Phosphatases that can dephosphorylate the C-terminal domain (CTD) are of great interest because of the role of the phosphorylation state of the CTD in controlling Pol II in transcription. The CTD pSer5 phosphatase Ssu72 recognizes the cis configuration of the pSer5–Pro6 peptide bond and also has pSer7 phosphatase activity. This study reports a structural and enzymatic analysis of Ssu72 and a 10-mer pSer7 CTD peptide and finds that the pSer7 peptide is bound in the active site with the opposite orientation of the pSer5 peptide. The result is a novel mode of substrate recognition by Ssu72 and an explanation of how the enzyme can dephosphorylate pSer5 and pSer7.
Keywords: RNA polymerase, protein phosphatase, protein structure
Abstract
Ssu72, an RNA polymerase II C-terminal domain (CTD) phospho-Ser5 (pSer5) phosphatase, was recently reported to have pSer7 phosphatase activity as well. We report here the crystal structure of a ternary complex of the N-terminal domain of human symplekin, human Ssu72, and a 10-mer pSer7 CTD peptide. Surprisingly, the peptide is bound in the Ssu72 active site with its backbone running in the opposite direction compared with a pSer5 peptide. The pSer7 phosphatase activity of Ssu72 is ∼4000-fold lower than its pSer5 phosphatase activity toward a peptide substrate, consistent with the structural observations.
Transcription of mRNA and noncoding RNA in eukaryotes is carried out by RNA polymerase II (Pol II), the activity of which is regulated in part by the phosphorylation state of the C-terminal domain (CTD) of its largest subunit (Komarnitsky et al. 2000; Schroeder et al. 2000; Meinhart et al. 2005; Phatnani and Greenleaf 2006; Buratowski 2009; Kim et al. 2009, 2010; Mayer et al. 2010; Tietjen et al. 2010). The CTD contains the consensus heptapeptide repeat Tyr1–Ser2–Pro3–Thr4–Ser5–Pro6–Ser7 (YSPTSPS), and phosphorylation of Ser5 and Ser2 has long been recognized for its importance in transcription and coupled RNA processing events. Phosphorylation of Ser7 is involved in snRNA transcription and 3′ end processing (Chapman et al. 2007; Egloff et al. 2007), and phosphorylation of Thr4 has been linked to histone mRNA 3′ end processing (Hsin et al. 2011). CTD kinases and phosphatases control the phosphorylation state of the CTD and thereby regulate Pol II activity. For example, Fcp1 preferentially dephosphorylates pSer2 over pSer5 (Hausmann and Shuman 2002; Hausmann et al. 2004; Ghosh et al. 2008), while Ssu72 is a pSer5 phosphatase (Krishnamurthy et al. 2004; Hausmann et al. 2005). Both are essential for viability in yeast.
Recently, it was reported that Ssu72 also has pSer7 phosphatase activity (Bataille et al. 2012; Zhang et al. 2012). However, such an activity for Ssu72 is unexpected from a structural perspective. Ssu72 recognizes the cis configuration of the pSer5–Pro6 peptide bond as a pSer5 phosphatase (Xiang et al. 2010; Werner-Allen et al. 2011). In comparison, pSer7 is followed by Tyr1 in the next repeat of the CTD (designated Tyr1′). If Ssu72 were to bind the pSer5 and pSer7 substrates in the same way, the pSer7–Tyr1′ peptide bond would need to be in the cis configuration, which is much less favorable energetically. Moreover, the bulkier Tyr side chain would be placed in the binding site for the Pro6 residue in the pSer5 substrate, which would clash with the enzyme.
