In this study, the authors dissect the mechanism of action of Pax7 in pituitary intermediate lobe melanotropes and find that it is a critical positive regulator of melantrope fate, acting as a selector switch between the two alternate pro-opiomelanocortin lineages dependent on the T-box factor Tpit for terminal differentiation. They conclude that Pax7 acts as a pioneer factor and selector gene for differentiation and may be a marker for pituitary adenomas causing Cushing's disease.
Keywords: differentiation, progenitors, cell cycle, POMC, Tpit, Cushing's disease
Abstract
The anterior and intermediate lobes of the pituitary gland derive from the surface ectoderm. They provide a simple system to assess mechanisms of developmental identity established by tissue determinants. Each lobe contains a lineage expressing the hormone precursor pro-opiomelanocortin (POMC): the corticotropes and melanotropes. The T-box transcription factor Tpit controls terminal differentiation of both lineages. We now report on the unique role of Pax7 as a selector of intermediate lobe and melanotrope identity. Inactivation of the Pax7 gene results in loss of melanotrope gene expression and derepression of corticotrope genes. Pax7 acts by remodeling chromatin and allowing Tpit binding to a new subset of enhancers for activation of melanotrope-specific genes. Thus, the selector function of Pax7 is exerted through pioneer transcription factor activity. Genome-wide, the Pax7 pioneer activity is preferentially associated with composite binding sites that include paired and homeodomain motifs. Pax7 expression is conserved in human and dog melanotropes and defines two subtypes of pituitary adenomas causing Cushing's disease. In summary, expression of Pax7 provides a unique tissue identity to the pituitary intermediate lobe that alters Tpit-driven differentiation through pioneer and classical transcription factor activities.
Selector genes were defined by genetic analyses of Drosophila development as cell-autonomously acting genes that provide early patterning and positional identity, typically within developmental domains or compartments such that later differentiation is specified as a result of selector gene action (Garcia-Bellido 1975; Irvine and Rauskolb 2001). The early positional identity provided by selector genes thus creates different cellular environments on which terminal differentiation mechanisms and regulators act to yield alternate fates. In mammals, the importance of selector genes has been particularly noted in brain and spinal cord development, where their regional expression restricts the differentiation potential of expressing domains. For example, the paired homeodomain (HD) transcription factors Pax3 and Pax7 confer dorsal identity in the spinal cord (Mansouri and Gruss 1998), whereas the related Pax6 defines a more ventral fate (Ericson et al. 1997).
The molecular mechanism of selector gene action has remained elusive, since this concept has rarely been brought down to specific cell differentiation events. Over the last decade, models of combinatorial transcription factor action have been proposed for the control of cell-specific differentiation-dependent genes (Heinz et al. 2010). These models have highlighted the concerted, if not cooperative, action of many transcription factors but do not readily account for the initiation of new gene expression programs. In contrast, the concept of pioneer transcription factors developed in recent years may well correspond to the genetic concept of selector genes, since pioneer factor recruitment to regions of less accessible chromatin is thought to increase accessibility to chromatin and targets of other transcription factors (Zaret and Carroll 2011).
The pituitary gland is a relatively simple endocrine organ that is composed of six secretory cell types. Critical transcription factors, such as Pit1, SF1, and Tpit, have been identified that direct terminal differentiation into different lineages through alternate binary cell fate choices (Drouin 2010). These cell fate choices are mutually exclusive, with the apparent exception of two lineages expressing the same hallmark hormone-coding gene: pro-opiomelanocortin (POMC). Indeed, POMC is expressed in one lineage of the anterior pituitary lobe (AL), the corticotropes, whereas the intermediate lobe (IL) is comprised of the other POMC-expressing lineage, the melanotropes. Beyond the expression of POMC, cells of both lineages have quite different identities, as they process the POMC protein precursor differently and regulate its expression in response to different signaling pathways. There is currently no known mechanism that accounts for the different identities of the two POMC-expressing lineages.
The two POMC-expressing lineages secrete different biologically active peptides because corticotropes express only proconvertase 1 (PC1), which processes POMC into adrenocorticotrophin (ACTH) among other peptides, whereas melanotropes express both PC1 and PC2 and further process ACTH into α-melanostimulin (αMSH). In addition to these differences, the two lineages are marked by the restricted expression of membrane receptors, such as the corticotropin-releasing hormone (CRH) and vasopressin (AVP) receptors in corticotropes and the dopamine D2 receptor (Drd2) in melanotropes (Cote et al. 1986; Papadimitriou and Priftis 2009; Drouin 2010).
Differentiation of the two POMC lineages does, however, depend on the same transcription factor, Tpit, but at different times in development (embryonic day 12.5 [E12.5] for the AL and E15.5 in the IL). We identified Tpit as a T-box-containing transcription factor that is highly restricted to POMC-expressing cells of the pituitary and is critical for POMC transcription (Lamolet et al. 2001). Tpit gene inactivation leads to near complete loss of POMC expression in both melanotropes and corticotropes (Pulichino et al. 2003b), and cells destined to become POMC cells fail to reach terminal differentiation. Most of these cells remain blocked in a noncycling precursor state (Bilodeau et al. 2009), although a few cells do appear to differentiate into POMC-expressing cells, and ∼10% differentiate into an alternate pituitary cell fate: gonadotropes. These alternate cell fates appear to be determined, at least in part, by the antagonistic actions of Tpit and SF1 (Pulichino et al. 2003b). TPIT mutations cause isolated ACTH deficiency in children, an often fatal condition that is fully rescued by hormone replacement therapy (Lamolet et al. 2001; Pulichino et al. 2003a; Vallette-Kasic et al. 2005), and this is faithfully mimicked by Tpit loss of function in mice.
A screen for transcription factors differentially expressed between the pituitary IL and AL led us to identify Pax7 as highly restricted to the IL. The present report assessed the role of Pax7 in IL melanotropes, and we found that Pax7 is a critical positive regulator of the melanotrope fate. Pax7 is not sufficient for melanotrope differentiation, as this requires Tpit. However, Pax7 is critical for activation of melanotrope-specific genes and repression of corticotrope-specific genes. Pax7 thus acts as a selector switch between the two alternate POMC lineages that are otherwise both dependent on Tpit for terminal differentiation. While Pax7 drives pituitary progenitors into differentiation, its action appears independent of progenitor cell cycle exit. At melanotrope-specific genes, Pax7 increases the chromatin accessibility of the regulatory sequences (enhancers) allowing for Tpit recruitment and action, thus acting as a pioneer factor and selector gene for differentiation.
Results
Pax7 is specifically expressed in pituitary melanotropes
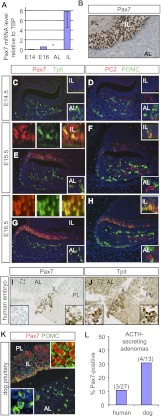
Expression profiling of transcription factors during pituitary ontogenesis led us to identify Pax7 for its preferential expression in the IL. Analysis of mouse pituitary RNA by RT-qPCR confirmed the highly restricted expression of Pax7 in the adult IL compared with ALs (Fig. 1A). Furthermore, Pax7 mRNA was detectable in E16 pituitaries but not at E14, in agreement with IL melanotrope differentiation within that developmental interval. By immunohistochemistry (IHC), Pax7 was found only in IL melanotropes and no other pituitary cells (Fig. 1B). In order to correlate Pax7 expression with cell differentiation in the developing pituitary, its expression was assessed by immunohistofluorescence together with Tpit. At E14.5 of mouse embryonic development, Tpit is already expressed in AL corticotropes, but neither Tpit nor Pax7 is expressed in ILs (Fig. 1C). By the next day (E15.5), most IL nuclei express Pax7, and a subset of those express Tpit, indicating that Pax7 expression precedes Tpit (Fig. 1E). At E16.5, most IL cells coexpress Pax7 and Tpit (Fig. 1G). The expression of these transcription factors is correlated with expression of POMC in both the AL and IL (Fig. 1D,F,H). In addition, IL expression of PC2 starts at E15.5, following expression of Tpit (Fig. 1F). PC2-positive cells of the AL at E14.5–E16.5 are presumptive gonadotropes. The onset of Pax7 expression in IL melanotropes thus precedes the terminal differentiation revealed by Tpit, POMC, and PC2 expression.
Figure 1.
Pax7 expression is restricted to pituitary melanotropes. (A) RT-qPCR analysis of Pax7 mRNA (relative to TBP) in mouse embryonic (E) and adult AL or IL pituitaries. (*) Below detection. Data are presented as mean ± SEM (n = 3). (B) Pax7 expression in the frontal section of the adult mouse pituitary revealed by IHC. Nuclear Pax7 protein is only detected in IL melanotropes. (PL) Posterior lobe. (C–H) Pax7 expression during pituitary development (E14.5, E15.5, and E16.5) revealed by immunofluorescence and colabeling for Tpit, PC2, or POMC as indicated. (I,J) Expression of immunoreactive human Pax7 (I) in Tpit-positive cells (J) of IL remnants of the 25-wk human pituitary. (Insets) At this stage, the human IL is regressing, but the remaining tissue marked by Tpit is positive for Pax7, whereas Tpit-positive cells of the AL are negative for Pax7. (K) Immunohistochemical localization of nuclear Pax7 (red) in ILs of dog pituitaries that are also positive for POMC, whereas AL POMC-positive cells (green) are Pax7-negative. (L) Frequency of Pax7-positive cells in corticotrope adenomas from Cushing's disease patients or dogs.
