Abstract
Background. The dengue virus serotype 3 (DENV-3) Indian subcontinent strain emerged in Puerto Rico in 1998 after a 21-year absence. The rapid expansion of DENV-3 on the island correlated with the withdrawal of the other serotypes for 7 years. The DENV-3 prevalence declined in 2008 and remains undetected.
Methods. We sequenced complete genomes of 92 DENV-3 clinical isolates to characterize the molecular evolution and phylogeography throughout 10 years of continued sampling (1998–2007).
Results. We documented 8 distinct lineages that emerged simultaneously and evolved independently. Two of the 8 lineages were highly associated with transient introductions of foreign viruses, and 2 of the 3 endemic lineages covered the entire study period. We found evidence of temporal-geographical clustering only within the 3 endemic lineages. The phylogeography analysis combined with serotype-specific incidence data showed that transmission of a DENV serotype in a given location and time is usually correlated with the absence of the other serotype.
Conclusions. Our study shows the cotransmission of DENV-3 lineages through a complex dissemination pattern dissimilar to the evolutionary dynamics of the other serotypes in the island. High virus genetic diversity and a large naive population were underlying factors in the expansion and collapse of DENV-3 in Puerto Rico.
The global spread of dengue virus (DENV) has dramatically increased in recent decades, with >2.5 billion people now at risk of infection by DENV [1]. Increased urban expansion and population growth, travel, and climatic changes have contributed to the expansion of the ecological niche favorable for the propagation of DENV vectors, Aedes species mosquitoes, resulting in intensification of DENV epidemics [2]. The Americas have experienced increased incidence of DENV throughout the last 30 years concomitant with establishment of the 4 DENV serotypes (DENV-1–4) in most countries. Although sociological and environmental factors have undoubtedly influenced transmission of DENV in the region, 2 major introductions of DENV strains seem to have contributed to increased endemicity and the potential to cause outbreaks: introduction of South East Asian DENV-2 in the 1980s and introduction of Indian subcontinent (subtype 3) DENV-3 in the mid-1990s. In the Americas, the Indian DENV-3 genotype was initially detected in Nicaragua and rapidly disseminated throughout Central America, the Caribbean basin, and South America [3, 4]; this serotype had been rarely reported during the previous 15 years. It appears that the expansion of the Indian subcontinent genotype of DENV-3 could have been favored by limited competition from the American genotype, including an immunological gap in the human population.
Puerto Rico, a Caribbean island of 9104 square kilometers and a population of 3.9 million, is densely populated and has experienced continued high rates of transmission of DENV for several decades [5–7]. Dengue virus serotype 3 reemerged in Puerto Rico in 1998 during an epidemic season in which DENV-1 and DENV-4 predominated [8]. This introduction resulted in shifts in serotype distribution among DENV cases over the next 6 years, with DENV-1 and DENV-4 no longer being detected, only a low proportion of cases being DENV-2, and DENV-3 being the predominant serotype [7, 9]. Similar patterns of DENV-3 introduction and predominance have also been reported in other South American countries [10]. Despite this ability to penetrate and disseminate, DENV-3 declined in 2008 and remained undetected by the island-wide DENV surveillance system, even during the 2010 epidemic, in which there were >24 000 reported cases of DENV (Centers for Disease Control and Prevention [CDC] unpublished data).
Phylogenetic or phylogeographic studies of DENV-3 in the Caribbean and Americas are limited; therefore little is known about its dynamics in DENV-endemic countries. This study examines the genetic complexity of DENV-3 in Puerto Rico by sequencing the complete genomes of 92 isolates obtained from human sera collected between 1998 and 2007. Our findings improve our understanding of the rapid propagation, persistence, and possibly the sudden decline of DENV-3 in Puerto Rico.
MATERIALS AND METHODS
Virus Isolates
The DENV-3 isolates in this study were obtained from cases of DENV reported to the Puerto Rico Department of Health and the CDC's Passive Dengue Surveillance System. This system records the number of confirmed cases in the 78 municipalities of Puerto Rico, including serotype identification by tissue culture and reverse transcription polymerase chain reaction (RT-PCR). One hundred and fifty serum samples (5 samples with positive DENV-3 diagnosis from the 3 municipalities with the highest DENV-3 reporting per year between 1998 and 2007) were originally selected (Figure 1C). All samples were handled in accordance with the institutional review boards of the CDC (protocol 4797) and the Broad Institute. All isolates were obtained as previously described by McElroy et al [9]. Briefly, serum was inoculated into C6/36 (Aedesalbopictus) cell culture maintained in Dulbecco's minimal essential medium [11] and incubated at 33°C for 5 days [12], and and the presence of DENV was confirmed by indirect immunofluorescence assay. Isolates were then passaged 1–2 times under the same conditions to generate stock material for sequencing analysis.
Figure 2.
