate the binding of transcription factors to their binding sites, a process which is accompanied by disruption of the histone octamer (65). Although hints for a role of histone chaperones such as NAP-1 in the activation of transcription are accumulating, it is not known how histone chaperones may be targeted to actively transcribing promoter regions.
The papillomavirus (PV) E2 protein is a transcription factor which regulates PV transcription and is also required for the replication of viral DNA (14, 18, 46). For bovine PV type 1 (BPV1), E2 strongly activates several BPV1 promoters by binding to its high-affinity palindromic recognition sequence, ACCGN4CGGT, present in multiple copies within the PV genome (60). The activation of transcription by viral E2 proteins involves the binding of the activation domain (AD) to transcription factors TFIIB and TBP and cofactors AMF1/Gps2 and p/CAF. All of these interactions seem to be important for activation by E2 (7, 9, 36, 44, 70). Furthermore, E2 interacts with p300/CBP (37, 49, 56). It was shown that the coexpression of p300 can potentiate activation by E2, indicating that binding to p300 may be a rate-limiting step for activation by E2 (49).
Here, we describe a yeast two-hybrid system that uses transcriptionally competent BPV1 E2 protein. This system allowed the identification of human NAP-1 (hNAP-1) as an interaction partner for E2. We present data demonstrating that a direct interaction between E2 and hNAP-1 is important for the activation of transcription by BPV1 E2. However, additional functions located within the N terminus of hNAP-1 are required. We show that hNAP-1 and E2 can bind simultaneously to p300 and that this stable ternary complex efficiently contributes to the activation of transcription. Furthermore, p53 also directly binds to hNAP-1. Our data suggest that the binding to NAP-1 may be an essential step for sequence-specific transcriptional activators that use p300 as a coactivator to activate transcription.
MATERIALS AND METHODS
Plasmid constructions.
All yeast plasmids were shuttle vectors, and cloning was performed with Escherichia coli strain XL1Blue. The yeast reporter construct expressing the lacZ gene under the control of the synthetic promoter containing four E2 binding sites in front of the TATA box of human PV (HPV) type 18 (HPV18) P105 was obtained in several steps. The SalI-BglII promoter fragment was isolated from construct p4E223T105 (24), ligated to an oligonucleotide encoding the junction between the promoter and the lacZ gene and followed by a BspEI restriction site, and cloned into the SalI-BspEI restriction site of pBR322. Subsequently, this promoter fragment was inserted into the XhoI-BspEI restriction sites of pΔUAS (34), resulting in the displacement of the CYC promoter by the 4E223P105 promoter. Finally, the HindIII-BamHI promoter fragment isolated from this construct was inserted into the HindIII-BamHI vector fragment of placZi (Clontech) to give rise to 4E223P105lacZi. 5E2-pHISmin was constructed by inserting oligonucleotides encoding E2 binding sites into the XbaI binding site of pHIS-i1 (Clontech). The yeast E2 expression vector pGBT9-E2 was obtained by removing the GAL4 DNA binding domain (DBD) from pGBT9 (Clontech) by digestion with BamHI and partial digestion with HindIII. The E2-encoding HindIII-BamHI fragment was isolated from pTZE2mHIII (34) and cloned into pGBT9. The HindIII-BamHI fragment derived from pTZE2Δ192- 282 (42) was inserted in a similar way to obtain pGBT9-E2Δ192- 282.
To express the AD of GAL4 fused to the DBD of E2, the StuI-BamHI fragment of E2 derived from pTZE2mHIII (42) was cloned into vector pGAD424 (Clontech). The full-length hNAP-1 clone was isolated from the keratinocyte cell line HaCaT cDNA library (expressing cDNA-encoded proteins; Clontech) by amplification with specific primers and cloned in frame with the hemagglutinin (HA) tag into vectors pXJ41 (68) and pcDNA3.1 (Invitrogen). Plasmids expressing a glutathione S-transferase (GST)- hNAP-1 fusion protein were obtained by cloning PCR products encoding full-length hNAP-1 into pGEX-5X-2 and pGEX-2T (Pharmacia Biotech) or deletion mutants into pGEX-2T. Some of the hNAP-1 deletion constructs were subsequently transferred into eukaryotic vector pXJ41 to express them fused to the HA tag. To express the E2 proteins of BPV1, HPV8, and HPV18 fused to a FLAG tag, the corresponding open reading frames were amplified by PCR, followed by cloning into vector pCMV2-FLAG (Kodak), as were BPV1 E2 lacking the AD and E2 expressing the AD. Expression vector pC59, expressing BPV1 E2 under the control of the simian virus 40 (SV40) early promoter, was described previously (69). Expression vectors for HPV8 E2 and HPV18 E2 and the HPV8 noncoding region-luciferase reporter construct also were described previously (49), as was the expression vector for p300 (13). The regulatory region of BPV1 from nucleotides 6958 to 475 (long control region [LCR]) was amplified by PCR and cloned into pALuc (12) to obtain BPV1 LCR-Luc. p53 expression vector pCp53wt was described by Nigro et al. (54), and the synthetic p53-responsive reporter construct was described by Funk et al. (17). The pG5-Luc reporter construct and the vector expressing the DBD of GAL4 (pM) were derived from a mammalian two-hybrid system (Clontech). PCR products encoding the AD of TEF-1 or HPV8 E2 were cloned into vector pM.
Yeast two-hybrid system.
Yeast maintenance, transformation, and storage were performed according to the instructions of the manufacturer of the yeast two-hybrid system (Clontech). Yeast strain YM954 (67) (genotype: MATa ura3-52 his3-200 ade2-101 lys2-801 leu2-3,112 trp1-901 gal4-542 gal80-538) was kindly provided by P. Bartel, Stony Brook, N.Y. β-Galactosidase (β-Gal) activity was determined by a filter lift assay and liquid β-Gal assays according to protocols from Clontech.
Cell culture and transfection.
RTS3b, a keratinocyte cell line established from an HPV-negative skin lesion from a renal transplant recipient (57), was cultivated in E medium (43). For transient transfection, 105 RTS3b cells were used to seed one six-well plate on the day before transfection. Transfection was performed with FuGene reagent according to the manufacturer's recommendations (Roche Diagnostics), and the cells were harvested 48 h later. Luciferase activities were determined as described previously (49). To generate a stable cell line expressing the luciferase reporter gene under the control of the BPV1 LCR integrated into the cellular genome, RTS3b cells were transfected with the BPV1 LCR-Luc construct. After 48 h, the cells were placed in medium containing 800 μg of G418/ml. After another 48 h, the medium was replaced with medium containing 400 μg of G418/ml; this medium was changed every second day for 2 to 3 weeks until resistant colonies had grown. The resistant colonies then were pooled. The presence of the intact BPV1 LCR-Luc cassette within the cells was confirmed by PCR (data not shown). At 24 h after transfection of this stable cell line with the E2 or hNAP-1 expression plasmids, the medium was replaced with medium supplemented with 400 μg of trichostatin A (TSA)/ml; incubation was continued for another 24 h before luciferase activities were determined as described above. To detect the effect of hNAP-1 on the activation of p21 expression by p53, RTS3b cells were transiently transfected with the corresponding expression vectors (see Fig. 6). A total of 90 μg of total cell extract was loaded onto a sodium dodecyl sulfate-15% polyacrylamide gel and analyzed for p21 by Western blotting with an antibody directed against p21 (Pharmingen).
FIG. 6.
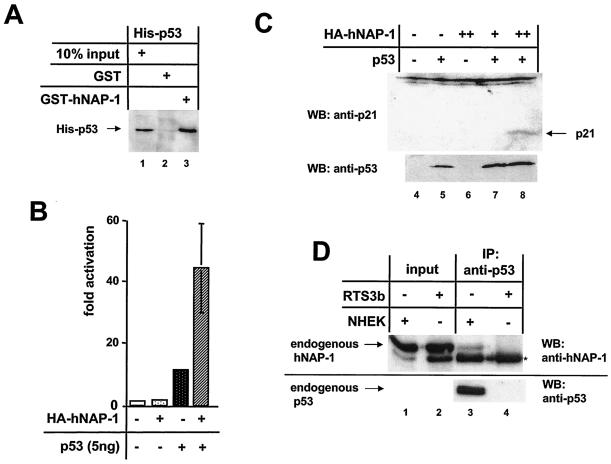
p53 interacts directly with hNAP-1. (A) The GST- hNAP-1 fusion protein or GST was incubated with His-tagged, bacterially expressed, purified p53. After the beads were washed with a buffer containing 200 mM KCl, bound p53 was revealed by an antibody directed against the His tag. (B) The p53-negative cell line RTS3b was cotransfected with a synthetic p53-responsive luciferase reporter construct, 5 ng of an expression vector for p53, and 400 ng of the vector for HA-tagged hNAP-1. The luciferase activity in the presence of the vector only was set at 1. Error bars are shown. (C) Extracts (90 μg) from RTS3b cells transiently transfected with an expression vector for p53 (lanes 5, 7, and 8) or for HA-tagged hNAP-1 in two different amounts (lanes 6 to 8) were used for Western blotting (WB) to detect endogenous p21. The blot was reprobed with an antibody against p53 to detect p53 protein levels. (D) Nuclear extracts from p53-negative RTS3b cells (lanes 2 and 4) and from neonatal human epidermal keratinocytes (NHEK) (lanes 1 and 3), which express wild-type p53, were subjected to immunoprecipitation (IP) with a p53 antibody. Bound endogenous hNAP-1 was analyzed by Western blotting with a monoclonal antibody against hNAP-1 (lanes 3 and 4). In lanes 1 and 2, one-sixth of the input of nuclear extracts was loaded to detect the level of expression of hNAP-1 in both types of cells. Lanes 3 and 4 were reprobed with the p53 antibody to confirm the pattern of expression of p53. A cross-reacting cellular protein is indicated by an asterisk.