Pax7 in human/dog pituitary and corticotrope adenomas (Cushing's disease)
The human adult pituitary does not have an IL because this tissue regresses after the 15th week of gestation; it is nonetheless interesting to find out whether Pax7-expressing pituitary cells are present in humans, since their POMC cells expand to form adenomas that cause Cushing's disease, and this results in excessive and uncontrolled ACTH and glucocorticoid (Gc) production (Drouin et al. 2007). Typically, these adenomas are resistant to Gc feedback, and hence they may be of IL or AL origin if corticotropes have developed Gc resistance (Bilodeau et al. 2006). We identified Pax7 and Tpit-positive cells in IL remnants of a 25-wk human embryo (Fig. 1I,J), indicating that the human fetal IL normally expresses Pax7. Cushing's disease is more frequent in dogs than in humans, and hence we showed that the dog pituitary has a distribution of Pax7-expressing IL cells similar to the mouse (Fig. 1K). Using a panel of human and dog Cushing adenomas that we previously investigated for mechanisms of Gc resistance (Bilodeau et al. 2006; Roussel-Gervais et al. 2010), we show that ∼10% of human and 30% of dog Cushing adenomas express Pax7 with varying penetrance (Fig. 1L; Supplemental Fig. S1). It will be interesting to investigate the clinical significance of Pax7 expression in this subset of adenomas and, in particular, its putative usefulness for the development of therapeutic strategies that rely on melanotrope-specific functions, such as expression of dopamine receptors.
Pax7-dependent expression of melanotrope-specific markers
The highly restricted expression profile suggests that Pax7 might be an important regulator of the melanotrope cell fate. We addressed this question using Pax7 knockout mice. These mice die during the perinatal period if bred in a C57bl/6 background. On a 129/sv background, however, they survive up to 30 d after birth but die shortly after for as-yet-to-be-determined reasons (Kuang et al. 2006). At that age, homozygous (−/−) mice are much smaller than their wild-type or heterozygous (+/−) littermates and, accordingly, display reduced body weight. This general size reduction affected the pituitary roughly within the same proportion (Supplemental Fig. S2A).
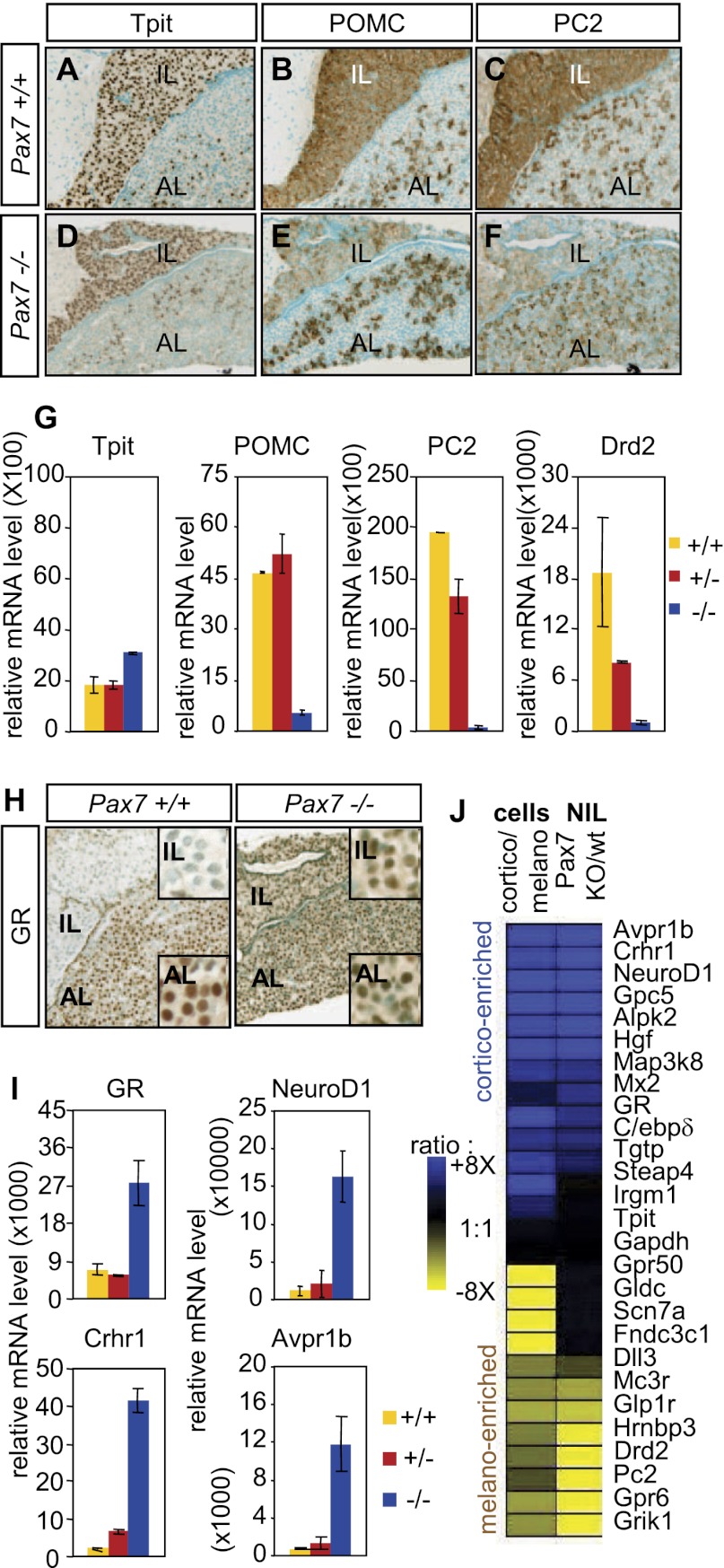
Lineage markers were used in order to assess the differentiation status of IL cells in Pax7−/− pituitaries. Expression of the POMC cell-restricted factor Tpit is unaffected by the Pax7 mutation, suggesting that IL cells are still directed toward a POMC lineage (Fig. 2, cf. D and A). However, expression of POMC as assessed by IHC is markedly reduced in the IL but not in AL corticotropes (Fig. 2B,E). Similarly, expression of PC2 is down to background level in IL Pax7−/− pituitaries, while its expression is unaffected in AL gonadotropes (Fig. 2C,F). We used postnatal day 24 (P24) dissected neurointermediate lobes (NILs) from normal and Pax7−/− pituitaries in order to quantify by RT-qPCR the effect of Pax7 deficiency on melanotrope markers (Fig. 2G). These analyses confirm that Tpit expression is not altered by the Pax7 mutation. In contrast, POMC mRNA levels are decreased 8.6-fold in Pax7−/− NILs compared with either wild-type or Pax7+/− pituitaries. An even greater loss of PC2 expression (56-fold) was observed in Pax7−/− NILs with a slight decrease in PC2 mRNA levels in Pax7+/− NILs. And similarly, Drd2 expression is 13-fold lower in Pax7−/− NILs compared with wild type. Collectively, these analyses indicate a loss of melanotrope-specific gene expression (POMC, PC2, and Drd2) but without affecting expression of the lineage marker Tpit.
Figure 2.
POMC cell type switch in the Pax7 knockout intermediate pituitary. Loss of melanotrope (A–G) and gain of corticotrope (H–J) markers. (A–F) Immunohistochemical analysis of adult (P24) Pax7−/− pituitaries. (A,D) Nuclear Tpit expression is maintained in Pax7−/− IL cells. (B,E) Expression of POMC is strongly reduced in Pax7−/− ILs but unchanged in ALs. (C,F) PC2 is decreased to background level in Pax7−/− IL. (G) RT-qPCR quantification (±SEM, n = 3) of melanotrope marker mRNAs in the neurointermediate pituitary (NIL) of the indicated Pax7 genotypes (relative to GAPDH). Pax7 inactivation results in decreased POMC, proconvertase PC2, and Drd2 mRNAs (P < 0.05 for each +/+ vs. −/− comparison). (H) GR expression is up-regulated in the Pax7−/− IL as assessed by immunoperoxydase histochemistry. (I) RT-qPCR quantification of NIL mRNA content (relative to GAPDH) for corticotrope markers GR, NeuroD1, Crhr1 and Avpr1b (P < 0.05 for each +/+ vs. −/− comparison). (J) The transcriptional signature of the Pax7−/− NIL is similar to a corticotrope, rather than melanotrope, profile. Genes were selected from expression array data according to their preferential expression in either melanotropes (yellow) or corticotropes (blue), and their expression was assessed by RT-qPCR in Pax7−/−NILs (pools of four animals per genotype). Expression ratios (color-coded) are shown for Pax7 knockout over wild-type ILs and compared with expression ratios for the same genes in FACS-sorted POMC-EGFP corticotropes over melanotropes.