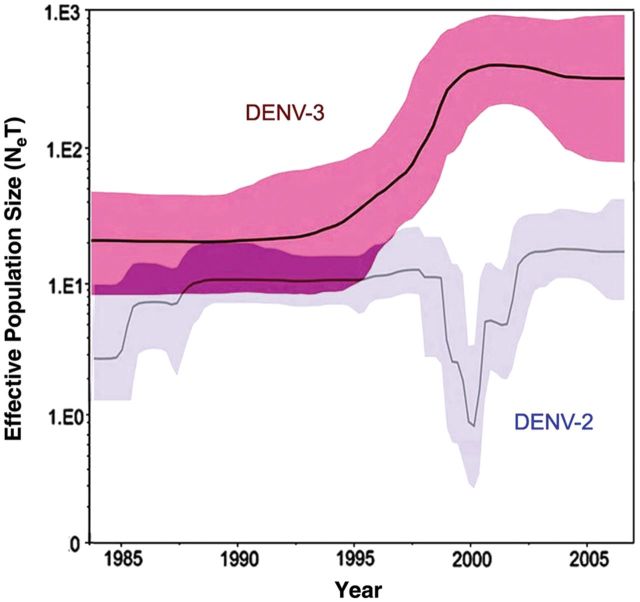
Genetic diversity of dengue virus serotype 2 (DENV-2) and serotype 3 (DENV-3) viral genomic sequences from Puerto Rico, 1998–2007. Bayesian skyline plots showing changes in relative genetic diversity through time (Neτ) and 95% highest probability density for DENV-3 (n = 92) and DENV-2 (n = 140). Neτ, product of the effective population size and generation length in years (relative genetic diversity); black line, median estimate; blue shadow, 95% highest probability density.
Sequencing
We performed RNA extraction, amplification, and genome sequencing as previously described by McElroy et al [9]. Briefly, viral RNA was extracted from tissue culture supernatant using an M48 or Universal BioRobot system (QIAGEN). Complementary DNA (cDNA) was generated using the SensiScript RT kit (QIAGEN) with random hexamers (Applied Biosystems) and Pfu UltraII (Stratagene) or iTaq DNA (Bio-Rad) polymerase-driven PCR confirmed the presence of cDNA. We performed bidirectional Sanger sequencing on pooled cDNA or PCR amplicons at the Broad Institute of MIT & Harvard using an ABI 3730 automated sequencer. We trimmed the resulting sequence at the ends to remove both low-quality and primer sequence and assembled resulting reads using Broad Institute's AV454 assembly algorithm [13]. Consensus assemblies were annotated by the Broad Institute using an in-house annotation algorithm. All sequences encompassing the complete open reading frames (10173 nt) and parts of the 5′ and 3′ untranslated regions were deposited in GenBank (Supplementary Table 1).
Sequence Analyses
We generated a multiple sequence alignment of 124 complete coding sequences using the ClustalWmodule available in MEGA 4 (http://www.megasoftware.net). Aligned sequences included the 92 Puerto Rico isolates from this study, an additional 22 South/Central American strains sequenced as part of the Broad Institute's Genome Resources in Dengue Consortium, and an additional 10 international strains obtained from GenBank. We generated maximum-likelihood trees using PAUP* [14] and MEGA 4 incorporating the GTR + I + Γ4 model of nucleotide substitution, as determined by MODELTEST version 3.07 [15]. Bootstrap analysis (repetitions = 1000) was performed using the maximum composite likelihood method in MEGA 4. We estimated mutation rates of substitution, effective population size, and maximum clade credibility tree using a Bayesian Markov Chain Monte Carlo approach as implemented in BEAST version 1.4.7 (http://mbe.oxfordjournals.org/content/25/7/1459) [16]. For all BEAST analyses, the year of sampling was utilized, and the model was parameterized using the GTR + I + Γ4 model of nucleotide substitution, a strict molecular clock, and a Bayesian Skyline tree prior. All Markov Chain Monte Carlo chains were run for sufficient length (60 M generations sampled every 10 000) to ensure stationary parameters, with statistical error reflected in values of the 95% highest probability density with 10% removed as burn-in. Tree topology across the phylogenetic methods and algorithms utilized were consistent. Selective pressures on the complete coding sequence or individual genes, indicated by nonsynonymous-to-synonymous nucleotide substitution ratios and number of positively or negatively selected sites, were evaluated by single likelihood ancestor counting [17], random effects likelihood [17], and internal fixed effects likelihood [18] methods using HYPHY accessed through the data monkey server [19]. Geographical and temporal clustering analysis was performed using SaTScan software (http://www.satscan.org) and associated to phylogeny using Bayesian tip-association significance testing (http://evolve.zoo.ox.ac.uk/evolve/BaTS.html) to infer any relationship between sequence data and geographical source of the isolate with the posterior sample of trees calculated by BEAST. For the parsimony score, association index, and monophyletic clade size, we considered P < .05 to be significant.