Coimmunoprecipitation and pull-down assays.
GST pull-down assays were performed as previously described (49). To detect a direct protein-protein interaction, His-tagged proteins were expressed in bacteria, purified as described previously (25), incubated with immobilized GST- hNAP-1 fusion protein or GST, and further treated as described elsewhere (49). To detect an interaction between various E2 proteins and hNAP-1 in vivo, 293T cells were transfected with an expression vector for FLAG-tagged BPV1 E2 protein (see Fig. 2E) or with vectors for various FLAG-tagged E2 proteins (see Fig. 2D) and for HA-tagged hNAP-1. After 48 h, the cells were washed in ice-cold phosphate-buffered saline, followed by four freeze-thaw cycles in LSDB buffer (50 mM Tris-HCl [pH 7.9], 10% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol, 0.2% NP-40) containing 100 mM KCl. Cell debris was removed by centrifugation for 10 min at 4°C. The supernatants were incubated with 20 μl of anti-FLAG M2 affinity resin (Kodak). The samples were incubated at 4°C for 2 h with gentle mixing, followed by four washes in LSDB buffer with 200 mM KCl or 250 mM KCl and one wash in LSDB buffer with 100 mM KCl. The presence of hNAP-1 was analyzed by Western blotting with a monoclonal antibody directed against the HA epitope (Roche Diagnostics) or with a monoclonal antibody directed against hNAP-1 (kindly provided by Y. Ishimi) to detect an interaction between E2 and endogenous hNAP-1. E2 proteins were detected with anti-FLAG M5 antibody. To detect an interaction between endogenous p53 and endogenous hNAP-1 in vivo, 85 μg of nuclear extracts from normal neonatal human epidermal keratinocytes (purchased from Clonetics) and 85 μg of nuclear extracts from RTS3b cells were incubated with 5 μl of anti-p53-agarose conjugate (DO-1; Santa Cruz) at 4°C for 2 h with gentle mixing, followed by four washes in LSDB buffer with 200 mM KCl and one wash in LSDB buffer with 100 mM KCl. The presence of the endogenous proteins was analyzed by Western blotting with antibodies directed against hNAP-1 and p53 (DO-1).
FIG. 2.
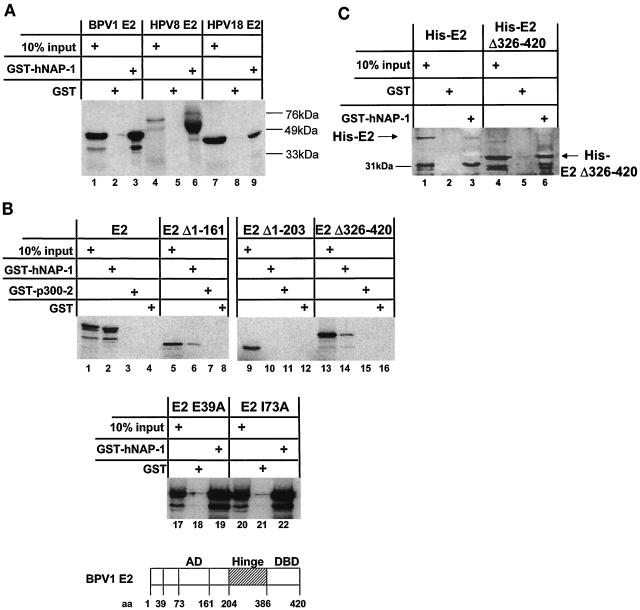
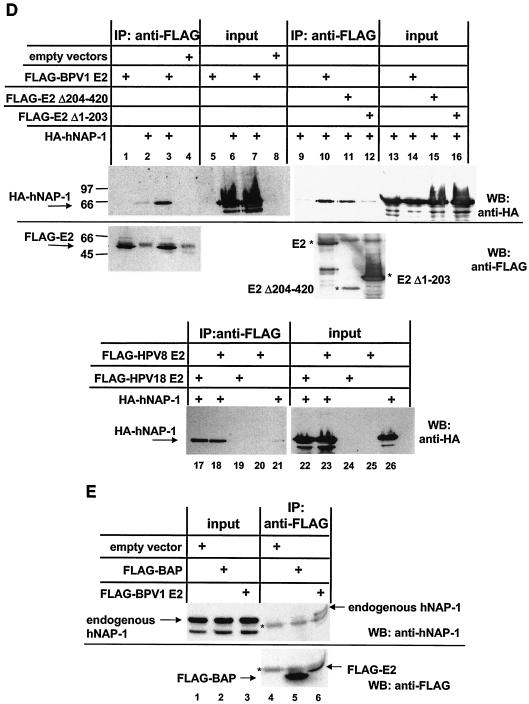
E2 proteins and hNAP-1 interact in vitro. (A) GST pull-down assays were carried out with purified GST, full-length hNAP-1 fused to GST (GST-hNAP-1), and 35S-labeled E2 proteins of BPV1, HPV8, and HPV18, obtained by in vitro translation with a rabbit reticulocyte lysate. Ten percent of the input is shown in lanes 1, 4, and 7. The positions of the marker proteins are indicated. (B) (Top panel) 35S-labeled full-length E2 of BPV1 (E2), two activation-deficient N-terminal deletion mutants (E2Δ1-161 and E2Δ1-203), and a C-terminal deletion mutant lacking the DBD (E2Δ326-420) were incubated with GST, GST-hNAP-1 or, as a negative control, GST-p300-2, expressing a fragment of p300 which was shown previously not to interact with E2 (49). Ten percent of the E2 protein and its derivatives used in one interaction assay is shown in the lanes labeled 10% input. (Middle panel) E2 proteins with point mutations in the AD, E2 E39A, which is replication deficient, and E2 I73A, which is impaired in transcription (14), were used in a GST pull-down assay. (Bottom panel) Structure of the E2 protein showing the positions of the amino acids (aa) in the various domains. (C) His-tagged, bacterially expressed, purified E2 protein of BPV1 (His-E2) or His-tagged E2Δ326-420 was incubated with purified GST or GST-hNAP-1. Bound E2 proteins were detected by Western blotting with an antibody directed against the His tag (Qiagen). The position of the 31-kDa marker protein is indicated. (D) (Upper panel) 293T cells were transfected with an expression vector expressing a FLAG-tagged full-length E2 protein of BPV1 (lanes 1, 3, 5, 7, 10, and 14), a FLAG-tagged BPV1 E2 mutant lacking the N-terminal AD (FLAG-E2Δ1-203; lanes 12 and 16), or FLAG-tagged AD (FLAG-E2Δ204-420; lanes 11 and 15) and an expression vector for HA-tagged hNAP-1 (lanes 2, 3, 6, 7, and 9 to 16). Cell extracts were incubated with the FLAG affinity gel, and bound hNAP-1 was detected with an antibody directed against the HA epitope (lanes 1 to 4 and lanes 9 to 12). In lanes 5 to 8 and lanes 13 to 16, 1/40 the input cellular extract was included, and the expression of HA-tagged hNAP-1 was analyzed with the HA antibody. The presence of FLAG-tagged E2 proteins was determined by reprobing of the blot shown in lanes 1 to 4 and lanes 10 to 12 with the FLAG M5 antibody. IP, immunoprecipitation; WB, Western blotting. (Lower panel) Western blot developed with the antibody directed against the HA epitope in a coimmunoprecipitation similar to that shown above but with cell extracts from 293T cells that had been transfected with an expression vector for FLAG-tagged HPV18 E2 (lanes 17, 19, 22, and 24) or HPV8 E2 (lanes 18, 20, 23, and 25) or an expression vector for HA-tagged hNAP-1 (lanes 17, 18, 21, 22, 23, and 26). (E) (Upper panel) Cell extracts of 293T cells that had been transfected with an expression vector for FLAG-tagged BPV1 E2 (lanes 3 and 6) or FLAG-tagged bacterial alkaline phosphatase (BAP) as a negative control (lanes 2 and 5) or with an empty vector (lanes 1 and 4) were incubated with the FLAG antibody coupled to Sepharose (lanes 4 to 6). Bound, endogenous hNAP-1 was detected by Western blotting with an antibody directed against hNAP-1. In lanes 1 to 3, 1/160 the cellular extract used for immunoprecipitation was loaded as an input control. (Lower panel) Part of the blot in the upper panel was reprobed with the FLAG antibody to detect the presence of FLAG-tagged proteins. A cellular protein cross-reacting with both antibodies is indicated by an asterisk.
Density gradient analysis of the ternary complex.
hNAP-1, E2 with a deletion of amino acids 326 to 420 (E2Δ326-420), and a fragment of p300 from amino acids 1195 to 1761 (p300 1195 to 1761) were expressed with a tag of six histidines in bacteria and purified. Totals of 800 ng of p300 1195 to 1761, 600 ng of hNAP-1, and 200 ng of E2Δ326-420 were incubated for 2 h in the presence of 25 mM HEPES-KOH (pH 7.9)-0.1 mM EDTA-12.5 mM KCl-10% glycerol-100 mM KCl in various combinations (see Fig. 5C). Next, the proteins were loaded onto a 2-ml 7.5 to 30% glycerol gradient prepared as described by Tanese et al. (61) and then centrifuged for 8 h at 50,000 rpm with a TL100 tabletop ultracentrifuge (Beckman) and a TLS-55 swinging-bucket rotor. Finally, 36 fractions of 60 μl each were carefully removed beginning from the top of the gradients. The presence of various proteins was analyzed by Western blotting.