Results and Discussion
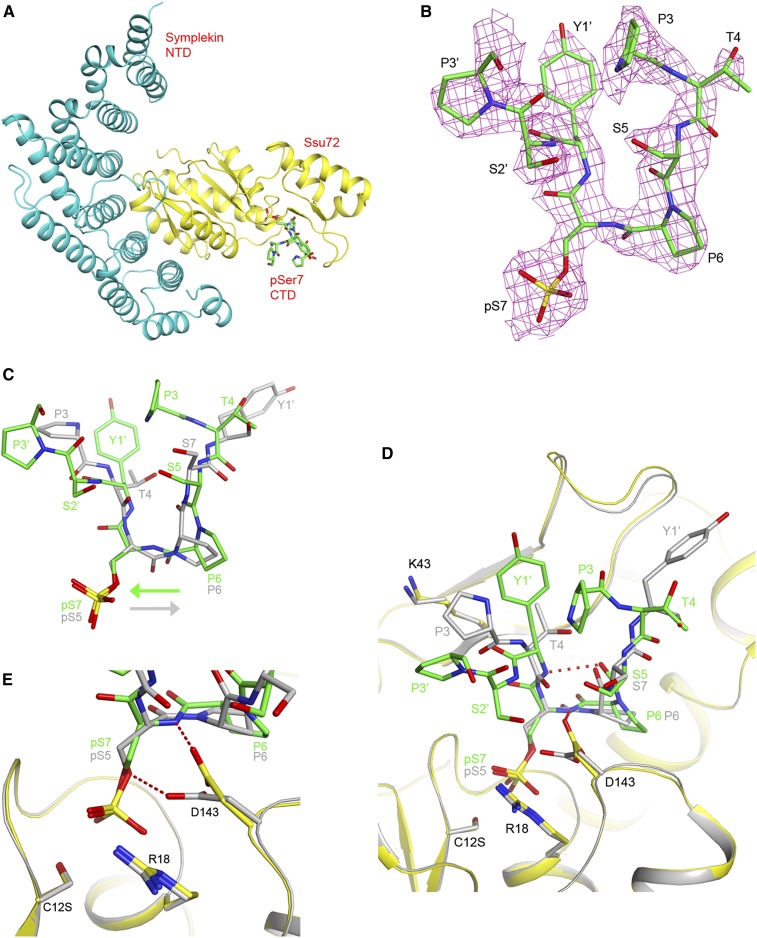
To understand the structural basis for how Ssu72 functions as a pSer7 phosphatase, we determined the crystal structure at 2.2 Å resolution of a ternary complex of a human symplekin N-terminal domain (NTD, residues 30–360), human Ssu72 (C12S mutant), and a 10-mer CTD peptide phosphorylated at Ser7 (Ser2–Pro3–Thr4–Ser5–Pro6–pSer7–Tyr1′–Ser2′–Pro3′–Thr4′, with the prime indicating the next repeat of the CTD) (Fig. 1A). Symplekin is a scaffold protein in the pre-mRNA 3′ end processing machinery (Takagaki and Manley 2000) and is known to interact with and enhance the activity of Ssu72 (Xiang et al. 2010). It was included here because it is important for the crystallization of human Ssu72 (Xiang et al. 2010). Crystals of the symplekin NTD–Ssu72 binary complex were soaked with the phosphopeptide, following the same protocol as used earlier for the pSer5 peptide (Xiang et al. 2010). Electron density was observed for most of the peptide based on the crystallographic analysis, except for one residue at either end of the peptide (Fig. 1B). In addition, the side chain of Tyr1′ has weak electron density.
Figure 1.
The Pol II CTD pSer7 peptide has a novel binding mode in the active site of Ssu72. (A) Schematic drawing of the structure of the ternary complex of the human symplekin NTD (cyan), human Ssu72 (C12S mutant; yellow), and the pSer7 CTD peptide (green). (B) Omit 2Fo − Fc electron density for the pSer7 peptide at 2.2 Å resolution, contoured at 0.8σ. (C) Overlay of the binding modes of the pSer7 peptide (in green) and the pSer5 peptide (in gray) (Xiang et al. 2010) in the active site of Ssu72. The directions of the peptide backbone are indicated by the arrows. (D) Overlay of the active site region of Ssu72 in the pSer7 complex (yellow) and the pSer5 complex (in gray). (E) Close-up of the active site region showing the structural differences in the pSer residues and Asp143. All of the structure figures were produced with PyMOL (http://www.pymol.org).