RESULTS
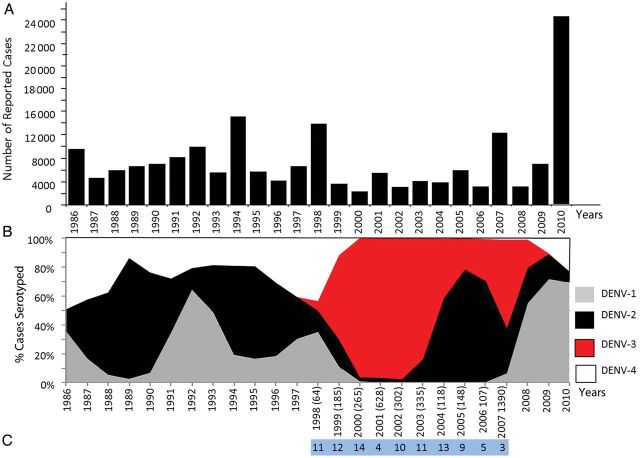
Major epidemics of DENV occurred in Puerto Rico during 1986, 1992, 1994, 1998, 2007, and 2010 (Figure 1A). In order to declare an epidemic, the historical average of reported cases between 1986 and the present year is calculated, and the epidemic threshold, which includes the 75th percentile of the distribution of cases in the same years, must be exceeded. The number of reported cases ranged from 2000 to approximately 24 000; however, most epidemics feature >8000 reported cases. Major fluctuations in serotype circulation have been observed during both epidemic and endemic periods of DENV transmission (Figure 1B). Over the 20 year period for which there are adequate data on the proportion of each DENV serotype circulating among DENV cases each year, a number of patterns are evident. From 1986 to 1998, DENV-1, DENV-2, and DENV-4 cocirculated in the island, with DENV-2 accounting for >50% of serotyped cases. Dengue virus serotype 3 emerged in Puerto Rico in 1998, a period marked by high prevalence of DENV-1 and DENV-4 and the sudden decline of DENV-2 [8]. By 1999, DENV-3 symptomatic cases expanded rapidly across the island and displaced DENV-1 and DENV-4 (Figure 1B); however, DENV-2 cases persisted but were restricted to the eastern part of the island between 2000 and 2002 [9]. The prevalence of DENV-3 cases transiently declined during a DENV-2 resurgence, only to reassume dominance during the 2007 epidemic (Figure 1B). Similar differences in symptomatic transmission dynamics have been documented elsewhere [20].
Figure 1.
Historic overview of dengue virus transmission in Puerto Rico. Transmission of dengue virus (DENV) in Puerto Rico documented from 1986 to 2010 using data obtained from the Puerto Rico Department of Health and the Centers for Disease Control and Prevention Dengue Branch Passive Dengue Surveillance System. A, Total number of reported cases, including all serotypes reported to the system since 1986. B, Proportion (percent) of DENV isolates by serotype during the period 1986–2010: DENV-1 (gray), DENV-2 (black), DENV-3 (red), and DENV-4 (white). X-axis indicated years. Numbers in parenthesis indicate the number of DENV-3 identifications per year. C, Number of isolates successfully sequenced by year in this study.
We investigated the population genetics of DENV-3 to elucidate how the virus adapted and evolved in the island. The genetic diversity of DENV-3 showed a steep increase that correlated with its emergence and expansion (Figure 2). This increase reached a plateau by year 2000, which is when approximately 95% of all DENV cases were caused by DENV-3. This diversity remained throughout the peak of the DENV-3 predominance of cases (2000–2002). However, the amplitude of diversity, indicated by the 95% high probability density, increased immediately after the proportion of DENV-3 cases began to decline, correlating with the onset of the 2007 island-wide epidemic. Conversely, the genetic diversity of DENV-2 was considerably lower than DENV-3 during the study period, and there was a steep decrease in diversity concomitant with the peak of DENV-3 predominance among cases.
The mean evolutionary rate for each gene ranged 5.33 × 10−4–1.30 × 10−4 nucleotide substitutions per site per year (Table 1), which is comparative with the rates reported by Twiddy and Araújo [21, 22]. Single likelihood ancestor counting and internal fixed effects likelihood analyses showed that no codons were under positive selective pressure based on dN/dS ratios <0.1 with a P value <.05. No evidence of intraserotypic recombination events was detected.
Table 1.
Estimated Mean Nucleotide Substitution Rates per Site per Year for Dengue Virus Serotype 3 Genes Calculated from the Complete Sequence Data of All Puerto Rican Isolates
| Gene | Mean Substitution Rate | 95% HPD CIs | Mean dN/dS |
|---|---|---|---|
| C | 3.51 × 10−4 | 5.85 × 10−4–1.55 × 10−4 | 0.092 |
| prM | 3.28 × 10−4 | 4.78 × 10−4–1.94 × 10−4 | 0.038 |
| E | 2.28 × 10−4 | 3.38 × 10−4–1.12 × 10−4 | 0.052 |
| NS1 | 3.23 × 10−4 | 4.66 × 10−4–1.89 × 10−4 | 0.120 |
| NS2a | 5.33 × 10−4 | 7.28 × 10−4–3.43 × 10−4 | 0.107 |
| NS2b | 4.34 × 10−4 | 6.42 × 10−4–2.43 × 10−4 | 0.063 |
| NS3 | 2.10 × 10−4 | 2.92 × 10−4–1.22 × 10−4 | 0.036 |
| NS4a | 3.46 × 10−4 | 5.41 × 10−4–1.66 × 10−4 | 0.066 |
| NS4b | 2.59 × 10−4 | 3.84 × 10−4–1.41 × 10−4 | 0.014 |
| NS5 | 1.30 × 10−4 | 7.25 × 10−5–1.90 × 10−4 | 0.058 |
Abbreviations: CI, confidence interval; HPD, high probability density.