FIG. 5.
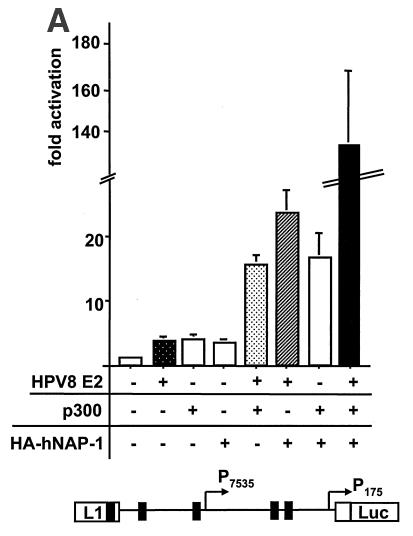
hNAP-1 and E2 can form a ternary complex with p300. (A) (Top panel) A reporter construct expressing the luciferase gene under the control of the regulatory region of HPV8 was cotransfected with expression vectors for HPV8 E2, p300, and HA-tagged hNAP-1, either alone or in combination. The luciferase activity in the presence of the empty expression vector was set at 1, and the fold activation of any of the proteins was calculated. The graph represents the averages of five independent experiments; error bars are shown. (Bottom panel) Structure of the regulatory region of HPV8, including the two promoters and the E2 binding sites (black boxes). (B) GST or GST-p300-4 was incubated with increasing amounts of His-tagged, bacterially expressed, purified hNAP-1 for 2 h at 4°C. After several washes to remove unbound His-tagged hNAP-1, incubation with 35S-labeled BPV1 E2 or hNAP-1 for another 2 h followed. The binding of radiolabeled E2 or hNAP-1 was analyzed by autoradiography. The radioactive signals were quantified with a PhosphorImager; the percentages bound to GST-p300-4 were calculated. Ten percent of the input of 35S-labeled E2 or 20% of the input of 35S-labeled hNAP-1 was loaded as an input control. (Bottom panel) The Western blot (WB) reveals the binding of hNAP-1 to GST-p300-4 in the absence (lanes 22 and 23) or presence (lanes 26 and 27) of 35S-labeled BPV1 E2. (C) Glycerol density sedimentation analysis performed as described previously (61) with 600 ng of His-tagged hNAP-1, 200 ng of His-tagged E2Δ326-420 (E2), and 800 ng of p300 1195 to 1761. Samples were subjected to 7.5 to 30% glycerol gradient centrifugation, and a total of 36 fractions were collected, starting from the top of the gradient. Since an initial analysis revealed that all proteins fractionated in fractions 10 to 31, only these fractions are shown here after analysis by Western blotting with an antibody against the His tag. The presence of E2, hNAP-1, and p300 in the various fractions is indicated on the right.
RESULTS
Identification of cellular factors binding to PV E2 proteins by the yeast two-hybrid system.
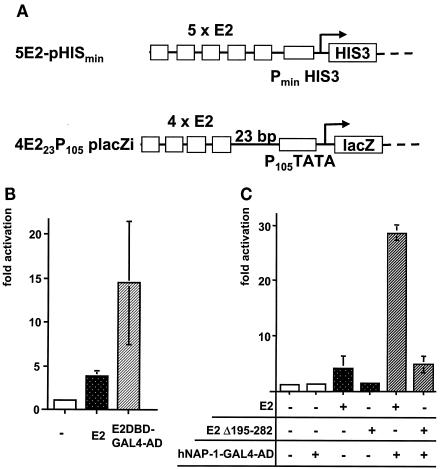
To identify cellular factors which may play a role in E2-mediated activation of transcription, we set up a yeast two-hybrid system. Since we wished to screen with wild-type E2 protein, which is a strong activator of transcription in yeast cells (34), we modified the classical yeast two-hybrid system. Previously, Ham et al. showed that in mammalian cells, E2 could not efficiently activate the transcription of a minimal promoter composed of E2 binding sites and a TATA box only but was an efficient activator of more complex promoters (24). In contrast, most cellular sequence-specific activator proteins, among them GAL4, are able to stimulate such a minimal promoter (66). To test whether E2 would be unable to activate transcription from this minimal promoter in yeast cells, we inserted a fragment expressing such a minimal promoter into vector placZi (Fig. 1). E2 yielded a three- to fourfold activation of the expression of the lacZ gene from reporter construct 4E223P105placZi integrated into the cellular genome. A fusion protein composed of the DBD of E2 and the AD of GAL4 led to a 14-fold activation (Fig. 1B), indicating that the mode of activation mediated by the two ADs is distinct in yeast cells also. Furthermore, E2 only weakly activated the HISmin promoter located behind five E2 binding sites and controlling the expression of the HIS gene in plasmid 5E2-pHISmin (Fig. 1A). Therefore, plasmid 5E2-pHISmin was used to select for yeast growth in the presence of 100 to 300 mM 3-amino-1,2,4-triazole, an inhibitor of the HIS gene product. It was necessary to include this additional selection to suppress yeast cell growth mediated by the weak activation of pHISmin by E2.
FIG. 1.
Identification of hNAP-1 as an interaction partner for the PV E2 protein. (A) Schematic representation of two reporter constructs. 5E2-pHISmin expresses the yeast HIS3 gene under the control of its minimal promoter, pHISmin, and five E2 binding sites. 4E223P105placZi expresses the lacZ gene under the control of a synthetic promoter composed of the TATA box of the early promoter of HPV18 (P105) and four E2 binding sites located 23 bp further upstream. For the yeast two-hybrid system, both reporter constructs were integrated into the cellular genome of YM954. (B) Yeast expression vector pGBT9-E2, expressing the E2 protein of BPV1 (E2), or pGBT9-E2DBD-GAL4AD (E2DBD-GAL4-AD), expressing the AD of GAL4 fused to the DBD of BPV1 E2, or the vector alone (−) was transformed into the yeast strain harboring the two reporter constructs shown in panel A. β-Gal activities of the respective strains were determined. The activity in the presence of the vector alone (−) was assigned an arbitrary value of 1, and the fold activation was calculated. The columns represent the averages of three independent experiments performed with two different clones in each case; error bars are shown. (C) β-Gal activities of parental yeast strain YM954 transformed with a yeast expression vector for E2 or E2Δ195-282 (an E2 protein lacking the internal hinge region) and with plasmid pACT-c-ΔNAP-1, expressing hNAP-1 fused to the AD of GAL4, were determined. The latter was isolated by the yeast two-hybrid screen. The fold activation in relation to that of the strain transformed with the expression vector was calculated as described for panel B. The columns represent the averages for three independent clones; error bars are shown.
A cDNA library derived from keratinocyte cell line HaCaT and allowing the expression of cDNA-encoded proteins fused to the GAL4 AD was transformed into the yeast strain harboring the two reporter genes in addition to expression vector pGBT9-E2. Out of 107 yeast clones, the two reporter genes were activated significantly in 41 clones. The cDNA-encoded proteins of two independent clones were identical to the 300 C-terminal amino acids of hNAP-1, which has 391 amino acids in total. The interaction between hNAP-1 and E2 could be confirmed with yeast strains which were retransformed with the corresponding plasmids (Fig. 1C). The interaction involved the N-terminal AD of BPV1 E2, since the C-terminal 94 amino acids of BPV1 E2, containing the DBD, did not reveal increased β-Gal activity in the presence of hNAP-1 (data not shown). However, E2Δ195-282, which lacks the internal hinge region, retained similar cooperativity with hNAP-1 (Fig. 1C). The reduced absolute levels of activation of this deletion mutant correlated with reduced protein levels (data not shown).
E2 proteins bind to hNAP-1 in vitro and in vivo.
To confirm an interaction between E2 and h-NAP-1 in vitro, the hNAP-1 fragment isolated by the yeast two-hybrid system and encoding hNAP-1 lacking the N-terminal 91 amino acids was inserted into vector pGEX-5X-2. A GST-hNAP-1 fusion protein retained the full-length BPV1 E2 protein obtained by in vitro translation with a rabbit reticulocyte lysate (Fig. 2A). The E2 proteins of different PV types are rather conserved within the N-terminal AD and the C-terminal DBD, in contrast to the variable hinge region, which was suggested to function as a linker between the AD and the DBD of E2 (20, 42). In vitro-translated E2 proteins of HPV8 and HPV18 bound to GST- hNAP-1 (Fig. 2A), indicating that the interaction with hNAP-1 may be conserved among E2 proteins of different PV types.
In correlation with the data obtained with yeast cells, the AD of BPV1 E2 is required for binding to hNAP-1 in vitro. E2Δ1-161, which lacks the N-terminal 161 amino acids and which is a naturally occurring N-terminally truncated mutant that represses activation by the full-length E2 protein (35), still interacted with hNAP-1; in contrast, E2Δ1-203, which lacks the entire AD, did not bind at all. An E2 protein which lacks the C-terminal DBD, E2Δ386-420, retained binding, as shown in Fig. 2B. The various E2 proteins average 30% amino acid identity within the AD. Many of the conserved residues throughout the AD are important for both replication and transcription. However, two amino acid substitutions clearly separated these two capacities. Changing Ile at position 73 to Ala (I73A) destroyed transcriptional activation while leaving replication function intact, whereas replacing Glu at position 39 with Ala (E39A) had the inverse phenotype (14). Neither amino acid change affected the binding of BPV1 E2 to GST-hNAP-1, as shown in Fig. 2B.