Surprisingly, the pSer7 peptide is bound in the Ssu72 active site with its peptide backbone running in the opposite direction compared with the pSer5 peptide (Fig. 1C). As a result, the binding sites for Pro6 and pSer7 are equivalent to those for Pro6 and pSer5 in the pSer5 peptide (Xiang et al. 2010). Residues Ser5–Pro6–pSer7–Tyr1′ assume the conformation of a type I β reverse turn, so that the overall shapes of the pSer7 and pSer5 peptides are similar when bound to Ssu72 (Fig. 1C). However, such a turn is wider than the cis proline structure, and a difference was observed in the conformation and position of Pro6 in the two peptides. On the other hand, the Pro6–pSer7 peptide bond and, in fact, all of the peptide bonds in the pSer7 peptide are in the trans configuration. The Tyr1′ residue in the pSer5 substrate is important for phosphatase activity (Hausmann et al. 2005) and shows interactions with Ssu72 (Fig. 1D; Xiang et al. 2010). Equivalent interactions are absent in the pSer7 substrate complex (Fig. 1D).
Recognition of both orientations of peptide substrates or ligands has rarely been observed with other proteins. Reported examples include the SH3 domain (Feng et al. 1994; Lim et al. 1994), stromelysin-1 (Becker et al. 1995), thioredoxin (Qin et al. 1996), and the adaptor protein SspB of the AAA+ protease ClpXP (Levchenko et al. 2005). As far as we are aware, this is the first time that a protein phosphatase has been shown to bind both orientations of its peptide substrate.
The pSer7 residue is anchored in the catalytic site and shows interactions with Ssu72 similar to those of the pSer5 residue (Fig. 1D; Xiang et al. 2010). Most of the Ssu72 residues in the active site have the same conformation in the pSer7 and pSer5 peptide complexes. One notable difference is in the side chain of Asp143, the general acid of the phosphatase reaction. The oxygen atom that should donate a proton to the leaving group (Oγ of Ser7) is instead hydrogen-bonded to the main chain amide of Ser7 (Fig. 1E). In contrast, such a hydrogen bond is not possible with the pSer5 substrate, as the pSer5–Pro6 peptide bond does not have an amide hydrogen. Moreover, the Cβ–Oγ bond of Ser7 is at a right angle to the Asp143 side chain (Fig. 1E), possibly due to the reversal of the backbone direction, and therefore the Oγ atom may not be in the optimal conformation for receiving the proton from Asp143. Both of these differences may be detrimental for the phosphatase activity with the pSer7 substrate.