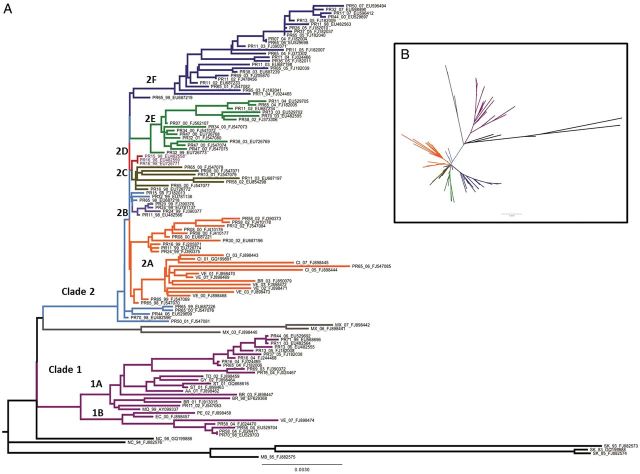
Maximum likelihood phylogenetic analysis of the complete coding sequence of the Puerto Rican isolates and international DENV-3 isolates showed 2 primary lineages in Puerto Rico, defined as clade 1 and clade 2 (Figure 3). Estimation of the time since the most recent common ancestor places the emergence of clade 1 around 1998 and the emergence of clade 2 around 1999. Clade 1 consisted of two subclades—1A and 1B—both of which were closely related with the international isolates. Clade 2 consisted of multiple subclades, all of which emerged rapidly and almost simultaneously from the parent population. In addition, several subclades—2A, 2E, and 2F—exhibited rapid, sustained diversification. Most clade 2 viruses were no longer detected by 2004, except for clade 2F, which was found until DENV-3 was no longer detected among Puerto Rican DENV cases. Among clade 2 strains, only clade 2A included isolates from international locations, and the Puerto Rican sequences were basal to these strains, suggesting common origins (Figure 3). The most recent common ancestor for both clade 1 and clade 2 found in GenBank was a Nicaraguan strain (Figure 3). Further analyses of clade 2 suggest that Puerto Rican viruses are basal in the phylogeny of DENV-3 South American lineages and most closely related to Mexican strains (Figure 3). International isolates were also found in both clade 1 subclades (Figure 3), and the phylogenetic relationships of the Puerto Rican strains suggest they emerged from an introduction event, became endemic, and continued to evolve in situ in genetic diversity across the island during the approximately 10-year period of DENV-3 presence.
Figure 3.
Phylogeny of dengue virus serotype 3 (DENV-3) in Puerto Rico. A Maximum likelihood phylogeny of Puerto Rican (n = 92) and international (n = 32) DENV-3 full genome sequences. The phylogenetic tree was rooted with isolates from Sri Lanka and Mozambique (black), which are basal to genotype 3 (Indian subcontinent). The Nicaragua 1994 and 1998 isolates (black) root the American lineage. Clade 1 (purple) consists of subclades1A and 1B. Clade 2consists of 6 subclades: 2A (orange), 2B (light purple), 2C (brown), 2D (red), 2E (green), and 2F (dark blue). Isolates associated with the parental strain are light blue. Nomenclature for each isolate consists of a 2-letter country code followed by a 2-digit collection year and GenBank accession number. For Puerto Rico (PR) isolates, a 2-digit number indicates the residence of the case (municipality). B, Maximum likelihood phylogeny of DENV-3 in star configuration. Subclade colors are the same as in A. See Supplementary Table 1 for more details.
Three amino acid differences exist between the 2 clades at NS2A-195 (clade 1, alanine; clade 2, threonine), NS2B-59 (clade 1, isoleucine; clade 2, valine), and NS4A-39 (clade 1, arginine; clade 2, lysine). Only 1 of these mutations, NS2A-195, is nonconservative, and the functional significance of these amino acid differences require further study.