Melanotropes switch to a corticotrope fate in the absence of Pax7
The Pax7−/− IL cells still express POMC but have lost important melanotrope features. They could be blocked in a partially differentiated state or may have switched to another cell fate. In order to address this, we first ruled out expression of other pituitary hormones in the Pax7−/− IL (Supplemental Fig. S2B). Alternatively, the POMC-expressing cells of the Pax7−/− IL may have adopted a corticotrope-like fate. AL corticotropes are marked by expression of the Gc receptor (GR) in contrast to the GR-negative melanotropes; we used IHC to reveal the GR in pituitary sections. As expected, the GR is widely expressed in the AL but not in the IL of normal pituitaries. In contrast, the GR is readily detectable in the Pax7−/− IL (Fig. 2H). We used RT-qPCR analysis of mRNA from wild-type and Pax7−/− NILs in order to quantify expression of corticotrope markers. These analyses indicated a fourfold up-regulation of the GR mRNA in the Pax7−/− NILs compared with wild type or heterozygotes (Fig. 2I). The basic helix–loop–helix (bHLH) transcription factor NeuroD1 is restricted to pituitary AL cells—in particular, corticotropes—during development (Drouin 2010). Interestingly, NeuroD1 expression was increased 14-fold in the Pax7−/− NIL, consistent with the idea that the Pax7−/− melanotropes might have acquired corticotrope features. This idea was further supported by assessment of CRH receptor 1 (CRH-R1) and AVP receptor 1b (AVP-R1b) expression in the mutant NIL (Fig. 2I). Expression of these two membrane receptors is normally restricted to AL corticotropes, and their significant up-regulation in the Pax7 mutant NIL supports the interpretation that Pax7−/− IL cells have switched to a corticotrope fate.
In order to better appreciate the extent of this fate change, we compared these results with expression data obtained from purified corticotropes and melanotropes. Expression profiles derived using FACS-sorted cells from POMC-EGFP transgenic mice (Lavoie et al. 2008) were used to select a panel of genes that are enriched in either corticotropes or melanotropes (Fig. 2J). Including the known markers mentioned above, we considered 26 expressed genes. Among these, 13 genes showed preferential expression in corticotropes (cortico-enriched) (blue in Fig. 2J). Concordantly, 11 of these showed higher expression in Pax7−/− than in wild-type ILs (Fig. 2J). Of the 12 genes that are preferentially expressed in melanotropes (melano-enriched) (yellow in Fig. 2J), eight displayed decreased expression in Pax7−/− ILs. Expression of the remaining four melanotrope-enriched genes (Gpr50, Gldc, Scn7a, and Fndc3cl) was not affected by loss of Pax7, suggesting that there may be other determinants of melanotrope identity in addition to Pax7. Other transcription factors or transcriptional coregulators may thus contribute together with Pax7 to the expression of melanotrope-specific genes; this may also account for the quantitative rather than absolute changes in expression profiles observed in the Pax7−/− IL. Notwithstanding, the switch of expression signature observed in the Pax7−/− IL supports the conclusion that Pax7 is an important determinant of melanotrope versus corticotrope cell identity.
Relative roles of Pax7 and Tpit in differentiation
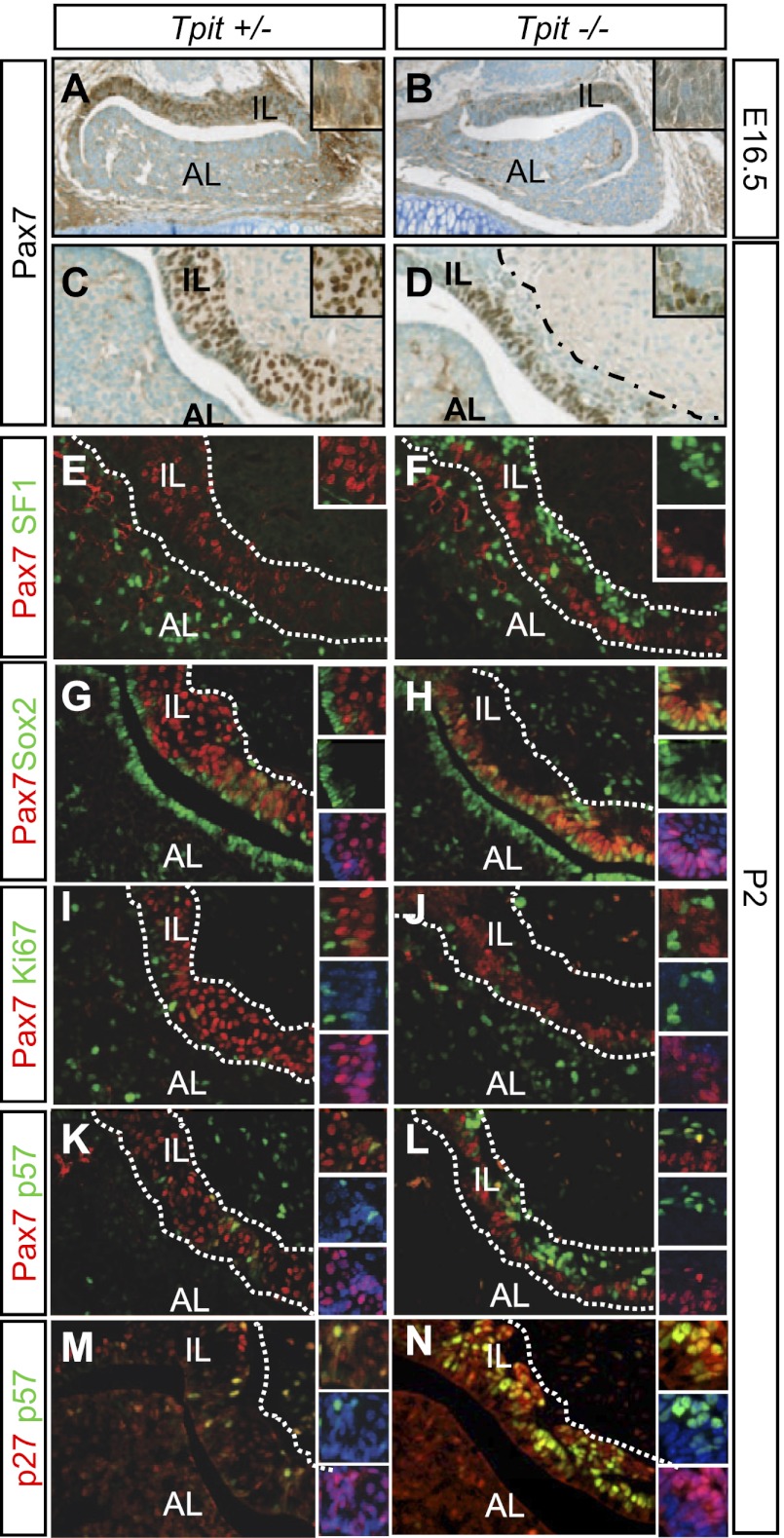
In contrast to the role of Pax7 as a positive factor for the melanotrope program and a negative regulator of the corticotrope fate, Tpit is essential for differentiation into both cell fates, since Tpit−/− pituitaries fail to express POMC, and cells destined for these lineages mostly remain undifferentiated (Pulichino et al. 2003b). An upstream action of Tpit would be consistent with its maintained expression in Pax7−/− pituitaries (Fig. 2A,D), and accordingly, expression of Pax7 is decreased, but still present, in developing ILs of Tpit−/− E16.5 pituitaries (Fig. 3A,B). However, Pax7 is expressed before Tpit in the developing IL (Fig. 1E). In postnatal (P2) Tpit−/− ILs, Pax7 expression is restricted to cells on the ventral/luminal side, with more dorsal cells appearing completely negative for Pax7 expression (Fig. 3, cf. D and C). This pattern of Pax7 expression is thus more consistent with independent activation of Pax7 and Tpit expression and with Tpit exerting a positive feedback on Pax7 expression.
Figure 3.
Altered differentiation and precursors in Tpit knockout ILs. (A–D) Pax7 expression in Tpit knockout pituitaries. (A,B) Pax7 is expressed, but less, in cells of the E16.5 Tpit−/− IL. (C,D) After birth (P2), Pax7 expression only marks the ventral half of the Tpit−/− IL. Dotted lines demarcate the IL. (E–J) Fate of Tpit−/− IL cells. (E,F) Colabeling for SF1 and Pax7 reveals IL SF1-positive cells only on the dorsal side of the Tpit−/− IL that is mutually exclusive with Pax7 labeling. (G,H) Colabeling for Pax7 and Sox2 of P2 pituitary sections from Tpit+/− (identical to +/+) and Tpit−/−. (I,J) Ki67-positive cells is rare in both control and Tpit mutant ILs. (M,N) The bulk of Tpit+/− IL cells express p27Kip1 and rarely p57Kip2, whereas many cells coexpress these two Cip–Kip cell cycle inhibitors in the dorsal Tpit−/− IL. (K,L) Whereas few cells express p57Kip2 in normal P2 ILs (some coexpressing Pax7), the bulk of dorsal Tpit−/− IL cells express high levels of p57Kip2, in contrast to the ventral Pax7-positive cells that rarely do so.