In order to exclude the possibility that the interaction between BPV1 E2 and hNAP-1 is mediated by a factor present within the reticulocyte lysate or in yeast cells, we incubated GST-hNAP-1 with His-tagged, bacterially expressed, purified full-length BPV1 E2 or E2Δ386-420. As shown in Fig. 2C, both E2 and E2Δ386-420 were specifically retained by GST-hNAP-1, indicating that the interaction is direct. In this experiment, the binding of full-length E2 was weaker than that of E2Δ386-420. This result may have been due to the lower concentration of full-length E2. We were not able to purify full-length E2 in the same amounts as E2Δ386-420. Furthermore, a faster-migrating product, which appears to be an N-terminal proteolytic form, since it retains the His tag, showed a stronger interaction with hNAP-1, supporting the notion that the interaction is mediated by the AD (Fig. 2C).
E2 and hNAP-1 also interact within the cell, as shown in Fig. 2D. 293T cells were transiently transfected with expression vectors for FLAG-tagged full-length BPV1 E2, an E2 protein that lacks the N-terminal AD (FLAG-E2Δ1-203), or the AD of BPV1 E2 (FLAG-E2Δ204-420) and an expression vector for HA-tagged full-length hNAP-1. Whereas in the presence of full-length BPV1 E2 or its AD, HA-tagged hNAP-1 could be coprecipitated by the FLAG antibody, E2Δ1-203 did not mediate this effect, confirming that the interaction within the cell is mediated by the AD of BPV1 E2 as well. The E2 proteins of HPV8 and HPV18 also were able to coprecipitate HA-tagged hNAP-1 (Fig. 2D).
Furthermore, E2 binds to endogenous hNAP-1. In all cervical carcinoma cell lines infected with PV, the HPV DNA is integrated into the cellular genome, resulting in the disruption of the open reading frame for E2. Therefore, cell lines expressing E2 at physiological levels during HPV infection do not exist, and to analyze the binding of E2 to endogenous hNAP-1, we overexpressed FLAG-tagged E2 in 293T cells. As shown in Fig. 2E, the FLAG antibody was able to precipitate endogenous hNAP-1 from extracts of cells transfected with the vector for FLAG-tagged BPV1 E2 and not from extracts of cells transfected with the empty expression vector or with a vector expressing a control protein, bacterial alkaline phosphatase fused to the FLAG epitope.
Taken together, the data shown in Fig. 1 and 2 demonstrate that E2 proteins from different PV types interact with hNAP-1. For BPV1 E2, we can demonstrate that this interaction is direct and involves the AD.
E2 and hNAP-1 cooperate in the activation of BPV1 gene expression.
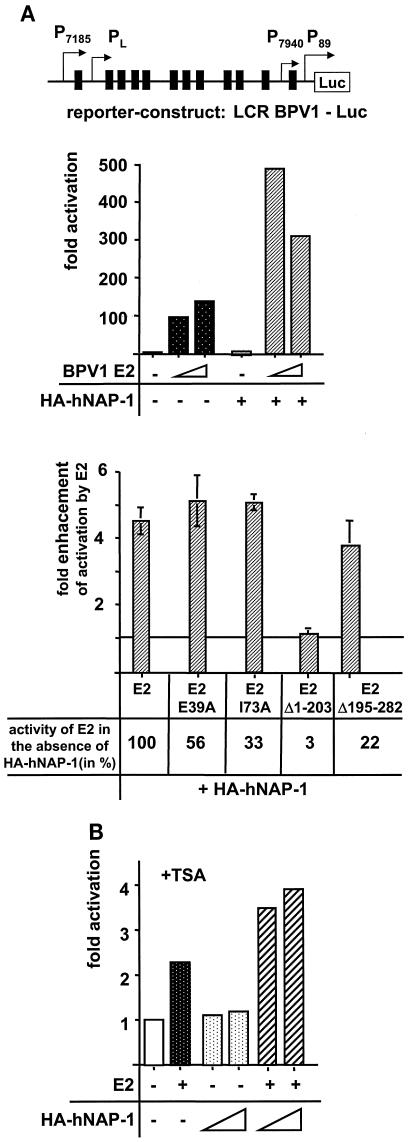
As already mentioned, NAP-1 and NAP-2 act as histone chaperones to which functions in both transcription and DNA replication have been ascribed (4, 26-28, 39, 50, 58). Here, we focused on an involvement of the interaction of E2 with hNAP-1, observed in vitro, in the activation of transcription. In BPV1, E2 strongly activates transcription from several BPV1 promoters by binding to the 12 E2 binding sites located within the regulatory region called the LCR (60). A reporter construct containing the entire LCR of BPV1 in front of the luciferase gene was cotransfected into human cell line RTS3b (57) derived from human keratinocytes, the natural target cells of PV, together with vectors for HA-tagged hNAP-1 and BPV1 E2. As shown in Fig. 3A, E2 could activate 100- to 130-fold, depending on the amount of expression vector transfected. The expression of hNAP-1 on its own stimulated promoter activity 2-fold; however, the coexpression of hNAP-1 and small amounts of E2 increased luciferase activity 500-fold. This cooperativity between hNAP-1 and BPV1 E2 was reduced in the presence of larger amounts of E2. However, at these high E2 concentrations, cooperativity between E2 and hNAP-1 was still observed.
FIG. 3.
E2 and hNAP-1 cooperate in the activation of gene expression. (A) (Top panel) RTS3b cells, immortalized skin keratinocyte cells (57), were transfected with a luciferase reporter construct containing the regulatory region of BPV1 called the LCR, the structure of which is shown. The different promoters are indicated, and the 12 E2 binding sites are depicted as black boxes. (Middle panel) Either 5 or 20 ng of expression vector for BPV1 E2, under the control of the SV40 promoter, was cotransfected together with 400 ng of expression vector for HA-tagged hNAP-1. The graph shows the results of one representative experiment. (Bottom panel) Transient transfection experiments with the BPV1 LCR-Luc reporter plasmid, an expression vector for HA-tagged hNAP-1, and expression vectors for full-length E2 (E2), E2 E39A (the replication-deficient mutant), E2 I73A (the mutant impaired in transcription), E2Δ1-203 (lacking the AD), and E2Δ195- 282 (lacking the internal hinge region). The activity of each of the E2 proteins in the absence of coexpressed HA-tagged hNAP-1 was arbitrary defined as 1. The fold enhancement of E2-mediated activation by hNAP-1 was calculated. The graph represents the averages of at least three independent experiments; error bars are shown. The activation of each E2 mutant protein in the absence of coexpressed hNAP-1, compared to that of the wild-type protein, which was set at 100%, is given below the graph. (B) RTS3b cells, containing the BPV1 LCR-Luc reporter construct integrated into the cellular genome, were cotransfected with expression vectors for HA-tagged hNAP-1 and for E2. At 24 h after transfection, TSA was added to the medium for another 24 h.
As shown in Fig. 2B, point mutations abolishing the replication or transcription function of E2 did not affect binding to hNAP-1 in vitro. In our experiments, the activation-deficient E2 I73A mutant retained 33% the activation capacity of wild-type E2 (data not shown), a finding which is in good agreement with previous findings (14). However, even this residual activation was stimulated about fivefold after cotransfection with hNAP-1, indicating that the step involving binding to hNAP-1 is not affected by this mutant. The same is true for the replication-deficient E2 E39A mutant (Fig. 3A). As expected, the activation of transcription by E2 is necessary for the effect of hNAP-1, since a mutant lacking the AD, E2Δ1-203, was not stimulated by hNAP-1 coexpression, in contrast to the mutant lacking the internal hinge region, E2Δ195-282 (Fig. 3A). The activity of the SV40 early promoter, which had driven the expression of BPV1 E2 in the previous transient transfection experiments, was not stimulated by coexpression of the amounts of E2 and hNAP-1 used here (data not shown). This result excludes the possibility that the enhancing effect is due to elevation of the expression of E2 by hNAP-1. These results demonstrate that E2 and hNAP-1 cooperate in the activation of transcription, which correlates with a direct interaction of the two proteins.
Because the role of NAP-1 as a histone chaperone is known, it is possible that hNAP-1, in cooperation with E2, also affects the state of chromatin during activation by E2. Until now, we have used an E2-responsive reporter construct, which has been transiently transfected. In order to test the effect of hNAP-1 on activation by E2 on reporter constructs organized into a higher-order chromatin structure, we established a cell line harboring reporter construct BPV1 LCR-Luc integrated into the cellular genome. After transiently transfecting this stable cell line with the expression vector for E2, we were not able to observe any activation, independent of whether the hNAP-1 vector was cotransfected or not (data not shown). Since we suspected that the tight packing of chromatin may inhibit the access of E2 to the DNA, we treated the cells at 24 h after transfection with TSA, an inhibitor of histone deacetylases. This treatment enabled E2 to stimulate luciferase activity weakly; this effect was slightly enhanced by the coexpression of hNAP-1 (Fig. 3B). These results demonstrate that with promoters organized in cellular chromatin, hNAP-1 also stimulates activation by E2, although to a much lesser extent than transiently transfected reporter constructs.
Full-length hNAP-1 is required for cooperation with E2.