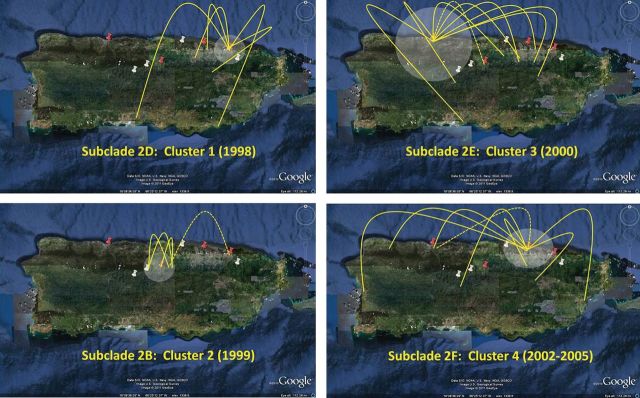
We determined the relationship between phylogenetics and the spatial–temporal distribution of DENV-3 cases throughout Puerto Rico using SaTScan under a Poisson-based model that included year of illness onset and place of residence (Figure 4). Spatial–temporal clustering of phylogenetically related viruses was observed with 4 clusters. There were 2 clusters of clade 2 viruses that were closely related to the parental strains: cluster 1 from subclade 2D occurred in 1998 around the municipalities of San Juan and Canóvanas, and cluster 2 from subclade 2B in 1999 was restricted to a small area surrounding the municipality of Corozal. Cluster 3 emerged from subclade 2E during the year 2000 around Arecibo, and cluster 4 emerged from subclade 2F between 2002 and 2005 in the San Juan metropolitan area. Bayesian tip-association significance testing analysis corroborated phylogenetic association of isolates in each clade and those that formed clusters (Table 2).
Figure 4.
Spatial–temporal distribution and clustering of dengue virus serotype 3 (DENV-3) dengue cases in Puerto Rico. Dates of illness onset and residence for each DENV-3 dengue case were analyzed using SatScan software package. Four clusters were identified (P < .05; white circles). Phylogenetic relationships for each cluster were analyzed using Bayesian tip-association significance testing. Yellow arches represent cases with phylogenetic descendants from a parent cluster in a new geographical location. Red pins indicate the cluster center, and white pins indicate the cluster radius. Numbers in parenthesis indicate year in which cluster was statistically significant P < .05. Isolates from subclade 2D generated cluster 1 (1998) = San Juan—Canóvanas region; subclade 2B generated cluster 2 (1999) = Corozal region; subclade 2E generated cluster 3 (2000) = Arecibo region; subclade 2F generated cluster 4 (2002–2005) = San Juan metro area.
Table 2.
Bayesian Tip-Association Significance Testing (BaTS) Analysis on Each Clade/Subclade Independently
| Group | No. of Isolates | Estimated BaTS (95% HPD CIs)a | P Value |
|---|---|---|---|
| Subclade 2B/C/D | 14 | 1.6 (1.0–2.1) | .01 |
| Subclade 2E | 15 | 2.2 (1.2–3.3) | .01 |
| Subclade 2F | 26 | 1.4 (1.0–2.0) | .01 |
| Subclade 2A | 18 | 1.6 (1.0–3.0) | .01 |
| Clade 2 | 16 | 1.6 (1.0–3.0) | .01 |
Abbreviations: CI, confidence interval; HPD, high probability density.
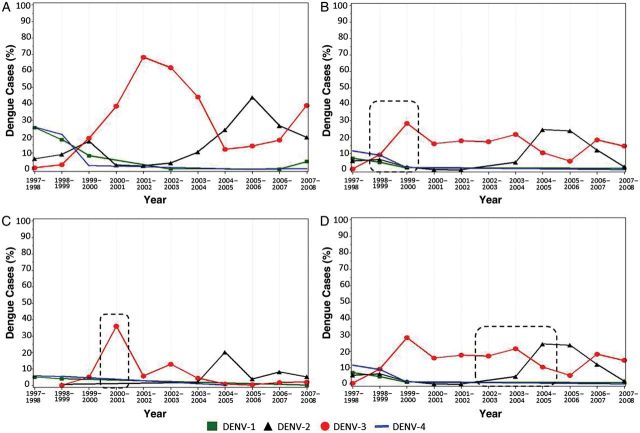
To further describe the spatial–temporal factors that may be associated with clustering of virus lineages in specific regions of the island, we determined the proportion of all DENV serotypes detected by RT-PCR during the season and geographical region of each DENV-3 cluster (Figure 5). These proportions were also supported by considerable high incidence of DENV-3 over the other serotypes in each cluster. During 1999–2006,Puerto Rico experienced an overall high proportion and incidence of DENV-3 cases, which peaked during the 2001–2002 DENV season (Figure 5A). Formation of cluster 1 and 3 was associated with a significant increase in DENV-3 cases preceding the peak of island-wide prevalence. Cluster 4 formed during the switch from DENV-3 to DENV-2. No sustained transmission of DENV-3 cases was found in the eastern region of the island during the years 2000–2002, which coincided with the refuge of DENV-2, as previously reported [9].
Figure 5.
Proportion of dengue virus (DENV) cases as a key factor in cluster formation. The proportion of each DENV serotype detected by reverse transcription polymerase chain reaction (RT-PCR) was determined every season of transmission from 1998 to 2007. Proportions were calculated based on the frequency of positive cases per serotype over the total number of RT-PCR cases in each specific region and time period. A, Proportion of DENV cases across the island. B, Proportion of dengue cases in San Juan—Canóvanas region. Dashed rectangle indicates year of cluster 1 formation (1998–1999). C, Proportion of dengue cases in Arecibo region. Dashed rectangle indicates year of cluster 3 formation (2000). D, Proportion of dengue cases in San Juan metropolitan region. Dashed rectangle indicates year of cluster 4 formation (2002–2005).