Prior work had shown that the bulk of adult Tpit−/− IL cells is blocked in a precursor state and that 10%–15% of the IL cells switch fate to become gonadotropes, as marked by expression of SF1 (Pulichino et al. 2003b; Bilodeau et al. 2009). At P2, cells that have undergone this fate switch are essentially observed on the dorsal side of the Tpit−/− IL, and SF1 expression appeared to be mutually exclusive with Pax7 (Fig. 3E,F). Thus, most dorsal cells of the Tpit−/− IL do not express Pax7 and have differentiated into gonadotropes. It is likely that these cells expressed Pax7 earlier, as most IL cells express Pax7 in early development, albeit at a lower level in Tpit−/− pituitaries (Fig. 3B).
Pax7 and progenitor cell cycle exit
The restriction of Pax7-positive cells on the luminal side of the Tpit−/− IL is intriguing, since a population of Sox2-expressing pituitary progenitors is localized around the lumen of the adult gland (Fauquier et al. 2008). We used coimmunofluorescence for Sox2 and Pax7 in order to define the relationship between pituitary stem cells and IL Pax7-positive cells. In P2 pituitaries, Sox2-positive cells are present around the lumen of the gland, as described previously, and Pax7-positive cells occupy the remainder of the IL (Fig. 3G). A few scattered cells are positive for both markers, suggesting that the transition from progenitor to putative precursor is accompanied by overlapping Sox2 and Pax7 expression. In the Tpit−/− IL, this overlap is widespread, with most Pax7-positive cells retaining Sox2 expression (Fig. 3H). Interestingly, the Pax7-negative cells of the Tpit−/− IL do not express Sox2. In order to assess whether Pax7/Sox2 double-positive cells are still in cell cycle, we performed colabeling for Pax7 and Ki67; this revealed few Ki67-positive cells and little overlap in either normal or Tpit−/− ILs (Fig. 3I,J), indicating that Pax7 cells have mostly exited the cell cycle in both genotypes.
In view of the apparent failure of IL cells to fully exit their progenitor state in Tpit−/− pituitaries (i.e., they maintain Sox2 in the presence of Pax7) and of the role of p57Kip2 as the cell cycle inhibitor driving pituitary progenitors out of cell cycle (Bilodeau et al. 2009), we assessed expression of Pax7 in relation to p57Kip2 and p27Kip1 in Tpit−/− ILs. p57Kip2 expression was mostly observed in the dorsal Pax7-negative cells of the Tpit−/− ILs (Fig. 3K,L) that have undergone a fate switch to SF1-positive gonadotropes (Fig. 3E,F). The Sox2/Pax7 double-positive cells do not appear to express either p57Kip2 or p27Kip1 (Fig. 3K–N). Interestingly, the distribution of p57Kip2, p27Kip1, and Sox2 expression was unaffected in the Pax7−/− ILs (Supplemental Fig. S3A), suggesting that Pax7, in contrast to Tpit, is not involved in cell cycle progression of pituitary cells. Furthermore, Pax7−/− IL cells do not exhibit changes in proliferation or apoptosis, as assessed by Ki67 or cleaved caspase3 labeling (Fig. 3I,J; data not shown). In normal development, Tpit is expressed later than Pax7 (Fig. 1E), and p57Kip2 expression starts earlier than either in the IL (Bilodeau et al. 2009). In support of this sequence, we observed some cells double-positive for p57Kip2 and Pax7 in the E16.5 IL (Supplemental Fig. S3B–D) but no p57Kip2/Tpit double-positive cells (Supplemental Fig. S3E–G). Thus, Tpit and Pax7 play different roles in the progression of pituitary progenitors toward differentiation and in the repression of the progenitor state.
Pax7 and pituitary differentiation
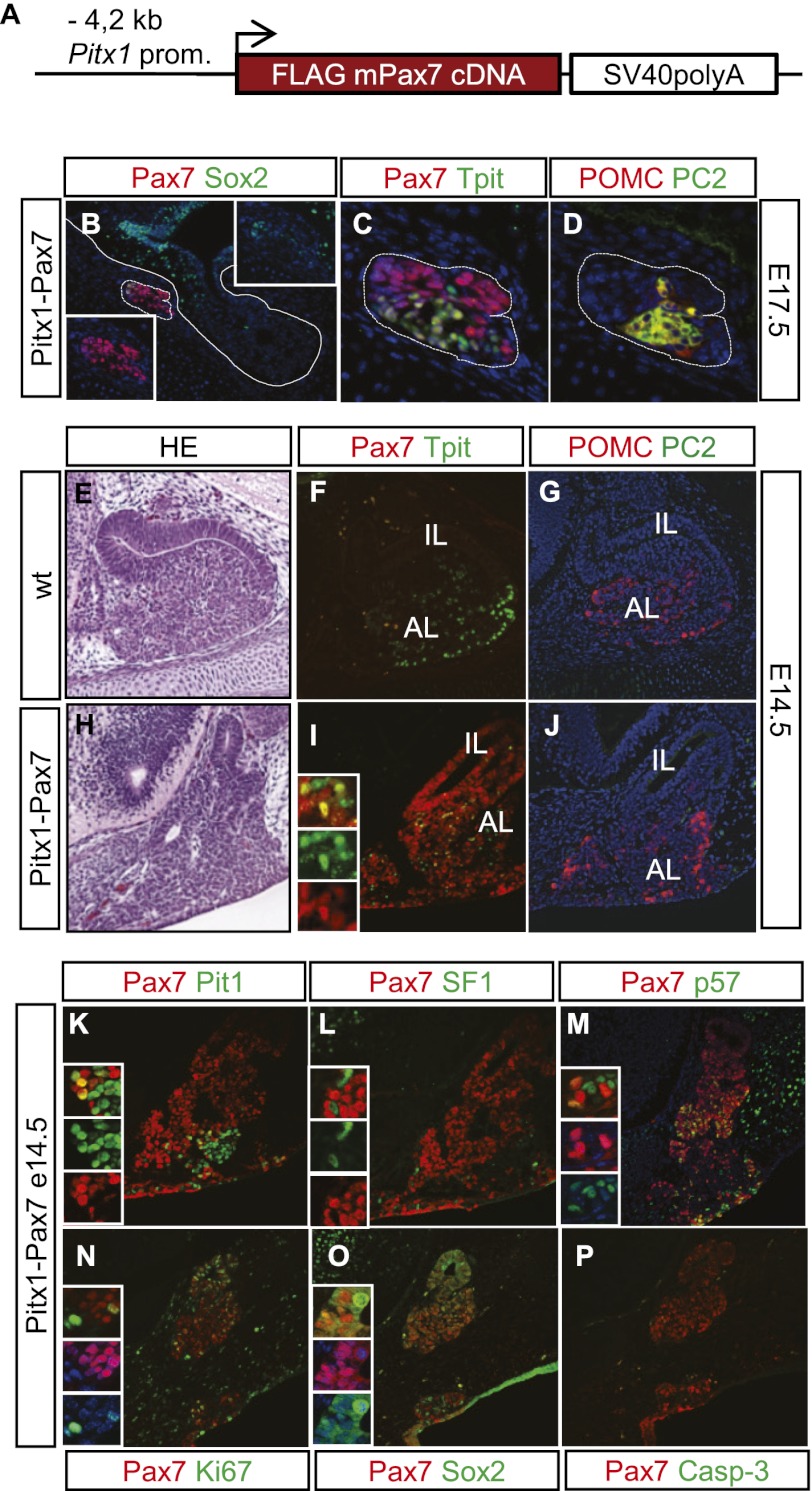
In order to explore further the role of Pax7 in engaging differentiation of pituitary progenitors, we designed a transgenic gain-of-function experiment where Pax7 expression is driven by the Pitx1 promoter (Fig. 4A). Pitx1 is expressed broadly within the oral ectoderm and pituitary primordium—much earlier than any pituitary differentiation event (Lanctôt et al. 1997). The Pitx1 promoter fragment used in these experiments exhibits appropriate expression, albeit with variable penetrance both spatially and temporally (Bilodeau et al. 2009). Of a total of 13 transgenic embryos, five showed transgene expression and phenotypes of varying severity. The most severely affected pituitary had very few cells remaining at E17.5 (Fig. 4B–D). Pax7 was expressed throughout this tissue (Fig. 4B,C), and some cells coexpressed Sox2 (Fig. 4B). Other cells coexpressed Tpit with Pax7 (Fig. 4C), and that is associated with expression of POMC and the melanotrope marker PC2 (Fig. 4D). Other transgenic pituitaries analyzed at either E14.5 or E17.5 revealed a mosaic pattern of Pax7 expression, indicating less penetrant transgene expression. While some transgenic pituitaries exhibited reduced pituitary tissue, one pituitary presented with expanded tissue extending all the way to the oral ectoderm (Fig. 4E,H) and extensive but not complete Pax7 expression (Fig. 4I). Both Pax7-positive and Pax7-negative cells exhibited cortico/melanotrope differentiation, as revealed by Tpit (Fig. 4I) and POMC (Fig. 4J) expression. Interestingly, transgenic expression of Pax7 in this E14.5 anterior pituitary did not cause premature expression of the melanotrope-specific PC2 (Fig. 4J). Some Pax7-positive and Pax7-negative cells have undergone somatotrope differentiation, as revealed by Pit1 (Fig. 4K) and GH expression (data not shown). Gonadotrope differentiation, marked by expression of SF1 (Fig. 4L) and LH (data not shown), is also observed, but SF1-positive cells never coexpress Pax7, suggesting that Pax7 expression is incompatible with gonadotrope differentiation; this is consistent with the mutually exclusive differentiation between POMC and gonadotrope lineages (Pulichino et al. 2003b).