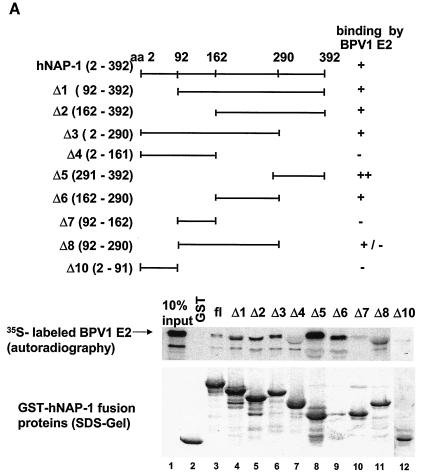
In order to gain insights into the mechanism of the cooperativity between hNAP-1 and E2 proteins, we determined the domains within hNAP-1 which are bound by E2 and analyzed their roles in the enhancement of activation by E2. A series of N- and C-terminal deletion mutants of hNAP-1 were expressed as GST fusion proteins. Although the N-terminal region from residues 1 to 162 possessed marginal binding activity (GST-hNAP-1Δ4, GST-hNAP-1Δ7, and GST-hNAP-1Δ10), an internal domain from residues 162 to 290 (GST-hNAP-1Δ6) and the C-terminal region from residues 291 to 392 (GST- NAP-1Δ5) bound with greater efficiency to 35S-labeled BPV1 E2 (Fig. 4A). These results suggest that E2 binds to at least two separable domains within hNAP-1, one internal from amino acids 162 to 290 and one C terminal from residues 291 to 392.
FIG. 4.
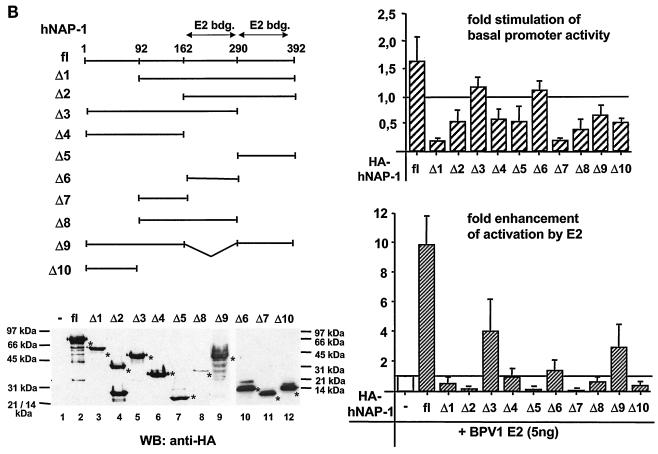
The binding of E2 to hNAP-1 is necessary but not sufficient for the stimulation of E2-mediated activation. (A) Different regions of hNAP-1 were fused to GST. Bacterially expressed, purified GST- hNAP-1 fusion proteins (shown in the sodium dodecyl sulfate [SDS] gel at the bottom, stained by Coomassie blue) were incubated with 35S-labeled BPV1 E2. Bound proteins were detected by autoradiography. The binding of E2 to hNAP-1 is summarized (−, no binding; +/−, weak binding; +, binding; ++, strong binding). In some cases, the presence of GST- hNAP-1 fusion proteins in the gel might have affected the migration of the E2 protein slightly. aa, amino acids; fl, full length. (B) (Upper left panel) RTS3b cells were cotransfected with the BPV1 LCR reporter construct and an expression vector for HA-tagged hNAP-1 or HA-tagged deletion mutants of hNAP-1, either alone or together with 5 ng of the E2 expression vector. (Upper right panel) Effects of the various hNAP-1 mutants on BPV1 promoter activity in the absence of E2. The activity of the BPV1 LCR in the absence of hNAP-1 was arbitrarily defined as 1. (Lower right panel) Effects of the hNAP-1 mutants on activation by E2. Here, the activity in the presence of E2 without hNAP-1 (lane −) was set at 1, and the effects of the various hNAP-1 mutants were calculated. Both graphs represent the averages of at least three independent experiments with two different plasmid DNA preparations; error bars are shown. (Lower left panel) Levels of expression of the various HA-tagged hNAP-1 deletion mutants analyzed by Western blotting (WB) with extracts of cells that had been transiently transfected with the corresponding expression vectors and a high-affinity antibody against the HA epitope. Asterisks indicate the positions of the respective hNAP-1 mutants. Lanes 1 to 9, 10% polyacrylamide gel; lanes 10 to 12, 15% polyacrylamide gel.
To analyze the contributions of the domains of hNAP-1 that interact with E2 to the stimulation of activation by E2, hNAP-1 deletion mutants (Fig. 4B) were fused at the N terminus to an HA tag and expressed. The results obtained in the cotransfection experiments shown in Fig. 4B demonstrate that the N-terminal 91 residues are indispensable for hNAP-1 to enhance activation by E2. All hNAP-1 mutants lacking the N-terminal 91 amino acids failed to cooperate with E2 in activation. Most of the mutants repressed activation by E2 from 2-fold (hNAP-1Δ8) to 10-fold (hNAP-1Δ1, hNAP-1Δ2, hNAP-1Δ5, and hNAP-1Δ7). This repression is not related to squelching due to unphysiologically high concentrations of these mutants compared to that of full-length hNAP-1 in transfected cells, since all hNAP-1 derivatives were present in similar amounts. The sole exception is mutant hNAP-1Δ8, which was produced at reduced levels, as shown in the Western blot in Fig. 4B. Furthermore, titration of these proteins revealed constant, dose-dependent repression (data not shown). Mutant hNAP-1Δ4, retaining the N-terminal domain but lacking an interaction with E2, had no effect on activation by E2. Mutants hNAP-1Δ3 and hNAP-1Δ9, containing the N-terminal domain and one E2 binding motif, respectively, were able to enhance activation by E2, although to a reduced extent compared to that obtained with wild-type hNAP-1. These results indicate that in addition to the N-terminal 91 amino acids, at least one E2 binding motif is required for hNAP-1 to stimulate activation by E2, although it seems that amino acids 162 to 290 of hNAP-1 are more important.
The various hNAP-1 mutants also modulated the activity of the BPV1 promoters in the absence of E2. However, there was no clear correlation between the effects of each mutant on activation by E2 and on basal promoter activity. For example, hNAP-1Δ5 and hNAP-1Δ9 both slightly reduced basal promoter activity, but hNAP-1Δ5 strongly repressed activation by E2 and hNAP-1Δ9 still weakly stimulated activation by E2. These results suggest that the effects of hNAP-1 may be specific for E2. Moreover, these data imply that hNAP-1 has another function in transcription, since some of the mutants, e.g., hNAP-1Δ7 and hNAP-1Δ10, which do not interact with E2 in vitro, strongly inhibited activation by E2; the latter result may be related to their inhibitory effects on promoter activity per se.
E2, hNAP-1, and p300 can form a ternary complex in vitro.
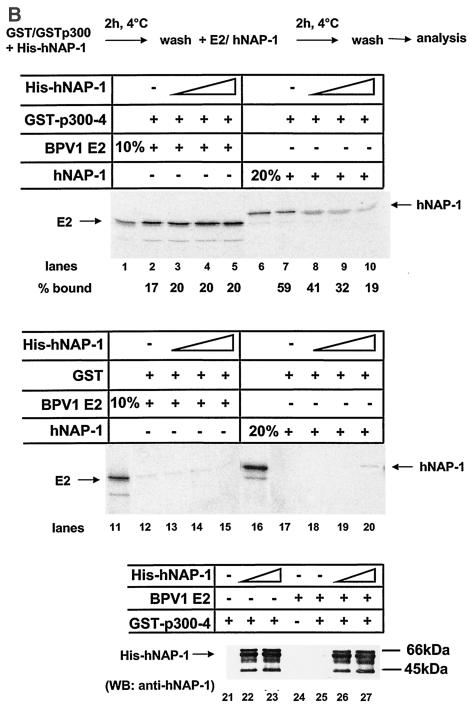
As already mentioned, NAP-1 and NAP-2 have been shown to interact with p300. Like that of E2, the binding of p300 to hNAP-1 may involve at least two domains, one from amino acids 123 to 230 and a second one C terminal to amino acid 290 (4, 58). Thus, the regions in hNAP-1 required for activation by E2 also include the p300 interaction domains, and we cannot rule out the possibility that the binding of hNAP-1 to p300 may be necessary as well. Since it has been demonstrated that E2 proteins of HPV8, HPV18, and BPV1 directly and functionally interact with p300 (37, 49, 56), as does hNAP-1, we were interested in the effects of hNAP-1 and p300 on E2-mediated activation. Unfortunately, coexpression of p300 with reporter construct BPV1 LCR-Luc, used in the experiments described so far, led to the inhibition of promoter activity (data not shown). However, Müller et al. showed previously that HPV8 E2 and coexpressed p300 cooperated in the activation of HPV8 gene expression (49). Since we demonstrated here that in addition to BPV1 E2, HPV8 E2 also bound to hNAP-1 (Fig. 2A and D) and coexpressed hNAP-1 enhanced activation by HPV8 E2 (data not shown; see Fig. 7), we used the HPV8 system to study the effects of hNAP-1 and p300 on E2-mediated activation. In agreement with previous observations, E2 and p300 cooperated, since they stimulated promoter activity 15.5-fold, compared to 4-fold by each of the proteins alone (Fig. 5A). As shown previously, the overexpression of p300 does not affect the expression of the E2 protein (49). hNAP-1 increased activation by either E2 or p300 by up to 23- or 17-fold, respectively. Thus, hNAP-1 can functionally interact with both proteins, in correlation with the direct binding of hNAP-1 to E2 (Fig. 2) and p300 (4, 58). The overexpression of all three proteins together resulted in 134-fold activation. This activation in the presence of E2, hNAP-1, and p300 was far beyond the sum of the effects of the single components, suggesting that a ternary complex of E2, p300, and hNAP-1 likely contributes very efficiently to the activation of transcription.
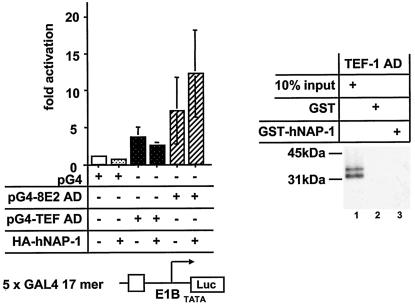
FIG. 7.