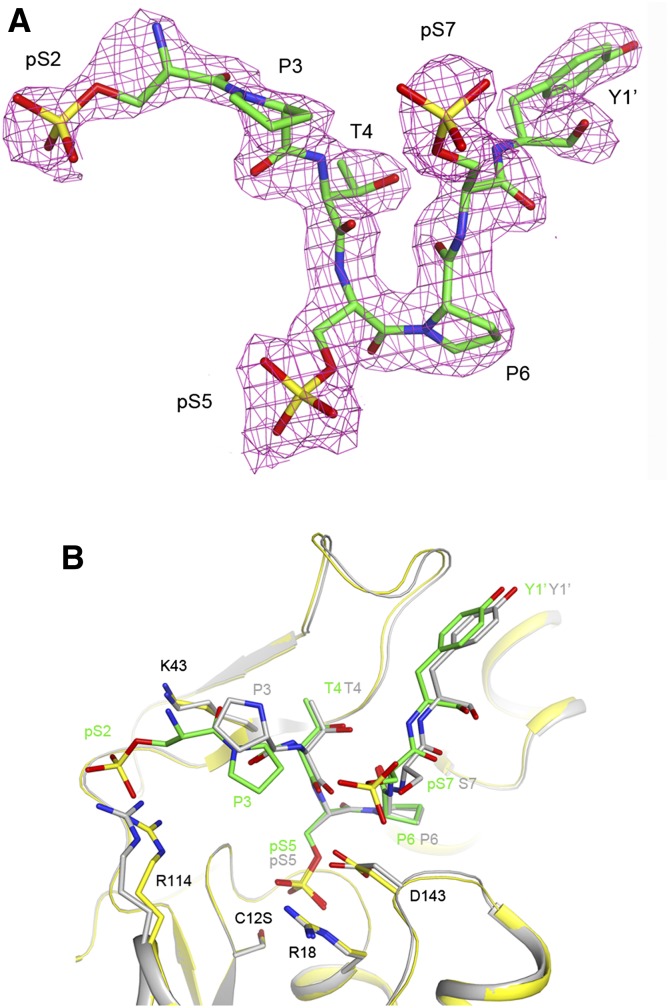
To understand how Ssu72 distinguishes between pSer2, pSer5, and pSer7 in the same CTD repeat, we determined the crystal structure at 2.0 Å resolution of another ternary complex using a 10-mer CTD peptide phosphorylated at all three positions, Ser7″–Tyr1–pSer2–Pro3–Thr4–pSer5–Pro6–pSer7–Tyr1′–Ser2′. The structure reveals binding of pSer5 in the catalytic site, while there is no evidence for pSer7 binding into this site (Fig. 2A). Instead, the phosphate group on Ser7 is exposed to the solvent, having no interactions with Ssu72 (Fig. 2B). The phosphate group on Ser2 may have ionic interactions with the side chain of Arg114 (Fig. 2B), although the electron density for pSer2 is rather weak (Fig. 2A). Overall, the binding mode of this triply phosphorylated peptide is similar to that of the pSer5 peptide reported earlier (Fig. 2B; Xiang et al. 2010). A conformational difference in the Pro3 residue of the peptide was observed, possibly linked to the interaction of pSer2 with the enzyme. We used another peptide with pSer7 in the center, Ser2–Pro3–Thr4–pSer5–Pro6–pSer7–Tyr1′–pSer2′–Pro3′–Thr4′–Ser5′, and theobserved electron density at 2.6 Å resolution was also consistent with pSer5 binding in the active site (data not shown). These observations indicate that Ssu72 prefers the binding of pSer5 and suggest that Ssu72 has higher affinity for pSer5 in the active site than for pSer7. This may also be detrimental for the phosphatase activity with the pSer7 substrate.
Figure 2.
Ssu72 prefers pSer5 in the active site. (A) Omit 2Fo − Fc electron density for the CTD peptide phosphorylated at Ser2, Ser5, and Ser7 at 2.0 Å resolution, contoured at 1σ. There is no indication of the binding of pSer7 into the catalytic site. (B) Overlay of the active site region of Ssu72 in the triply phosphorylated peptide complex (yellow) and the pSer5 complex (in gray) (Xiang et al. 2010).
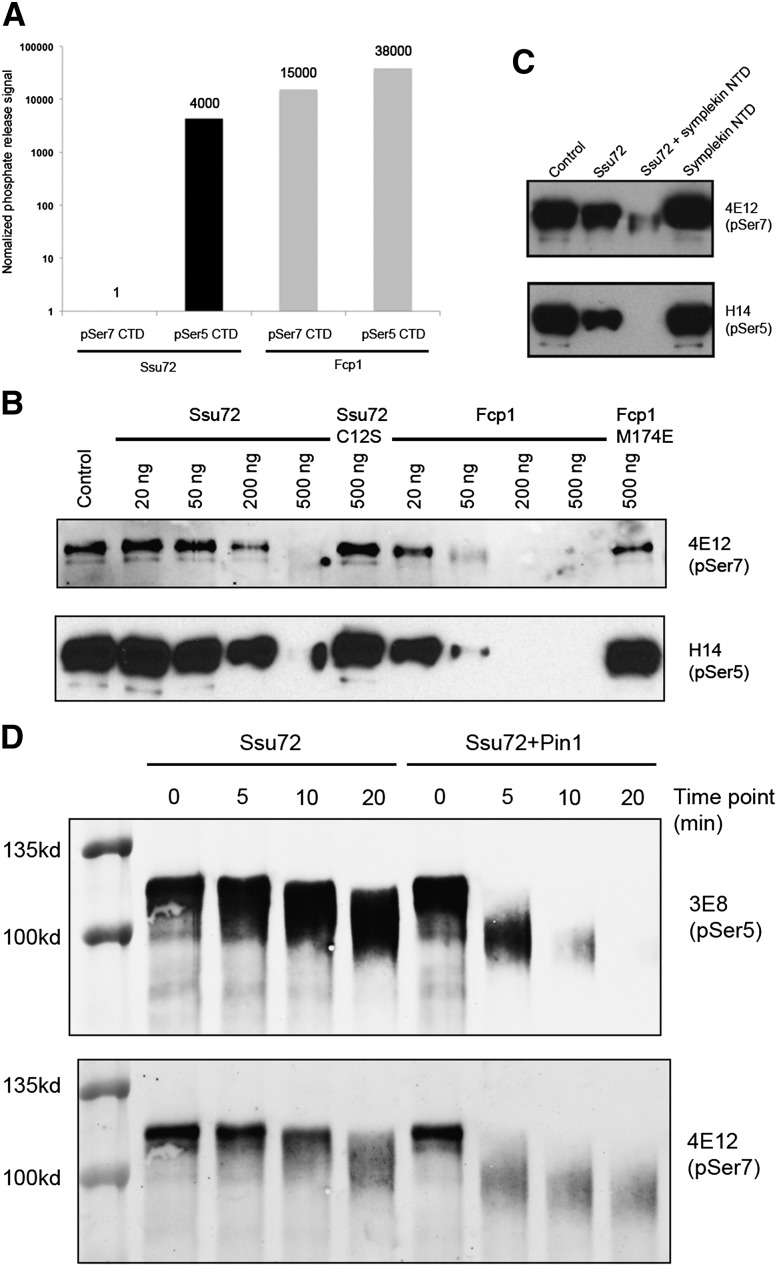