Figure 4.
Pax7 drives pituitary progenitors into differentiation. (A) Schematic representation of the Pitx1–Pax7 transgene. (B–D) Pitx1–Pax7 E17.5 transgenic pituitary exhibiting severe loss of pituitary tissue. Of four transgenic pituitaries that exhibited loss of pituitary tissue, this transgenic founder retains only a small fragment of Pax7-positive (red, B) tissue. Most cells are Pax7-positive (C), and a fraction also expresses Sox2 (green; B). A subset of cells coexpress Pax7 and Tpit (C) together with POMC (red) and PC2 (green; D). (E–P) Transgenic embryo (E14.5) showing expansion of Pax7-positive pituitary tissue extending all the way to the oral ectoderm. (E,H) Hematoxilin/eosin stain reveals expanded pituitary tissue that is largely positive for Pax7 expression (red in I compared with F). Tpit-positive cells are present throughout this expanded pituitary (green; I), and these cells express POMC (J). (K) The Pax7-positive expanded pituitary has Pit1-positive (green) cells that sometime coexpress Pax7 (yellow). (L) In contrast, SF1-positive (green) cells within this transgenic Pitx1–Pax7 tissue are never found to coexpress Pax7. Expression of p57Kip2 (green/yellow; M), Ki67 (N), and Sox2 (O) does not correlate with expression of Pax7. (P) The transgenic tissue does not exhibit apoptosis, as assessed by cleaved caspase3 immunoreactivity.
Collectively, the ectopic expression of Pax7 during pituitary development appears to engage progenitors into differentiation pathways. Although Pax7 expression itself did not lead to complete differentiation, it appears permissive for cortico/melanotrope and Pit1-dependent, but not gonadotrope, differentiation. Transgenic pituitaries with expanded pituitary tissue exhibited more Pax7-negative cells than those with impaired pituitary expansion, suggesting that an early timing of Pax7 expression may prevent expansion of progenitors, whereas its later or less-penetrant expression may contribute to their expansion. Consistent with this, we found Pax7-negative and Pax7-positive cells that are also positive for either p57Kip2 (Fig. 4M) or Ki67 (Fig. 4N). These transgenic pituitaries also contain a mixture of Sox2-positive cells that are either Pax7-negative or Pax7-positive (Fig. 4O), but no apoptotic cells (Fig. 4P). By engaging progenitors into differentiation pathways, Pax7 expression thus appears to exhaust the progenitor pool in the most severe transgenic pituitaries (Fig. 4B–D). These gain-of-function experiments further support the model of Pax7 acting as an upstream selector gene for modulation of Tpit-driven differentiation.
Pax7 exerts its selector function through chromatin reprogramming
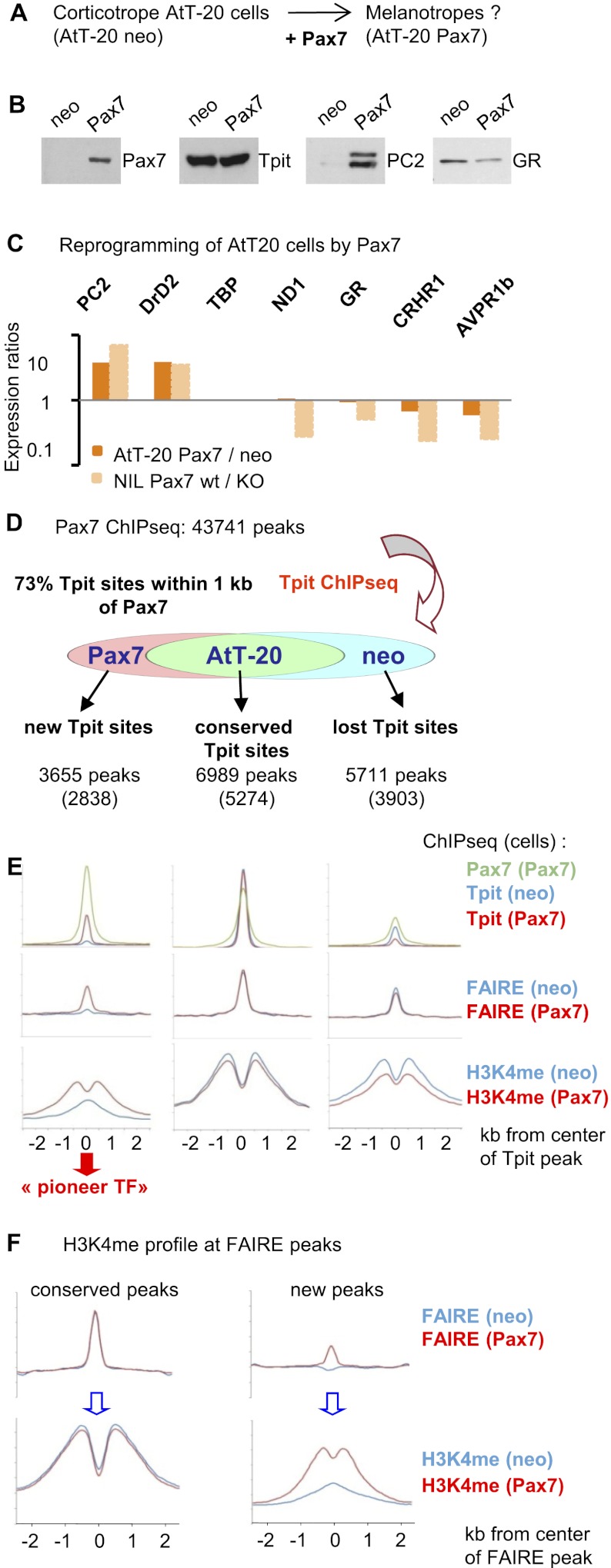
In order to determine how Pax7 may exert its selector function on cell differentiation, we expressed Pax7 in AtT-20 cells, an established tissue culture model of pituitary corticotrope cells. This was achieved using a retrovirus-mediated Pax7 vector and compared with cells infected with a similar empty vector conferring neomycin resistance. For each, pools of at least 103 colonies were expanded and characterized (Fig. 5A). These cells express Pax7, as ascertained by Western blot, and this expression does not affect Tpit levels but increased the melanotrope-specific PC2 and slightly decreased GR levels (Fig. 5B). In order to more broadly assess changes in gene expression elicited by Pax7, we used RT-qPCR to quantify expression of melanotrope- and corticotrope-specific genes in AtT-20Pax7 compared with control AtT-20neo cells (Fig. 5C). These analyses showed increased expression of melanotrope-specific genes such as PC2 and DrD2 and slight repression of corticotrope-specific markers such as CRH-R1, AVP-R1b, and GR. Pax7 expression thus exerts some reprogramming of AtT-20 cells with a major effect on activation of melanotrope genes and less repression of corticotrope genes when compared with their relative levels in the IL of Pax7−/− mice (Fig. 5C).
Figure 5.
Pax7 reprograms chromatin for Tpit action. (A) Experimental scheme. (B) Western blot analysis of pools of AtT-20 cells infected with empty retroviral vector (neo) or Pax7-expressing vector (Pax7) for expression of Pax7, Tpit, PC2, and GR as indicated. (C) RT-qPCR analysis of mRNA levels for the indicated melanotrope- or corticotrope-specific genes. Data are expressed as expression ratios in Pax7 over neo AtT-20 cells and are compared with the corresponding expression ratios in wild-type compared with Pax7−/− ILs. (D) Summary of relationships between Pax7 and Tpit ChIP-seq data. A large fraction (73%) of Tpit peaks is associated (within 1 kb) with Pax7 peaks. This association is similar for Tpit peaks that are conserved in AtT-20neo and AtT-20Pax7 cells and for Tpit peaks that are lost (neo) or gained in AtT-20Pax7 compared with neo cells. The total number of Tpit peaks is indicated below each subgroup, with the fraction showing association with Pax7 peaks indicated between parentheses. (E) Alignment of Pax7 and Tpit ChIP-seq, FAIRE-seq, and H3K4me1 ChIP-seq profiles for the three subgroups of Tpit peaks defined above. Data from AtT-20neo cells are shown in blue, and data for AtT-20-Pax7 cells are in red. Pax7 ChIP-seq profiles are shown in green. All data are aligned relative to Tpit peak positions, and the profiles include data for 2.5 kb on each side. Profiles were derived from the 500 best Tpit peaks in each group. (F) H3K4me1 profiles for FAIRE-seq peaks that are conserved (ratios between 0.8 and 1.2) in AtT-20neo compared with AtT-20Pax7 cells and for FAIRE-seq peaks that appear (ratio >3) in AtT-20Pax7 compared with AtT-20neo cells.