TEF-1 does not interact with hNAP-1. (Left panel) An expression vector for the DBD of GAL4 fused to either the AD of TEF-1 (pG4-TEF AD) (68) or the AD of HPV8 E2 (pG4-8E2 AD) was cotransfected either alone or together with the expression vector for HA-tagged hNAP-1 and the GAL4-responsive reporter construct, which is shown schematically below the graph. The data represent the averages of four experiments, and error bars indicate standard deviations. (Right panel) 35S-labeled TEF-1 AD was incubated with GST or GST-hNAP-1. After the samples were washed with a buffer containing 100 mM KCl, the reaction was analyzed by autoradiography. Ten percent of the input is shown in lane 1.
To analyze possible ternary complex formation among the three proteins in vitro, we used a fragment of p300 from amino acids 1453 to 1882 fused to GST (GST-p300-4). Müller et al. observed previously that E2 binds to this segment of p300, which colocalizes with the C/H3 domain (49). In addition to the KIX domain (4), NAP-2 and NAP-1 were shown to require amino acids 1572 to 1818 for binding to p300 (58), like E2. To confirm the formation of a ternary complex in vitro, we performed a competition experiment. As shown in Fig. 5B, GST or the GST-p300-4 fusion protein was incubated with increasing amounts of bacterially expressed, purified His-tagged hNAP-1 for 2 h. After several washing steps to remove unbound His-tagged hNAP-1, either 35S-labeled E2 or hNAP-1, each obtained by in vitro translation, was added; after incubation for another 2 h, binding was analyzed. The signals were quantified with a PhosphorImager. While in the absence of unlabeled His-tagged hNAP-1 59% of radiolabeled hNAP-1 was precipitated by GST-p300-4, preincubation of increasing amounts of unlabeled hNAP-1 reduced the binding of labeled hNAP-1 to 19% (Fig. 5B, lanes 6 to 10). This competition is consistent with the notion that hNAP-1 binding sites within p300-4 have been occupied by the nonlabeled protein. In contrast, preincubation of GST-p300-4 with the same increasing amounts of His-tagged hNAP-1 did not affect the binding of radiolabeled E2. A total of 17 to 20% of the input E2 proteins were precipitated by GST-p300-4, regardless of preincubation with increasing amounts of His-tagged hNAP-1 (Fig. 5B, lanes 1 to 5). As expected, purified His-tagged hNAP-1 also was bound by GST-p300-4 in the presence of E2, as revealed by Western blotting with a monoclonal antibody against hNAP-1 (Fig. 5B, lanes 21 to 27). These results suggest that E2 can still interact with p300 when hNAP-1 is bound and that a ternary complex among E2, p300, and hNAP-1 is likely.
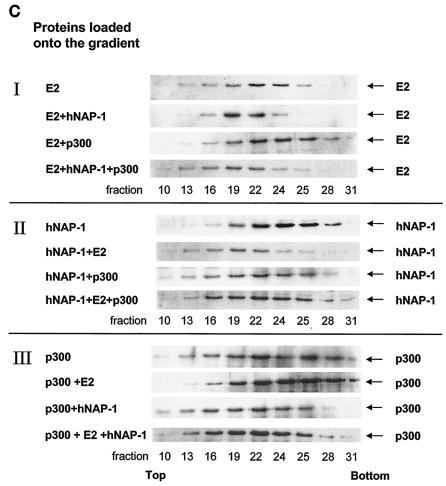
To further address the formation of a ternary complex among E2, p300, and hNAP-1 in vitro, we investigated the fractionation of the E2 protein in the presence of hNAP-1 and p300 in a density gradient. After incubation of His-tagged, bacterially expressed, purified E2Δ326-420 with His-tagged hNAP-1 and p300 1195 to 1761, the binding reaction was subjected to glycerol gradient centrifugation as described by Tanese (61). A total of 36 fractions were collected, and the presence of E2Δ326-420, hNAP-1, and p300 1195 to 1761 was monitored by Western blotting. Since all three proteins easily were distinguishable due to their characteristic sizes, we used an antibody directed against the His tag. To ensure that most of E2 was complexed by p300 and hNAP-1, we added E2 in limiting amounts to the binding reaction. An initial analysis revealed that all three proteins sedimented near the middle of the gradient within fractions 10 to 31. Therefore, only these fractions were used for the detailed analysis shown in Fig. 5C. Most of E2 was present in fractions 22 to 24. After incubation with hNAP-1, E2 sedimented more toward the top of the gradient, with the highest concentrations in fractions 19 to 22 (Fig. 5C, panel I, E2 vs. E2+hNAP-1). Thus, the interaction between E2 and hNAP-1 can be demonstrated in this glycerol gradient analysis, as indicated by the observation that hNAP-1 cofractionated (Fig. 5C, panel II, hNAP-1 vs. hNAP-1+E2). In contrast to the addition of hNAP-1, the addition of p300 to E2 had the consequence that E2 sedimented more toward the bottom of the gradient (Fig. 5C, panel I, E2+p300). In addition, E2 cofractionated with p300, which was detectable in higher fractions after incubation with E2, compared to free p300, confirming the interaction between E2 and p300 (Fig. 5C, compare panel I, E2+p300, with panel III, p300+E2). In the presence all three proteins, E2 again was shifted toward the top of the gradient (Fig. 5C, panel I, E2+hNAP-1+p300). However, the E2 pattern differed from that obtained after incubation with hNAP-1 alone, since now a larger portion of E2 was detectable in fractions 13 to 16, accompanied by a reduction in fractions 24 and 25. Significant amounts of p300 and hNAP-1 also were found in fractions 13 to 22 when all three proteins were incubated (Fig. 5C, panels II and III, all three proteins). Thus, the cofractionation of E2, hNAP-1, and p300 and the change in the sedimentation of E2 after simultaneous incubation with both hNAP-1 and p300 correlate with ternary complex formation among E2, hNAP-1, and p300.
Together, transient transfection, GST pull-down, and density gradient sedimentation experiments suggested that multiple protein-protein interactions (E2-p300, E2-hNAP-1, and hNAP-1-p300) may contribute to the formation of a stable ternary complex efficient in the activation of transcription.
A direct interaction between p53 and hNAP-1 also is involved in activation mediated by p53.
Previously, it was suggested that NAP-2, which is closely related to NAP-1, augments transcription by activators such as p53 and E2F, which use p300 as a coactivator, through a direct interaction with p300 (58). In order to test whether, in addition to p300- hNAP-1 binding, a direct interaction between p53 and hNAP-1 (like that for PV E2) may be involved, we incubated His-tagged, bacterially expressed, purified p53 with a GST-hNAP-1 fusion protein. As shown in Fig. 6A, p53 was precipitated by the GST-hNAP-1 fusion protein and not by GST. The interaction could be confirmed with GST-p53 and His-tagged, purified hNAP-1 (data not shown). These results demonstrate that the interaction between hNAP-1 and p53 is direct and not merely mediated by other factors, such as p300, as has been suggested elsewhere (58). When a synthetic reporter construct containing one p53 binding site upstream of the Hsp70 promoter was transiently transfected into the p53-negative cell line RTS3b, hNAP-1 also potentiated activation by p53 (Fig. 6B), confirming that the interaction is functional, as has been shown for NAP-2 (58). On the basis of these results, we suspected that hNAP-1 is also able to stimulate the activation by p53 of an endogenous p53-responsive promoter in the absence of the coexpression of p300. We analyzed the effect of the coexpression of p53 and hNAP-1 on the p21 level in cell line RTS3b. Under the Western blotting conditions shown in Fig. 6C, endogenous p21 was not visible. Cotransfection of the p53 expression vector or of small amounts of the hNAP-1 expression vector did not induce p21 expression to detectable levels. However, the coexpression of p53 and large amounts of hNAP-1 led to the appearance of the p21 protein. In addition, endogenous p53 and hNAP-1 can form a stable complex, as shown in Fig. 6D. Extracts from RTS3b cells, which do not express a detectable level of the p53 protein, and from primary neonatal human epidermal keratinocytes, which express high levels of p53 (Fig. 6D, lower panel), were subjected to immunoprecipitation with a monoclonal antibody against p53. Only in neonatal human epidermal keratinocytes could endogenous hNAP-1 be coprecipitated. The results shown in Fig. 6 imply that p53 and hNAP-1 interact directly with each other.
In order to test whether a functional interaction with hNAP-1 is a general strategy for activators or is restricted to activators interacting with p300, we used the AD of TEF-1. Various investigators have shown that TEF-1 neither binds to p300 in vitro nor cooperates with coexpressed p300 (49, 59). As shown in Fig. 7, the AD of TEF-1 failed to interact with GST-hNAP-1 in vitro. The TEF-1 AD fused to the DBD of GAL4 stimulated transcription from a reporter containing GAL4 binding sites about fourfold; this effect was not enhanced after the coexpression of hNAP-1. However, as a control, activation by a hybrid protein composed of the AD of HPV8 E2 and the DBD of GAL4 was further stimulated by coexpressed hNAP-1.