DISCUSSION
This is the first comprehensive analysis of DENV-3 molecular evolution and phylodynamics in Puerto Rico and the Caribbean using complete genome sequences with detailed epidemiological data. Dengue virus serotype 3 reemerged in Puerto Rico in 1998 shortly after the introduction of the Indian subcontinent genotype in the Americas and during one of the largest DENV epidemics recorded on the island. After a year, DENV-3 expanded across the island, displacing the other serotypes, with the exception of DENV-2, whose transmission was limited to a “refuge region” in the east of the island [9]. Transient serotype cross-protection may have a role in DENV transmission dynamics, influencing cluster formation and serotype exclusion. Cross-protection between serotypes and specific DENV-2 genotypes was recently reported in Nicaragua [23]. It seems plausible to hypothesize that the 21-year absence of DENV-3 rendered the population susceptible to infection by this serotype and that cross-reactive DENV-3 antibodies temporarily blocked transmission of other serotypes, creating a favorable condition for expansion. The reemergence of DENV-2 correlated with a reduction in DENV-3 symptomatic cases and possible loss of cross-protection even after DENV-3's attempt to reemerge during the 2007 epidemic. Symptomatic transmission of DENV-3 was interrupted by 2009. This study is limited to the analysis of symptomatic cases; viruses causing subclinical transmission must be addressed in order to fully understand DENV transmission and evolution. Additional studies incorporating the complete genome sequences from isolates of all 4 serotypes will be necessary to fully understand the evolutionary dynamics at work between the serotypes that drive their coexistence as well as displacement.
We expected to detect an increase in genetic diversity during the first few years of DENV-3 expansion; however, this increase in DENV-3 genetic diversity was concomitant with a decrease of DENV-2 genetic diversity. This inverse relationship suggests an antagonistic interaction between these serotypes, which hinders cotransmission. However, we do not know the contribution of DENV genetic diversity to various determinants of infection in a population, such as mosquito replication fitness or protection by heterotypic DENV antibodies.
Further studies on the genetic diversity of the Puerto Rican DENV-3 lineage has shown an evolutionary rate faster than those reported by others [21, 24]. This accelerated mutation rate could benefit the virus's ability to establish stable transmission even over nonpermissive hosts protected with heterotypic antibodies. The high proportion of conservative relative to nonconservative mutations and the low dN/dS ratios suggest that the expansion of DENV-3 in Puerto Rico is driven by stochastic events and strong purifying selection similar to the endemic DENV-2 [25]. Holmes and Twiddy suggest that positive selection may be acting on DENV-3 subject to immune pressures, a result in contrast with our observations; however, their conclusions were based on partial genome sequence and limited sample size, which could impact the detection of positive selection [26].
The evolutionary history depicted in this study is limited to the availability of clinical isolates and published genomic sequences; however, the short genetic distance between the Puerto Rican DENV-3 lineage and the Nicaragua 1994 isolate (Figure 3) could suggest an earlier, though undetected, presence of DENV-3 in Puerto Rico [27]. The maximum likelihood analysis shows an evolutionary divergence dissimilar to the linear evolution model followed by the other serotypes in the island. Soon after DENV-3 was detected, between 1998 and 1999, the virus diverged simultaneously into 2 main clades. Clade 2 viruses do not represent an organized population structure, but rather an array of multiple lineages scattered across the island. It seems that each clade 2 lineage evolved independently in exclusive regions of the island; however, all subclades but 2F are short lived and became extinct before the reemergence of DENV-2 in 2004–2005. Transmission of clade 1 and subclade 2F viruses persisted until 2007 among DENV cases. However, subclade 2F was the only endemic group that maintained transmission and was possibly the etiology on the 2007 DENV-3 epidemic. Clade 1 managed to persist on the island despite a high influence of foreign viruses, suggesting frequent importation–exportation events. This contrasts with DENV-2 evolution, in which the frequency of foreign viruses is much lower and transient [9]. Several South American sequences obtained from GenBank associated with subclade 2A and the isolation dates of these isolates suggest that this Puerto Rican lineage functioned as the springboard for the colonization of South America by DENV-3 [28].
Integration of phylogeny with spatial–temporal data identified clusters of virus microevolution from which fast virus migration occurred across the island, especially to regions of with a low proportion of DENV-2 cases. The factors that induced the formation of these clusters in specific geographical regions of the island remain unclear; however, this study identified disease incidence as a potential contributing element. The incidence and proportion of DENV-3 cases significantly exceeded the rate of all the other serotypes during cluster formation; therefore the area covered by the cluster may represent a permissive environment for DENV-3 to evolve. The regions adjacent to the cluster radius might have been protected by herd immunity to DENV-3 or cross-protection from other serotypes limiting cluster expansion. Two clusters in northern Puerto Rico showed elevated rates of DENV-3 cases in consecutive seasons but preceded the peak of prevalence and transmission of DENV-3 in the rest of the island. We could then speculate that the expansion of DENV-3 occurred through a transmission corridor that originated from the north of the island. The presence of a highly transited highway connecting major cities across northern Puerto Rico and the physical separation between northern and southern cities by a large mountain range could represent potential factors influencing the migration trajectory that our phylogeographic analysis suggests. Similar observations have been documented in Asia, where virus migrations follow human movement and urbanization [29]. We should also consider that cluster regions could be more susceptible to virus transmission facilitated by a higher density of Aedes aegypti pupae per person. Spatial and temporal patterns in the distribution of A. aegypti have been suggested to contribute to the focal nature of DENV transmission and clustering of DENV cases [30, 31].