We next characterized the pSer7 phosphatase activity of Ssu72 using in vitro assays. We first used an assay that monitored the release of free phosphate from CTD phosphopeptides, which we previously used successfully with the pSer5 peptide (Xiang et al. 2010). Very low activity was observed in this assay using the 10-mer pSer7 peptide as substrate. Appreciable phosphate production was observed only after overnight incubation of purified His-tagged Ssu72 (Supplemental Fig. 1) with the phosphopeptide (40 μM phosphate was released after 12 h of incubation of 200 μM Ssu72 with 2 mM peptide at room temperature). The low activity did not appear to be affected by the presence of the symplekin NTD (results not shown). Parallel experiments with the 10-mer pSer5 peptide (Xiang et al. 2010) showed that the pSer5 phosphatase activity was ∼4000-fold higher than the pSer7 phosphatase activity (160 μM phosphate was released after 45 min of incubation of 5 μM Ssu72 with 1 mM peptide at room temperature) (Fig. 3A, left panel). As a comparison, purified yeast Fcp1 (Supplemental Fig. 1) displayed high activity with both the pSer5 and pSer7 peptides (Fig. 3A, right panel). While the physiological role of Fcp1 in pSer7 dephosphorylation is unclear (e.g., see Bataille et al. 2012), this result at a minimum indicates that the pSer7 peptide used in our assays can be effectively dephosphorylated in vitro, strengthening the conclusion that Ssu72 activity toward this peptide substrate is low.
Figure 3.
Phosphatase activity of Ssu72 toward pSer5 and pSer7 substrates. (A) Relative phosphatase activity for human Ssu72 (black bars) and yeast Fcp1 (gray bars) toward the pSer5 and pSer7 CTD peptide substrates. The amount of phosphate released in each reaction is normalized by the reaction time, substrate concentration, and enzyme concentration. The activity for Ssu72 with the pSer7 substrate is set at 1. The vertical axis is shown in log scale. (B) Phosphatase activity for human Ssu72 and yeast Fcp1 toward the GST-CTD substrate phosphorylated with Cdk7. (C) Symplekin NTD stimulates Ssu72 phosphatase activity toward both pSer7 and pSer5. (D) The prolyl isomerase Pin1 stimulates Ssu72 phosphatase activity toward pSer7 and pSer5 in the GST-CTD substrate.