DISCUSSION
There is growing evidence demonstrating that hNAP-1 plays important roles during the activation of transcription. NAP-1 and other histone chaperones, such as ASF1 and chromatin assembly factor 1, have been shown to cooperate with ATP-dependent chromatin remodeling complexes (4, 28, 48, 50). Furthermore, NAP-1 and NAP-2 are part of the p300/CBP coactivator complex (4, 27, 58). Thus, it has been suggested that NAP-1 may serve as a point of interaction between transcriptional coactivators and chromatin. We identified hNAP-1 as a target of the AD of PV transcription factor E2 by a yeast two-hybrid screen. In contrast to a previous screen in which a transactivation-defective point mutant of E2 was applied (9), we were able to use wild-type E2 protein as bait, since our reporter construct was only weakly activated by E2 in comparison to the GAL4 AD. We were able to confirm the interaction between E2 and hNAP-1 by GST pull-down assays and coimmunoprecipitation experiments. Thus, using three different methods and proteins synthesized in different ways, i.e., in yeast, bacterial, and eukaryotic cells, we were able to demonstrate an interaction between E2 and hNAP-1. Moreover, E2 also precipitates endogenous hNAP-1. Therefore, it is unlikely that the interaction is artifactual as a result of protein misfolding, but is indeed specific.
In addition to the E2 protein of BPV1, the E2 proteins of two HPV types, HPV18 (infecting the genital mucosa and associated with cervical cancer) and HPV8 (infecting the skin), also contact hNAP-1 in vitro and in vivo, indicating that the binding to hNAP-1 may be common to E2 proteins. The interaction between BPV1 E2 and hNAP-1 is direct, since bacterially expressed purified proteins also coprecipitate. Furthermore, we show that another transcriptional activator, p53, also directly binds to hNAP-1. This finding demonstrates that the interaction does not have to be mediated by a bridging factor. p300 may be such a factor, since hNAP-1 interacts with p300, as do PV E2 and p53 (4, 5, 23, 49, 56, 58).
The binding of BPV1 E2 to hNAP-1 is mediated by its AD, as revealed by the yeast two-hybrid system, GST pull-down experiments, and coimmunoprecipitation. The concept of an interaction between E2 and hNAP-1 is reinforced by the ability of hNAP-1 overexpression to stimulate activation by E2. Even in the presence of saturating E2 concentrations, hNAP-1 and E2 cooperated, indicating that binding to hNAP-1 is a rate-limiting step for E2. The observation that the activation-compromised mutant E2 I73A (14) was stimulated by hNAP-1 to the same extent as wild-type E2 suggests that binding to hNAP-1 is not affected in this mutant, in correlation with the in vitro binding data shown in Fig. 2B. Thus, binding to hNAP-1 may be one of several steps, performed by E2, leading to the activation of transcription. These include contacts with components of the PIC, such as TBP and TFIIB (8, 44). Furthermore, E2 binds to coactivators such as AMF-1/Gps, CBP/p300, and p/CAF (36, 37, 49, 56). Interestingly, Lefebvre et al. showed previously that the activation of transcription by E2 correlates with a change in the chromatin structure in yeast cells (38), where the interaction with NAP-1 may play a role.
While E2 strongly activated transcription from reporter constructs which had been transiently transfected, it was not able to activate transcription when the enhancer-promoter region was integrated into the cellular genome. Transiently transfected plasmids are thought to assemble with histones into a nucleosome-like structure (10, 36). However, it is possible that the overall structure is not organized as cellular chromatin (10). The observation that treatment with TSA partially restores the ability of E2 to activate and to act in synergy with hNAP-1 strengthens the notions that a compact chromatin structure may inhibit the access of E2 to chromosomal DNA and that hNAP-1 can enhance E2-mediated activation only after E2 has bound. Thus, hNAP-1 may not be able to stimulate the binding of E2 to sites organized in chromatin, as has been shown for GAL4 in vitro (65).
How does hNAP-1 enhance activation by E2? With the use of deletion mutants, we demonstrated that all of hNAP-1 is required to stimulate E2 activity. The interaction of E2 with hNAP-1 may be necessary, since mutants hNAP-1Δ3, hNAP-1Δ4, and hNAP-1Δ9, which lack either one or both E2-interacting motifs, failed or showed a reduced capacity to stimulate activation by E2. In addition to the C-terminal 301 amino acids, which are bound by both E2 and p300, the N-terminal 91 amino acids of hNAP-1 are crucial. All N-terminally truncated mutants tested here not only lacked stimulating activity but also had a dominant-negative effect on activation by E2. Some of these N-terminally truncated overexpressed hNAP-1 mutants may compete with endogenous, wild-type hNAP-1 for binding to E2. Thus, the recruitment of N-terminally deleted hNAP-1 by E2 would lead to nonfunctional complexes, unable to support transcription. Moreover, the hNAP-1 domains expressed from these mutants may bind to and sequester other cellular factors necessary for the activation of transcription. This may be the case with hNAP-1 mutants still repressing activation by E2 but lacking binding to E2 in vitro (such as hNAP-1Δ7 or hNAP-1Δ10). The notion that targets other than E2 or p300 that are involved in transcription are bound by hNAP-1 is supported by the observation that some of these mutants not only repressed activation by E2 but also inhibited the basal activity of the promoters.
What is the role of the N-terminal part of hNAP-1? According to published data, it does not participate in binding to p300 (58). It has been suggested that the import of histones H2A and H2B into the nucleus is essential for NAP-1 function in the activation of transcription, since yeast NAP-1 mutations inactivating a leucine-rich nuclear export signal (NES) within the N terminus led to reduced transcription of some genes (47). In our study, mutations of corresponding conserved residues in hNAP-1 did not lead to such inhibition. In addition, the mutations also did not affect the localization of hNAP-1, indicating that the human counterpart may have an NES at a different position (M. Rehtanz and G. Steger, unpublished results). However, localization studies with our hNAP-1 mutants revealed stronger nuclear staining of all N-terminal deletion mutants than of full-length hNAP-1 or of C-terminal truncation mutants. A precise mutational analysis of the N-terminal 91 amino acids of hNAP-1 must show whether the nuclear import of H2A and H2B, which requires a functional NES motif, is essential for hNAP-1 to activate transcription. Furthermore, other functions which have been attributed to this domain, such as dimerization or multimerization of NAPs (58) or nucleosome assembly activity (16), may be important.
The recruitment of hNAP-1 to the promoter through fusion with the DBD of GAL4 did not lead to the activation of transcription (M. Rehtanz and G. Steger, unpublished results), indicating that hNAP-1 must act in concert with an AD. The observation that the enhancing effect of hNAP-1 on E2-mediated activation was much stronger in the presence of subsaturating amounts of E2 may be explained by the fact that hNAP-1 affects the interaction of E2 with another factor. p300 may be such a factor. p300 has been shown to bind to two separable domains within the C-terminal 300 amino acids of hNAP-1 (58), as does E2. Our data demonstrate that in vitro the formation of a ternary complex among hNAP-1, p300, and E2 is possible. First, a competition experiment revealed that hNAP-1 cannot displace E2 from p300 (Fig. 5B). Second, results from glycerol gradient density sedimentation correlated with ternary complex formation. Purified hNAP-1 and p300 1195 to 1761 affect the sedimentation of E2Δ386-420, a result which is consistent with direct interactions of both proteins with E2. In the presence of both hNAP-1 and p300, the sedimentation of E2 is changed again, indicating that E2 is complexed not only with either p300 or hNAP-1 but also with both proteins simultaneously. Moreover, E2, hNAP-1, and p300 cofractionate when incubated together (Fig. 5C). This ternary complex may be very efficient in the activation of transcription, since overexpressed E2, hNAP-1, and p300 strongly cooperate in the activation of HPV8 gene expression. Ito et al. demonstrated that the acetylation of histones by p300 helps transfer histones H2A and H2B from nucleosomes to NAP-1 (27). In vitro, the absence of H2A and H2B correlates with increased gene activity, probably by decreasing the level of chromatin folding (reviewed in reference 11). Thus, E2 may induce the recruitment of hNAP-1 and p300, which then participate in the creation of such an H2A- and H2B-free environment.
The importance of hNAP-1 seems not to be restricted to a single factor, since we provide further evidence that bacterially expressed, purified p53 can bind directly to hNAP-1 in vitro and that endogenous p53 and hNAP-1 interact as well. This interaction seems to be functional, a conclusion which is supported by the observation that overexpressed hNAP-1 alone can stimulate p53-mediated activation from a synthetic reporter construct and from a p53-responsive endogenous promoter, that of p21, which does not necessarily require the coexpression of p300, as has been suggested elsewhere (58). Since we were able to demonstrate that TEF-1, devoid of binding to p300, also lacks a functional interaction with hNAP-1, hNAP-1 may function specifically in concert with p300. However, future experiments will need to confirm whether binding to hNAP-1 is a phenomenon only for activators binding to p300. Our data demonstrate for the first time that in addition to p300, hNAP-1 is an essential target for activator proteins to activate transcription. The result may be sufficiently high local concentrations of hNAP-1 and p300 on actively transcribed promoters, which are essential for efficient activation.
Acknowledgments
We thank Y. Ishimi for the gift of the monoclonal antibody against hNAP-1, D. Livingston and M. Scheffner for providing plasmid DNAs, and P. Bartel for providing yeast strain YM954. We also thank Herbert Pfister and Andrew Barker for critical reading of the manuscript and Karin Schnetz for helpful support.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB274-A8, STE604/3-1, and STE604/3-2) and by the BMBF (Aktenzeichen 0312708F). G.S. was the recipient of a Lise-Meitner Habilitationsstipendium.