In order to investigate how Pax7 may reprogram AtT-20 cells, we conducted chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) for Pax7 in AtT-20Pax7 cells. Genome-wide, this led to the identification of 43,741 peaks of Pax7 recruitment with a P-value ≥10−6; their characterization is described below. Since Pax7 modulates POMC cell differentiation that is primarily dependent on Tpit, we assessed whether Pax7 altered Tpit action by performing Tpit ChIP-seq in both AtT-20neo and AtT-20Pax7 cells (Supplemental Fig. S4A). While the total number of Tpit sites (∼11,000) is similar in these cells, only 43% of all Tpit sites are the same (conserved sites) in both (Fig. 5D; Supplemental Fig. S4B). In contrast, ∼35% of the Tpit sites present in AtT-20neo cells were lost in AtT-20Pax7 cells: In general, these tend to be weaker sites of lesser P-value, although a subset of strong Tpit sites was completely lost. Finally, a last subset of new Tpit sites (22%) appeared in AtT-20Pax7 cells. Overall, 73% of Tpit sites are within 1 kb of a Pax7 site. In view of this close relationship, we investigated the effect of Pax7 on not only Tpit binding, but also chromatin surrounding the binding sites. We thus conducted FAIRE-seq (formaldehyde-assisted isolation of regulatory elements [FAIRE] and deep sequencing) (Giresi and Lieb 2009; Song et al. 2011) and ChIP-seq for histone H3 Lys 4 methyl-1 (H3K4me1) that constitutes chromatin marks of active regulatory sequences, particularly enhancers (Heintzman et al. 2007). While FAIRE reveals regions of accessible DNA, likely nucleosome-free, active enhancers are marked by a bimodal H3K4me1 signature. We assessed these chromatin marks at the different subgroups of Tpit peaks (Fig. 5E). Whereas conserved Tpit peaks present in both AtT-20neo and Pax7 cells exhibit the expected FAIRE-seq signal flanked by H3K4me1 ChIP-seq signals, similar but weaker signatures are also observed at Tpit sites that are lost in AtT-20Pax7 cells, consistent with the idea that these sites identify active regulatory sequences. The recruitment of Pax7 to these sites likely reflects a transcriptional modulatory role of Pax7. A similar chromatin signature (FAIRE peak with bimodal H3K4me1) appears at new Tpit sites induced by Pax7 (Fig. 5E). Strikingly, this signature is not present at these positions in AtT-20neo cells; in fact, they exhibit a single peak of H3K4me1 and no FAIRE signal (Fig. 5E). The appearance of active chromatin marks in Pax7-expressing cells is consistent with a pioneer transcription factor role of Pax7 that would open chromatin to allow Tpit binding. In contrast, these active chromatin marks are not erased at Tpit-binding sites that are lost in AtT-20Pax7, but their intensity is diminished, consistent with the greater ability of Pax7 to activate melanotrope genes than repress corticotrope genes (Fig. 5C). In order to assess genome-wide whether Pax7 leads to chromatin remodeling and whether chromatin opening revealed by FAIRE is generally correlated with the switch in H3K4me1 signature from single to double peaks, we investigated these marks at all FAIRE-seq peaks that are constant (i.e., ratio of 0.8–1.2 comparing AtT-20Pax7 over neo cells) and at positions where a FAIRE peak appears (ratio >3) in AtT-20Pax7 compared with neo cells. Thus, conserved FAIRE peaks are associated with constant H3K4me1 bimodal signatures, whereas the appearance of new FAIRE peaks is associated with a shift from single to bimodal H3K4me peaks (Fig. 5F). This switch in H3K4me1 profile is thus clearly associated with chromatin remodeling and, in this instance, Pax7-dependent pioneering action.
Pax7, a pioneer transcription factor
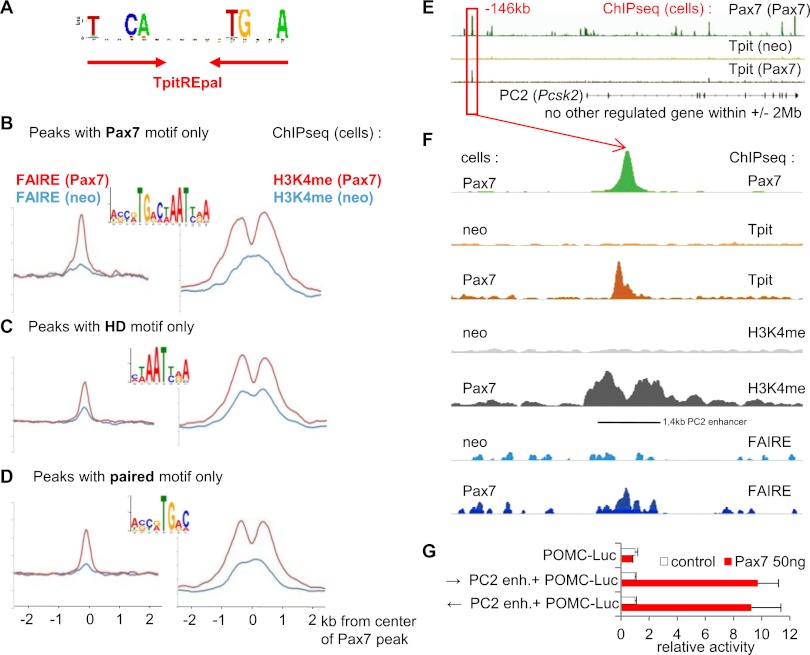
The action of Pax7 that allows new Tpit binding has the hallmark of so-called active pioneer factors (Zaret and Carroll 2011), since its activity is associated with remodeling of chromatin (Fig. 5E,F). In order to better define this activity, we analyzed our ChIP-seq data to identify Pax7 and Tpit DNA-binding sites. The search for conserved motifs present under the Tpit-binding peaks identified a predominant palindromic motif corresponding to the TpitREpal (Fig. 6A; Supplemental Fig. S4C) that we previously identified in the POMC gene −7-kb enhancer (Langlais et al. 2011). The TpitREpal contains two half-sites related to TCACACC separated by 2 base pairs (bp); a similar motif is present under 70% of Tpit-binding sites revealed by ChIP-seq (Supplemental Fig. S4D). The TpitREpal half-site corresponds to the previously described Brachyury-binding site (Lamolet et al. 2001).
Figure 6.
Pax7 acts as a pioneer transcription factor. (A) Sequence of the TpitREpal motif identified by de novo search of sequences (±50 bp) flanking the Tpit ChIP-seq peaks. (B) The full-length Pax7 motif identified by de novo search of Pax7 ChIP-seq data together with the associated FAIRE and H3K4me1 signals for the subgroup of peaks (±50 bp) that contain only the Pax7 motif. (C) The HD motif identified by de novo search of Pax7 ChIP-seq data together with FAIRE and H3K4me1 profiles present at peaks that contain only this motif and neither Pax7 nor a paired motif. (D) The paired motif identified by de novo search of Pax7 ChIP-seq data together with FAIRE and H3K4me1 profiles present at peaks that contain only the paired motif. (E) Overview of the PC2 locus showing Pax7 and Tpit ChIP-seq data in neo compared with Pax7 cells. The diagram represents ∼500 kb. (F) Blow-out of data around the −146-kb Pax7 peak upstream of the PC2 gene. ChIP-seq data are shown for Pax7 (green), Tpit (brown), H3K4me1 (gray), and FAIRE (blue) in neo and Pax7 cells. (G) Luciferase activity of a reporter containing a POMC promoter with/without the −146-kb PC2 enhancer (1.4-kb fragment) transfected in AtT-20 cells with/without the Pax7 expression vector as indicated.
The search for conserved motifs at Pax7 ChIP-seq binding sites revealed three motifs that are related to each other and to previously identified binding sites for other Pax factors (Soleimani et al. 2012). Indeed, a Pax7 motif (Fig. 6B; Supplemental Fig. S5A) comprised of juxtaposed paired and HD motifs is most prevalent at Pax7-only peaks (i.e., >3 kb away from Tpit sites) and at Pax7 sites associated with new Tpit sites (Supplemental Fig. S5B). As for the latter (Fig. 5E), the Pax7-only sites that have a Pax7 motif are globally associated with chromatin remodeling (and pioneer activity), as revealed by FAIRE and H3K4me1 profiles (Fig. 6B). A large number of Pax7 peaks do not have a complete Pax7 motif but rather either of its half-sites, as they contain only paired or HD motifs. Whereas Pax7 peaks that only have the HD motif are clearly associated with pre-existing active FAIRE and H3K4me1 marks (Fig. 6C), the peaks associated with paired-only sequences have weaker marks and less clearly bimodal H3K4me1 in AtT-20neo cells (Fig. 6D): This may reflect the weaker conservation of the HD half of the Pax motif and the associated difficulty for search algorithms to identify it. Hence, these data suggest that Pax7 interaction with the HD motif may not be sufficient for pioneer activity, but the conclusion is less clear for the paired motif. In contrast, the clear association of the full-length Pax7 motif with pioneer activity indicates that Pax7 interactions with DNA that involve both paired and HD domains represent the privileged mode of pioneering action.