We next tested whether the pSer7 peptide can compete with the pSer5 peptide for binding to Ssu72 and thereby inhibit its dephosphorylation. We included up to 20-fold higher concentration of the pSer7 peptide (2 mM) than the pSer5 peptide (0.1 mM) in the assay but did not observe any appreciable effect on the dephosphorylation of the pSer5 peptide (results not shown). This supports our structural observation that the affinity of Ssu72 for the pSer5 peptide is substantially higher than that of the pSer7 peptide.
Phosphorylation of CTD Tyr1 in yeast was recently reported to inhibit the recruitment of Pol II termination factors (Mayer et al. 2012). We tested the activity of Ssu72 toward a pSer7, pTyr1 doubly phosphorylated peptide, Ser2–Pro3–Thr4–Ser5–Pro6–pSer7–pTyr1′–Ser2′–Pro3′–Thr4′–Ser5′, and found that Tyr1 phosphorylation had essentially no effect on pSer7 dephosphorylation even though pTyr1 directly follows pSer7 in the primary sequence. This is consistent with our structural observations that the Tyr1 side chain has weak electron density (Fig. 1B) and does not make contact with Ssu72 (Fig. 1D).
We also used the entire Pol II CTD, purified as a GST fusion protein and phosphorylated with purified Cdk7 (Akhtar et al. 2009; Glover-Cutter et al. 2009), as the substrate in our assays. With this longer substrate and using antibodies capable of recognizing pSer7 and pSer5 in Western blots to monitor dephosphorylation, Ssu72 demonstrated comparable activity toward pSer7 and pSer5 (Fig. 3B), consistent with results reported earlier (Bataille et al. 2012; Zhang et al. 2012). Moreover, the symplekin NTD stimulated the pSer7 phosphatase activity of Ssu72 (Fig. 3C), as observed earlier with the pSer5 phosphatase activity (Xiang et al. 2010). We also found that the prolyl isomerase Pin1 modestly (about twofold based on reactivity with the anti-pSer7 antibody) (see the Materials and Methods) stimulated dephosphorylation of pSer7 in this GST-CTD substrate (Fig. 3D), consistent with an earlier study in yeast cells (Bataille et al. 2012). This apparent weak stimulation may reflect an indirect effect by which Pin1 promotes dephosphorylation of pSer5, which in turn makes pSer7 more accessible for dephosphorylation by Ssu72. Similar to results with the peptide substrates, Fcp1 showed stronger activity toward both pSer7 and pSer5 with the GST-CTD substrate than did Ssu72 (Fig. 3B). The possible significance of this remains to be determined.
The weak phosphatase activity of Ssu72 toward the pSer7 peptide substrate is consistent with our structural observations. The active site of Ssu72 likely has a higher affinity for the CTD phosphorylated on Ser5, with the pSer5–Pro6 peptide bond in the cis configuration. In the presence of pSer5, the enzyme appears not to bind pSer7 in the active site. In addition, the Asp143 general acid is misaligned in the pSer7 peptide complex, which would further reduce the activity of Ssu72 toward this substrate. On the other hand, with the full-length Pol II CTD substrate, Ssu72 appears to have similar activity toward both pSer5 and pSer7 (Bataille et al. 2012; Zhang et al. 2012). It might be possible that other regions of the CTD can indirectly affect the binding and/or catalysis of pSer7 in the Ssu72 active site. It is also notable that activity toward both pSer5 and pSer7, in the context of the intact CTD, was strongly enhanced by the symplekin NTD. This is consistent with the known requirement of this interaction for Ssu72 function in yeast (Ghazy et al. 2009) and may help to ensure that Ssu72 activity is constrained until the appropriate time during the transcription cycle. Overall, our results demonstrate a novel mode of substrate recognition by Ssu72 and explain how the enzyme can dephosphorylate both pSer5 and pSer7.