REFERENCES
- 1.Altman, R., and D. R. Kellogg. 1997. Control of mitotic events by NAP1 and the Gin4 kinase. J. Cell Biol. 138:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1 is required for tissue specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 3.Asahara, H., B. Santoso, E. Guzman, K. Du, P. A. Cole, I. Davidson, and M. Montminy. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol. Cell. Biol. 21:7892-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asahara, H., S. Tartare-Deckert, T. Nakagawa, T. Ikehara, F. Hirose, T. Hunter, T. Ito, and M. Montminy. 2002. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol. Cell. Biol. 22:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avantaggiati, M. L., V. V. Ogryzko, K. Gardner, A. Giordano, S. A. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53 dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 6.Becker, P. B. 2002. Nucleosome sliding: facts and fiction. EMBO J. 21:4749-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson, J. D., and P. M. Howley. 1995. Amino-terminal domains of the bovine papillomavirus type 1 E1 and E2 proteins participate in complex formation. J. Virol. 69:4364-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson, J. D., R. Lawande, and P. M. Howley. 1997. Conserved interaction of the papillomavirus E2 transcriptional activator proteins with human and yeast TFIIB proteins. J. Virol. 71:8041-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breiding, D. E., F. Sverdrup, M. J. Grossel, N. Moscufo, W. Boonchai, and E. J. Androphy. 1997. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol. Cell. Biol. 17:7208-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cervoni, N., and M. Szyf. 2001. Demethylase activity is directed by histone acetylation. J. Biol. Chem. 276:40778-40787. [DOI] [PubMed] [Google Scholar]
- 11.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 12.Dong, X.-P., F. Stubenrauch, E. Beyer-Finkler, and H. Pfister. 1994. Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int. J. Cancer 58:803-808. [DOI] [PubMed] [Google Scholar]
- 13.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson, M. K., and M. R. Botchan. 1996. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J. Virol. 70:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fryer, C. J., and T. K. Archer. 1998. Chromatin-remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:89-91. [DOI] [PubMed] [Google Scholar]
- 16.Fujii-Nakata, T., Y. Ishimi, A. Okuda, and A. Kikuchi. 1992. Functional analysis of nucleosome assembly protein, NAP-1. J. Biol. Chem. 267:20980-20986. [PubMed] [Google Scholar]
- 17.Funk, W. D., D. T. Pak, R. H. Karas, W. E. Wright, and J. W. Shay. 1992. A transcriptionally active DNA binding site for human p53 protein complexes. Mol. Cell. Biol. 12:2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillitzer, E., G. Chen, and A. Stenlund. 2000. Separate domains in E1 and E2 proteins serve architectural and productive roles for cooperative DNA binding. EMBO J. 19:3069-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giordano, A., and M. L. Avantaggiati. 1999. p300 and CBP: partners for life and death. J. Cell. Physiol. 181:218-230. [DOI] [PubMed] [Google Scholar]
- 20.Giri, I., and M. Yaniv. 1988. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 7:2823-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 22.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 23.Gu, W., X.-L. Shi, and R. G. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature 387:819-823. [DOI] [PubMed] [Google Scholar]
- 24.Ham, J., G. Steger, and M. Yaniv. 1994. Cooperativity in vivo between the E2 transactivator and the TATA box binding protein depends on core promoter structure. EMBO J. 13:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann, A., and R. G. Roeder. 1991. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 19:6337-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899-1907. [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, W., S. K. Nordeen, and J. T. Kadonaga. 2000. Transcriptional analysis of chromatin assembled with purified ACF and dNAP-1 reveals that acetyl-CoA is required for preinitiation complex assembly. J. Biol. Chem. 275:39819-39822. [DOI] [PubMed] [Google Scholar]
- 29.Kellogg, D. R., A. Kikuchi, T. Fujii-Nakata, C. W. Turck, and A. W. Murray. 1995. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 130:661-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 31.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP β isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 32.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert, P. F., N. Dostatni, A. A. McBride, M. Yaniv, P. M. Howley, and B. Arcangioli. 1989. Functional analysis of the papilloma virus E2 trans-activator in Saccharomyces cerevisiae. Genes Dev. 3:38-48. [DOI] [PubMed] [Google Scholar]
- 35.Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. A transcriptional repressor encoded by BPV1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 50:69-78. [DOI] [PubMed] [Google Scholar]
- 36.Lee, D., S. G. Hwang, J. Kim, and J. Choe. 2002. Functional interaction between p/CAF and human papillomavirus E2 protein. J. Biol. Chem. 277:6483-6489. [DOI] [PubMed] [Google Scholar]
- 37.Lee, D., B. Lee, J. Kim, D. W. Kim, and J. Choe. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2 dependent transcription. J. Biol. Chem. 275:7045-7051. [DOI] [PubMed] [Google Scholar]
- 38.Lefebvre, O., G. Steger, and M. Yaniv. 1997. Synergistic transcriptional activation by papillomavirus E2 protein occurs after DNA binding and correlates with a change in chromatin structure. J. Mol. Biol. 266:465-478. [DOI] [PubMed] [Google Scholar]
- 39.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 40.Li, Q., A. Imhof, T. N. Collingwood, F. D. Urnov, and A. P. Wolffe. 1999. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 18:5634-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Balbas, M. A., A. J. Bannister, K. Martin, P. Haus-Seuffert, M. Meisterernst, and T. Kouzarides. 1998. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 17:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride, A. A., J. C. Byrne, and P. M. Howley. 1989. E2 polypeptides encoded by the bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyers, C., M. G. Frattini, and L. A. Laimins. 1994. Tissue culture techniques for the study of human papillomaviruses in stratified epithelia. Academic Press, Inc., Orlando, Fla.
- 44.Miller Rank, N., and P. F. Lambert. 1995. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 69:6323-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mink, S., B. Haenig, and K.-H. Klempnauer. 1997. Interaction and functional collaboration of p300 and C/EBPβ. Mol. Cell. Biol. 17:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 47.Mosammaparast, N., C. S. Ewart, and L. F. Pemberton. 2002. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21:6527-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moshkin, Y. M., J. A. Armstrong, R. K. Maeda, J. W. Tamkun, C. P. Verrijzer, J. A. Kennison, and F. Karch. 2002. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16:2621-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller, A., A. Ritzkowsky, and G. Steger. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 76:11042-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakagawa, T., M. Bulger, M. Muramatsu, and T. Ito. 2001. Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin and remodeling factor. J. Biol. Chem. 276:27384-27391. [DOI] [PubMed] [Google Scholar]
- 51.Nakajiama, T., C. Uchida, S. F. Anderson, J. D. Parvin, and M. Montminy. 1997. Analysis of a c-AMP-responsive activator reveals a two component mechanism for transcriptional induction via signal dependent factors. Genes Dev. 11:738-743. [DOI] [PubMed] [Google Scholar]
- 52.Neely, K. E., A. H. Hassan, C. E. Brown, H. LeAnn, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nigro, J. M., S. J. Baker, A. C. Preisinger, J. M. Jessup, R. Hostetter, K. Cleary, S. H. Bigner, N. Davisdson, S. Baylin, P. Devilee, T. Glover, F. S. Collins, A. Weston, R. Modali, C. C. Harris, and B. Vogelstein. 1989. Mutations in the p53 gene occur in diverse human tumor types. Nature 342:705-708. [DOI] [PubMed] [Google Scholar]
- 55.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 56.Peng, Y.-C., D. E. Breiding, F. Sverdrup, J. Richard, and E. J. Androphy. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 74:5872-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purdie, K. J., C. J. Sexton, C. M. Proby, M. T. Glover, A. T. Williams, J. N. Stables, and I. M. Leigh. 1993. Malignant transformation of cutaneous lesions in renal allograft patients: a role for human papillomavirus. Cancer Res. 53:5328-5333. [PubMed] [Google Scholar]
- 58.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. M. Smith, C.-W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20:8933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slepak, T. I., K. A. Webster, J. Zang, H. Prentice, A. O′Dowd, M. N. Hicks, and N. H. Bishopric. 2001. Control of cardiac-specific transcription by p300 through myocyte enhancer factor-2D. J. Biol. Chem. 276:7575-7585. [DOI] [PubMed] [Google Scholar]
- 60.Spalholz, B. A., P. F. Lambert, C. L. Lee, and P. M. Howley. 1987. Bovine papillomavirus transcriptional regulation: localization of the E2-responsive elements of the long control region. J. Virol. 61:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanese, N. 1997. Small-scale density gradient sedimentation to separate and analyze multiprotein complexes. Methods 12:224-234. [DOI] [PubMed] [Google Scholar]
- 62.Tyler, J. 2002. Cooperation between histone chaperones and ATP-dependent nucleosome remodeling machines. Eur. J. Biochem. 269:2268-2274. [DOI] [PubMed] [Google Scholar]
- 63.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallberg, A. E., K. E. Neely, A. H. Hassan, J.-A. Gustafsson, J. L. Workman, and A. P. Wright. 2000. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol. Cell. Biol. 20:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walter, P. P., T. A. Owen-Hughes, J. Cote, and J. L. Workman. 1995. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol. 15:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster, N., J. R. Jin, S. Green, M. Hollis, and P. Chambon. 1988. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL 4 trans-activator. Cell 52:169-178. [DOI] [PubMed] [Google Scholar]
- 67.Wilson, R. B., A. A. Brenner, T. B. White, M. J. Engler, J. P. Gaughran, and K. Tatchell. 1991. The Saccharomyces cerevisiae SRK1 gene, a suppressor of bcy1 and ins1, may be involved in protein phosphatase function. Mol. Cell. Biol. 11:3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao, J. H., I. Davidson, H. Matthes, J.-M. Garnier, and P. Chambon. 1991. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65:551-568. [DOI] [PubMed] [Google Scholar]
- 69.Yang, Y.-C., H. Okayama, and P. M. Howley. 1985. Bovine papillomavirus contains multiple transforming genes. Proc. Natl. Acad. Sci. USA 82:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao, J.-M., D. E. Breiding, and E. J. Androphy. 1998. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 72:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]