The collapse of DENV-3 transmission could have been influenced by the reemergence of DENV-2; however, we should also consider that the eruptive expansion of DENV-3 might have resulted as a self-limiting factor, rendering the population immune and rapidly eliminating susceptible hosts. Our previous study [9] and these observations show that geographical partitioning of lineages may correlate with serotype incidence. The virus then appears to localize within subsets of the population with low levels of homologous and heterologous immunological protection through time. Our data support that some of the lineages that emerged in Puerto Rico may have more epidemic potential than others, a theory concurrent with others [32]. Elucidating the factors that induced cluster formation will require further investigation.
The multiple lineage divergence pattern documented in this study differed significantly from the single lineage, low divergence, linear evolution of DENV-3 previously reported by others in South America [20, 26]. The large effective population size, the high proportion of foreign strains, and the coexistence of 8 simultaneous, independent lineages evidence the great genetic diversity of DENV-3. Although we did not detect any furrows in genetic diversity of the general virus population, it is possible that extinction of most lineages might have generated population bottlenecks, which have been shown to impact DENV evolutionary dynamics through genetic drift [33]. The high mutation rates and rapid replication of DENV-3 might have produced a highly heterogeneous population structure at each lineage; however, these transient populations appeared to have suffered from reduced genetic plasticity and adaptability. We can then suggest that the endemic strain failed to establish a reservoir of variants with a phenotype adapted to the environmental changes and consequently drove toward its own collapse. This type of virus emergence, expansion, and subsequent collapse represents a concept that must be further defined in population dynamics. Our study confirms the utility of genomic sequencing in large-scale surveillance and provides a novel approach to determine molecular evolution and population dynamics.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Vance Vorndam, Mark Verduin, Candimar Colón, and Edgardo Vergne for providing laboratory support. International isolates were provided by the Caribbean Epidemiology Centre, Trinidad and Tobago; Instituto Nacional de Diagnóstico y Referencia Epidemiológicos, Mexico; and Laboratorio Regional de Diagnóstico e Investigación del Dengue y Otras Enfermedades Virales, Venezuela. We thank Eddie Holmes for comments on the manuscript, and also the Broad Sequencing Platform and Biological Samples Repository for assistance in data generation.
Financial support. This project was funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (under contract HHSN272200900006C [to B. W. B.] and HHSN272200900018C [to B. W. B.]).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Dengue and dengue hemorrhagic fever. 2009 Fact sheet no. 117 http://www.who.int/mediacentre/factsheets/fs117/en/index.html . [Google Scholar]
- 2.Lambrechts L, Scott TW, Gubler DJ. Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl Trop Dis. 2010;4:e646. doi: 10.1371/journal.pntd.0000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman MG, Vazquez S, Martinez E, et al. Dengue in Nicaragua, 1994: reintroduction of serotype 3 in the Americas. Bol Oficina Sanit Panam. 1996;121:102–10. [PubMed] [Google Scholar]
- 4.Gubler DJ. Dengue and dengue hemorrhagic fever in the Americas. P R Health Sci J. 1987;6:107–11. [PubMed] [Google Scholar]
- 5.Morens DM, Rigau-Perez JG, Lopez-Correa RH, et al. Dengue in Puerto Rico, 1977: public health response to characterize and control an epidemic of multiple serotypes. Am J Trop Med Hyg. 1986;35:197–211. doi: 10.4269/ajtmh.1986.35.197. [DOI] [PubMed] [Google Scholar]
- 6.Dietz V, Gubler DJ, Ortiz S, et al. The 1986 dengue and dengue hemorrhagic fever epidemic in Puerto Rico: epidemiologic and clinical observations. P R Health Sci J. 1996;15:201–10. [PubMed] [Google Scholar]
- 7.Tomashek KM, Rivera A, Munoz-Jordan JL, et al. Description of a large island-wide outbreak of dengue in Puerto Rico, 2007. Am J Trop Med Hyg. 2009;81:467–74. [PubMed] [Google Scholar]
- 8.Rigau-Perez JG, Ayala-Lopez A, Garcia-Rivera EJ, et al. The reappearance of dengue-3 and a subsequent dengue-4 and dengue-1 epidemic in Puerto Rico in 1998. Am J Trop Med Hyg. 2002;67:355–62. doi: 10.4269/ajtmh.2002.67.355. [DOI] [PubMed] [Google Scholar]