Materials and methods
Protein expression, purification, and crystallization
The experimental protocols followed those described earlier (Xiang et al. 2010). Briefly, human symplekin NTD and human Ssu72, both His-tagged, were expressed in Escherichia coli and purified separately using nickel affinity and gel filtration chromatography. The symplekin NTD–Ssu72 complex was purified by gel filtration after mixing the two proteins. Crystals of this binary complex were transferred to a soaking solution containing 100 mM Tris (pH 8.5), 10 mM NaCl, 25% (w/v) PEG 3000, and either the pSer7 peptide (10 mM) for 48 h or the pSer2, pSer5, and pSer7 triply phosphorylated peptide (30 mM) for 16 h. The crystals were then transferred to the same soaking solution supplemented with 25% (v/v) ethylene glycol for 30 sec and flash-frozen in liquid nitrogen.
His-tagged yeast Fcp1 (168-606) was overexpressed in E. coli BL21 (DE3) Star cells and purified by nickel affinity and gel filtration chromatography, following a published protocol (Kamenski et al. 2004).
Data collection and structure determination
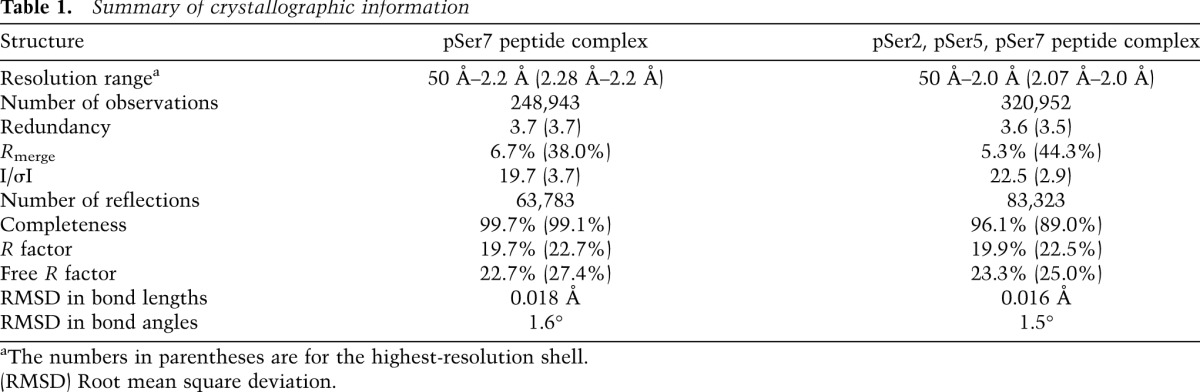
X-ray diffraction data were collected on an ADSC charge-coupled device at the X29A beamline of National Synchrotron Light Source (NSLS). The diffraction images were processed and scaled with the HKL package (Otwinowski and Minor 1997). The structure refinement was carried out with the program Refmac (Murshudov et al. 1997), and manual rebuilding of the model was performed with the program Coot (Emsley and Cowtan 2004). The statistics on the structure refinement are summarized in Table 1.
Table 1.
Summary of crystallographic information
CTD peptide phosphatase assays
Reaction mixtures (25 μL) containing 50 mM HEPES (pH 7.0), 100 mM NaCl, 10 mM MgCl2, 500 μM to 2 mM CTD peptide, and 5 μM to 200 μM Ssu72 were incubated for 45 min (for pSer5 peptide) or 12 h (for pSer7 peptide) at room temperature and then quenched by adding 0.5 mL of malachite green reagent (BIOMOL Research Laboratories). Assays with Fcp1 were performed similarly, using 5 μM Fcp1 and 500 μM peptides. Phosphate release was determined by measuring A620 and comparing it with a phosphate standard curve.
GST-CTD phosphatase assays
Purified GST-CTD fusion protein was phosphorylated in vitro by Cdk7 complex as described (Glover-Cutter et al. 2009). CTD phosphatase assays were performed in a total volume of 20 μL under the standard phosphatase condition (50 mM Bis-Tris at pH 6.5, 20 mM KCl, 10 mM MgCl2, 5 mM DTT) containing 200 ng of phosphorylated GST-CTD and the indicated amount of Ssu72 or Fcp1. Reaction mixtures were incubated for 1 h at 30°C and stopped by adding 5 μL of 5× SDS loading buffer, and 2.5 μL from each reaction was resolved on an 8% SDS-PAGE gel. pSer7 levels were detected by Western blot using the 4E12 antibody (Millipore), and pSer5 levels were detected using the H14 antibody (Covance).