The PC2 locus provides an interesting example of Pax7 action that allows Tpit recruitment and expression. The PC2 gene lies within a 4-Mb region that is devoid of Tpit-binding sites in AtT-20 cells and contains no other gene that has altered expression in the Pax7−/− IL (Fig. 6E). Pax7 binds several positions within this locus, but one Pax7-binding site located at −146 kb relative to the PC2 transcription start site (TSS) is very close to an induced Tpit-binding site (Fig. 6F). In fact, the well-conserved Pax7 site present at the Pax7 peak is 337 bp away from the TpitREpal present under the Tpit peak; there is also a smaller Tpit peak that colocalizes with the Pax7 peak. Consistent with a pioneering action of Pax7, binding of Pax7 results in the appearance of a FAIRE peak flanked by H3K4me1 peaks, indicative of chromatin remodeling (Fig. 6F). Since Tpit does not occupy its −146-kb target sequence in the PC2 locus in AtT-20neo cells, this suggests that Tpit does not have pioneer activity, in contrast to Pax7.
We assessed the putative enhancer properties of the 1.4-kb fragment that is delimited by the H3K4me1 profile in AtT-20Pax7 cells and that corresponds to conserved DNA sequences. When inserted upstream of the POMC promoter, this fragment was inactive, but Pax7 enhanced expression of the luciferase reporter, indicating that its activity requires Pax7 (Fig. 6G). These data suggest that Pax7 has at least two roles for transcription of the PC2 gene: first, a pioneer activity that opens chromatin structure at the −146-kb enhancer, and second, a canonical transcription factor activity when Pax7 is bound to the enhancer.
Discussion
Pax7, a selector gene that modulates Tpit-driven differentiation
The role of Pax7 in the differentiation of the two pituitary POMC lineages is unique in comparison with many other transcription factors that have been implicated in cell fate decisions. The role of Pax7 in this context may be the first example of the selector gene concept developed to define developmental fields >35 years ago by Garcia-Bellido (1975). In order to delineate this role, it is interesting to first discuss what Pax7 does not do in comparison with the large number of transcription factors that have been implicated in differentiation and cell fate decisions. First and foremost, Pax7 is not a pituitary differentiation gene, since its presence in the Tpit−/− IL is not sufficient to drive differentiation. Also, Pax7 does not play a maintenance role in the pituitary, as it does in muscle satellite cells (Oustanina et al. 2004; Lepper et al. 2009). Thus, the IL Pax7-positive cells are engaged in the differentiation pathway but need Tpit to complete the process. In contrast, Tpit is truly a differentiation gene, since Tpit−/− IL cells remain either nondifferentiated (Sox2/Pax7 double-positive) or differentiate into the alternate gonadotrope cell fate.
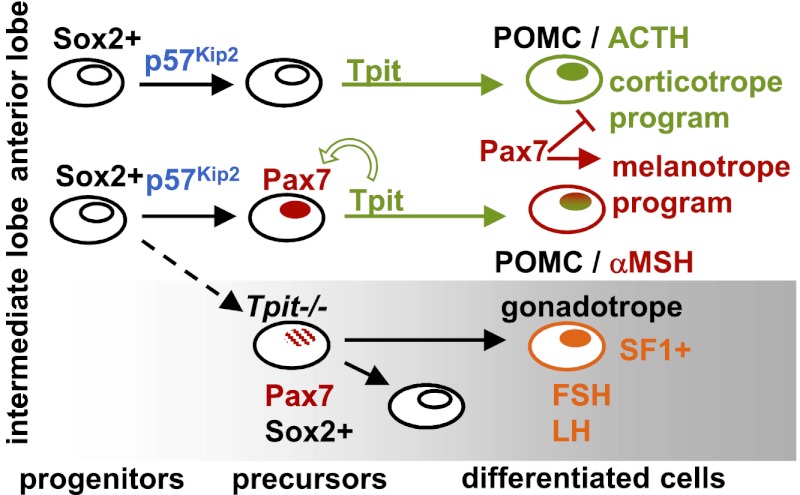
In normal IL development, progenitors would exit the cell cycle under the action of p57Kip2 and switch on Pax7 expression followed by Tpit (Fig. 7). Tpit exerts a positive feed-forward effect on Pax7 expression, and together, these factors set up a melanotrope-specific gene expression program. In the absence of Pax7, this program shifts to a corticotrope-like profile, in agreement with the importance of Tpit for AL corticotrope differentiation (Pulichino et al. 2003b). Pax7 thus has a dual role of activating the melanotrope and repressing the corticotrope programs (Fig. 7). In the absence of the terminal differentiation function provided by Tpit, the IL accumulates Pax7/Sox2 double-positive cells that either remain in an intermediate precursor state or switch fate to become SF1-positive gonadotropes (Pulichino et al. 2003b).
Figure 7.
Pax7, a selector gene that modulates for Tpit-driven differentiation. Schematic representation of differentiation for pituitary POMC lineages: the IL melanotropes (αMSH) and AL corticotropes (ACTH). Pax7 is expressed slightly before Tpit in the IL, thus marking this tissue compared with the anterior pituitary. Differentiation driven by Tpit then leads to expression of a melanotrope-specific genetic program in the IL, whereas Tpit differentiation leads to expression of corticotrope-specific genes in the anterior pituitary. In IL melanotropes, Pax7 is both a positive regulator of melanotrope-specific genes and a repressor of corticotrope genes. Tpit exerts a positive feedback effect for the maintenance of Pax7 expression in the IL. In Tpit−/− pituitaries, cells destined to become POMC either switch fate to acquired gonadotrope identity or remain blocked in a Pax7/Sox2 double-positive state in the intermediate lobe.
Pax7 thus fits well within the original description of selector genes as cell-autonomously acting genes that provide positional identity and define developmental fields. The highly restricted expression of Pax7 in the pituitary IL thus defines the identity of this tissue relative to the other pituitary lobes: the anterior and the posterior pituitaries. However, the contribution of Pax7 to the establishment of a unique cell-specific genetic program is critically dependent on and only realized after Tpit-driven terminal differentiation. In promoting melanotrope-specific genes and repressing corticotrope-specific genes, Pax7 provides tissue identity to the cell differentiation process, as postulated for early-acting developmental selector genes. The action of Pax7 is thus very different from that of terminal differentiation genes, such as Tpit. The role of Pax7 in IL pituitary cells represents the first well-defined selector role for a transcription factor that specifies compartment identity and modulates the differentiation process.
Selector function is exerted through pioneer action
A major effect of Pax7 as a selector gene is the activation of melanotrope-specific genes such as PC2 and DrD2. Interestingly, the mechanism underlying these actions is consistent with the concept of a pioneer transcription factor (Zaret and Carroll 2011). Indeed, Pax7 expression in AtT-20 cells results in many new Tpit-binding sites; for example, at the PC2 and DrD2 loci (Fig. 6E,F). These new Tpit-binding sites contain TpitREpal sequences that are indistinguishable from other Tpit-binding sites active in AtT-20 cells. The pioneer action of Pax7 at these putative regulatory sequences creates a chromatin environment that is permissive for Tpit binding. This environment includes a FAIRE signal (similar to DNase-1-hypersensitive regions) that has been associated with nucleosome-free regions (Song et al. 2011). Another hallmark of these regions is the presence of flanking peaks of H3K4me1 (Heintzman et al. 2007). This signature appeared in Pax7-expressing cells at putative enhancers that were previously silent but marked by a single shallow peak of H3K4me1 (Fig. 5E,F). This single peak may be a previously unrecognized mark of silent enhancers, such as those revealed by the presence of p300 (Heintzman et al. 2007). The shift in the profile of H3K4me1 observed when putative enhancers switch from silent to active may reflect an important role of the flanking H3K4me1 peaks in defining an “open” region of chromatin. The switch of the H3K4me profile was correlated with chromatin opening, as revealed by FAIRE independently of Pax7 or Tpit peak positions (Fig. 5F), highlighting their genome-wide correlation.
Pax factors have the ability to bind DNA through either or both of their DNA-binding domains: the paired and HD domains. It was recently reported that Pax7 may have a higher affinity for HD motifs than the related Pax3 and that this may account for the greater number of Pax7 targets (Soleimani et al. 2012). We found both paired and HD motifs associated with Pax7 peaks (in agreement with this study in muscle cells) but a high frequency of juxtaposed sites that define the Pax7 motif (Fig. 6B). It is noteworthy that many peaks have multiple copies of these motifs but with no specific spacing (Supplemental Fig. S5C). However, when we focused our analyses on peaks that contain only one of these motifs (based on search algorithms that are not fail-safe), we found the highest frequency of Pax7 motifs associated with peaks that exhibit chromatin opening (Fig. 6B) compared with Pax7 peaks that contain only HD motifs (Fig. 6C) and that have FAIRE and H3K4me1 profiles of accessible chromatin.
These data thus suggest that the Pax7 motif has a greater pioneering potential than HD and possibly paired domains on their own. Binding of Pax7 to these latter motifs may thus be associated with canonical transcription factor action, consistent with such a role for Pax7 action on the PC2 gene −146-kb enhancer (Fig. 6G).