- 9.McElroy KL, Santiago GA, Lennon NJ, Birren BW, Henn MR, Munoz-Jordan JL. Endurance, refuge, and reemergence of dengue virus type 2, Puerto Rico, 1986–2007. Emerg Infect Dis. 2011;17:64–71. doi: 10.3201/eid1701.100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nogueira RM, de Araujo JM, Schatzmayr HG. Dengue viruses in Brazil, 1986–2006. Rev Panam Salud Publica. 2007;22:358–63. doi: 10.1590/s1020-49892007001000009. [DOI] [PubMed] [Google Scholar]
- 11.Gubler DJ, Kuno G, Sather GE, Velez M, Oliver A. Mosquito cell cultures and specific monoclonal antibodies in surveillance for dengue viruses. Am J Trop Med Hyg. 1984;33:158–65. doi: 10.4269/ajtmh.1984.33.158. [DOI] [PubMed] [Google Scholar]
- 12.Kuno G, Oliver A. Maintaining mosquito cell lines at high temperatures: effects on the replication of flaviviruses. In vitro cellular & developmental biology. Journal of the Tissue Culture Association. 1989;25:193–6. doi: 10.1007/BF02626177. [DOI] [PubMed] [Google Scholar]
- 13.Henn MR BC, Charlebois P, Lennon NJ, et al. Whole denome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathogens. 2011;3:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0604s00. Feb; Chapter 6:Unit 6.4. [DOI] [PubMed] [Google Scholar]
- 15.Bennett SN, Holmes EC, Chirivella M, et al. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J Gen Virol. 2006;87:885–93. doi: 10.1099/vir.0.81309-0. [DOI] [PubMed] [Google Scholar]
- 16.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–92. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 17.Kosakovsky Pond S, Frost S. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–22. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 18.Kosakovsky Pond S, Frost S, Grossman Z, Gravenor M, Richman D, Leigh-Brown A. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol. 2006;2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Data monkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–7. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79:14680–7. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Twiddy SS, Holmes EC, Rambaut A. Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol. 2003;20:122–9. doi: 10.1093/molbev/msg010. [DOI] [PubMed] [Google Scholar]
- 22.Araujo JM, Nogueira RM, Schatzmayr HG, Zanotto PM, Bello G. Phylogeography and evolutionary history of dengue virus type 3. Infect Genet Evol. 2009;9:716–25. doi: 10.1016/j.meegid.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 23.OhAinle M, Balmaseda A, Tellez Y, et al. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med. 2011;114:114ra128. doi: 10.1126/scitranslmed.3003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez A, Fajardo A, Moros Z, et al. Evolution of dengue virus type 3 genotype III in Venezuela: diversification, rates and population dynamics. Virol J. 2010;7:329. doi: 10.1186/1743-422X-7-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myat Thu H, Lowry K, Jiang L, Hlaing T, Holmes EC, Aaskov J. Lineage extinction and replacement in dengue type 1 virus populations are due to stochastic events rather than to natural selection. Virology. 2005;336:163–72. doi: 10.1016/j.virol.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Twiddy SS, Woelk CH, Holmes EC. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J Gen Virol. 2002;83:1679–89. doi: 10.1099/0022-1317-83-7-1679. [DOI] [PubMed] [Google Scholar]
- 27.Amarilla AA, de Almeida FT, Jorge DM, et al. Genetic diversity of the E protein of dengue type 3 virus. Virol J. 2009;6:113. doi: 10.1186/1743-422X-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt DJ, Pickett BE, Camacho D, et al. A phylogenetic analysis using full-length viral genomes of South American dengue serotype 3 in consecutive Venezuelan outbreaks reveals a novel NS5 mutation. Infect Genet Evol. 2011;11:2011–9. doi: 10.1016/j.meegid.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabaa MA, Ty Hang VT, Wills B, Farrar J, Simmons CP, Holmes EC. Phylogeography of recently emerged DENV-2 in southern Viet Nam. PLoS Negl Trop Dis. 2010;4:e766. doi: 10.1371/journal.pntd.0000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammen MP, Pimgate C, Koenraadt CJ, et al. Spatial and temporal clustering of dengue virus transmission in Thai villages. PLoS Med. 2008;5:e205. doi: 10.1371/journal.pmed.0050205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seng CM, Setha T, Nealon J, Socheat D. Pupal sampling for Aedes aegypti (L.) surveillance and potential stratification of dengue high-risk areas in Cambodia. Trop Med Int Health. 2009;14:1233–40. doi: 10.1111/j.1365-3156.2009.02368.x. [DOI] [PubMed] [Google Scholar]
- 32.Gubler DJ, Suharyono W, Lubis I, Eram S, Gunarso S. Epidemic dengue 3 in central Java, associated with low viremia in man. Am J Trop Med Hyg. 1981;30:1094–9. doi: 10.4269/ajtmh.1981.30.1094. [DOI] [PubMed] [Google Scholar]
- 33.Bennett SN, Drummond AJ, Kapan DD, et al. Epidemic dynamics revealed in dengue evolution. Mol Biol Evol. 2009;4:811–8. doi: 10.1093/molbev/msp285. [DOI] [PMC free article] [PubMed] [Google Scholar]