While pioneer activity has been ascribed to Fox transcription factors (Zaret and Carroll 2011) and associated with epigenetic changes (Serandour et al. 2011), others have argued that combinatorial action of lineage-determining factors may suffice for cell fate determination (Heinz et al. 2010). Given that transcription factors may have both pioneer and canonical transcription activator functions, the association of preferred DNA motif-binding preferences and open chromatin profiles may be required to demarcate these two properties and their gene targets.
Materials and methods
Mice, histology, and IHC
Tpit mutant and POMC-EGFP transgenic mice were described previously (Pulichino et al. 2003b; Lavoie et al. 2008). Pax7 mutant mice (Mansouri and Gruss 1998) were provided by Mona Nemer. IHC was performed on formalin-fixed paraffin-embedded tissue sections, as described in Lamolet et al. (2001). Antibody sources and dilutions are indicated in Supplemental Table 1. For IHC, biotinylated anti-mouse or anti-rabbit secondary antibodies (Vector Laboratories) were revealed with HRP-coupled streptavidin (Perkin Elmer). Immunofluorescence was performed under the same conditions using fluorescent (Alexa 488 or Alexa 546) anti-mouse, anti-rabbit, or anti-guinea pig antibodies obtained from Molecular Probe.
Animal experimentation was approved by the IRCM Animal Ethics Review Committee. Studies using human samples were approved by the IRCM Human Ethics Review Committee.
RT-qPCR and ChIP
For analysis of Pax7 knockout NIL mRNAs, mice were sacrificed at P24, and RNA was prepared from dissected ALs and NILs. RNA was extracted using RNeasy columns according to the manufacturer's recommendations (Qiagen), and cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and purified using QIAquick columns. qPCR was performed using PerfeCta reagent (Quanta) on a MX-3005 device (Stratagene), and results were analyzed using the accompanying software. All quantifications were relative to GAPDH or TBP mRNA as stated in the figure legends. The primers used are provided in Supplemental Table 2.
ChIP and ChIP-seq were performed and analyzed as described (Langlais et al. 2012).
ChIP-seq peak-finding analysis
Sequence reads mapping to the mouse mm9 genome assembly were performed with Bowtie using the following settings: bowtie −t−trim5 1−best mm9 −S−chunkmbs 1024 (Langmead et al. 2009). About 95% of the sequence reads were mapped to the reference genome. To identify significant binding events, we processed the mapped sequence reads with MACS version 1.4.1 using a matching number of IgG control reads. We used the following setting in MACS: −to-small−bw 250-g mm−wig (Zhang et al. 2008). The MACS option –mfold was determined with the prior diag option. The complete number of sequence reads and peaks are listed in Supplemental Figure S4A. We kept peaks with a P-value >10−5 and a length <5 kb.
To compare Tpit genomic recruitment in AtT-20 cells versus AtT-20Pax7 cells, we used a similar number of reads (∼67 million reads after filtering) for each sample. To analyze the best subgroups of new, conserved, and lost Tpit sites, we ranked the peaks in each sample based on P-values and then compared their ranking.
We generated the density profiles with MACS using these attributes: −wig,−space 50.
De novo motif analyses
We extracted 100 bp of DNA sequence surrounding the binding peaks from the University of California at Santa Cruz Web site (Kent et al. 2002). The sequences of each cluster were processed using MEME (Bailey et al. 2006) and HOMER (Heinz et al. 2010). The following settings were used in MEME: −dna −nmotifs 5 −mod zoops −minsites 25. Graphical representation of the position weight matrices obtained from these analyses were generated with WebLogo (http://demo.tinyray.com/weblogo) (Crooks et al. 2004).
Transcription factor-binding motif searches
We searched for sequence motifs in 60-bp fragments surrounding ChIP-seq binding peaks using HOMER. The resulting occurrence for each motif was compared with the number of predicted binding sites in a set of 7311 randomly chosen genomic sequences.
Acknowledgments
We thank Guillaume Bourque (McGill/Genome Québec Innovation Center) for his advice on bioinformatic analyses, Dr. Michel Vekemans (Hôpital Necker, Paris) for human pituitary sections, and all members of the laboratory for their support and thoughtful comments. We are thankful to K. Morohashi (Fukuoka, Japan) and N.G. Seidah (IRCM) for providing the SF1 and PC2 antisera, respectively. The Pax7 monoclonal antibody developed by Dr. A. Kawakami was obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa. We thank E. Massicotte and M. Dupuis of the IRCM FACS Facility for sorting of POMC-EGFP cells. This work was supported by grants (to J.D.) from the Canadian Institutes of Health Research.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.200436.112.
References
- Bailey TL, Williams N, Misleh C, Li WW 2006. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 doi: 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Vallette-Kasic S, Gauthier Y, Figarella-Branger D, Brue T, Berthelet F, Lacroix A, Batista D, Stratakis S, Hanson J, et al. 2006. Role of Brg1 and HDAC2 in GR trans-repression of pituitary POMC gene and misexpression in Cushing disease. Genes Dev 20: 2871–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Roussel-Gervais A, Drouin J 2009. Distinct developmental roles of cell cycle inhibitors p57Kip2 and p27Kip1 distinguish pituitary progenitor cell cycle exit from cell cycle re-entry of differentiated cells. Mol Cell Biol 29: 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote TE, Felder R, Kebabian JW, Sekura RD, Reisine T, Affolter HU 1986. D-2 dopamine receptor-mediated inhibition of pro-opiomelanocortin synthesis in rat intermediate lobe. Abolition by pertussis toxin or activators of adenylate cyclase. J Biol Chem 261: 4555–4561 [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin J. 2010. Pituitary development. In The pituitary (ed. S Melmed), pp. 3–19. Elsevier-Academic Press, London. [Google Scholar]
- Drouin J, Bilodeau S, Vallette-Kasic S 2007. Of old and new diseases: Genetics of pituitary ACTH excess (Cushing) and deficiency. Clin Genet 72: 175–182 [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J 1997. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90: 169–180 [DOI] [PubMed] [Google Scholar]
- Fauquier T, Rizzoti K, Dattani M, Lovell-Badge R, Robinson IC 2008. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proc Natl Acad Sci 105: 2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A 1975. Genetic control of wing disc development in Drosophila. Ciba Found Symp 0: 161–182 [DOI] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD 2009. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods 48: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine KD, Rauskolb C 2001. Boundaries in development: Formation and function. Annu Rev Cell Dev Biol 17: 189–214 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA 2006. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol 172: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamolet B, Pulichino AM, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J 2001. A pituitary cell-restricted T-box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell 104: 849–859 [DOI] [PubMed] [Google Scholar]
- Lanctôt C, Lamolet B, Drouin J 1997. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development 124: 2807–2817 [DOI] [PubMed] [Google Scholar]
- Langlais D, Couture C, Sylvain-Drolet G, Drouin J 2011. A pituitary-specific enhancer of the POMC gene with preferential activity in corticotrope cells. Mol Endocrinol 25: 348–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais D, Couture C, Balsalobre A, Drouin J 2012. The Stat3/GR interaction code: Predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell 47: 38–49 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie PL, Budry L, Balsalobre A, Drouin J 2008. Developmental dependence on NurRE and EboxNeuro for expression of pituitary POMC. Mol Endocrinol 22: 1647–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM 2009. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Gruss P 1998. Pax3 and Pax7 are expressed in commissural neurons and restrict ventral neuronal identity in the spinal cord. Mech Dev 78: 171–178 [DOI] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T 2004. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J 23: 3430–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou A, Priftis KN 2009. Regulation of the hypothalamic-pituitary-adrenal axis. Neuroimmunomodulation 16: 265–271 [DOI] [PubMed] [Google Scholar]
- Pulichino AM, Vallette-Kasic S, Couture C, Gauthier Y, Brue T, David M, Malpuech G, Deal C, Van Vliet G, De Vroede M, et al. 2003a. Human and mouse Tpit gene mutations cause early onset pituitary ACTH deficiency. Genes Dev 17: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulichino AM, Vallette-Kasic S, Tsai JPY, Couture C, Gauthier Y, Drouin J 2003b. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev 17: 738–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel-Gervais A, Bilodeau S, Vallette S, Berthelet F, Lacroix A, Figarella-Branger D, Brue T, Drouin J 2010. Cooperation between cyclin E and p27Kip1 in pituitary tumorigenesis. Mol Endocrinol 24: 1835–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, et al. 2011. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani VD, Punch VG, Kawabe Y, Jones AE, Palidwor GA, Porter CJ, Cross JW, Carvajal JJ, Kockx CE, van Ijcken WF, et al. 2012. Transcriptional dominance of pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev Cell 22: 1208–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. 2011. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21: 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallette-Kasic S, Brue T, Pulichino AM, Gueydan M, Barlier A, David M, Nicolino M, Malpuech G, Déchelotte P, Deal C, et al. 2005. Congenital isolated adrenocorticotropin deficiency: An underestimated cause of neonatal death, explained by TPIT gene mutations. J Clin Endocrinol Metab 90: 1323–1331 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS 2011. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]