Pin1 stimulation assay
GST-Pin1 was prepared as previously described (Xiang et al. 2010). A 35-μL reaction containing 100 μM Ssu72 with or without 200 nM GST-Pin1 was incubated at 10°C. Eight microliters was taken out at the indicated time points, mixed with 2 μL of 5× SDS sample buffer, and boiled for 5 min. Two microliters was resolved in an 8% SDS-PAGE gel. pSer5 and pSer7 levels were detected by Western blot with 3E8 (Millipore) and 4E12 antibodies, respectively, and quantitated with LI-COR. The same experiment was also performed at room temperature, with similar results.
pSer7 peptide inhibition of pSer5 peptide dephosphorylation
Ssu72 (10 μM) was preincubated with 0, 100 μM, 200 μM, 500 μM, 1 mM, or 2 mM pSer7 CTD peptide in a buffer containing 50 mM HEPES (pH 7.0), 100 mM NaCl, and 20 mM MgCl2 for 5 min at room temperature. A final concentration of 100 μM pSer5 CTD peptide was added in each reaction. The reaction was incubated for 45 min at 37°C and stopped by adding malachite green reagent. After another 30 min, OD620 was recorded for each reaction to determine phosphatase release.
Acknowledgments
We thank Patrick Cramer for the yeast Fcp1 expression plasmid; Stephane Larochelle and Robert P. Fisher for Cdk7 baculovirus; Jing-Ping Hsin and David Zhang for discussions; and Neil Whalen, Stuart Myers, Rick Jackimowicz, and Howard Robinson for access to the X29A beamline at the NSLS. This research was supported in part by grants from the NIH to L.T. (GM077175) and J.L.M. (GM028983).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.198853.112.
References
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 34: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques P-E, Laramee L, Fortin M-E, Forest A, Bergeron M, Hanes SD, Robert F 2012. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 45: 158–170 [DOI] [PubMed] [Google Scholar]
- Becker JW, Marcy AI, Rokosz LL, Axel MG, Burbaum JJ, Fitzgerald PMD, Cameron PM, Esser CK, Hagmann WK, Hermes JD, et al. 1995. Stromelysin-1: Three-dimensional structure of the inhibited catalytic domain and of the C-truncated proenzyme. Protein Sci 4: 1966–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan KD 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Feng S, Chen JK, Yu H, Simon JA, Schreiber SL 1994. Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3–ligand interactions. Science 266: 1241–1247 [DOI] [PubMed] [Google Scholar]
- Ghazy MA, He X, Singh BN, Hampsey M, Moore C 2009. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensible for pre-mRNA 3′-end processing. Mol Cell Biol 29: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Shuman S, Lima CD 2008. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol Cell 32: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL 2009. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 29: 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Shuman S 2002. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J Biol Chem 277: 21213–21220 [DOI] [PubMed] [Google Scholar]
- Hausmann S, Erdjument-Bromage H, Shuman S 2004. Schizosaccharomyces pombe carboxyl-terminal domain (CTD) phosphatase Fcp1: Distributive mechanism, minimal CTD substrate, and active site mapping. J Biol Chem 279: 10892–10900 [DOI] [PubMed] [Google Scholar]
- Hausmann S, Koiwa H, Krishnamurthy S, Hampsey M, Shuman S 2005. Different strategies for carboxyl-terminal domain (CTD) recognition by serine 5-specific CTD phosphatases. J Biol Chem 280: 37681–37688 [DOI] [PubMed] [Google Scholar]
- Hsin J-P, Sheth A, Manley JL 2011. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 334: 683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenski T, Heilmeier S, Meinhart A, Cramer P 2004. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol Cell 15: 399–407 [DOI] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore CL, Hampsey M 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394 [DOI] [PubMed] [Google Scholar]
- Levchenko I, Grant RA, Flynn JM, Sauer RT, Baker TA 2005. Versatile modes of peptide recognition by the AAA+ adaptor protein SspB. Nat Struct Mol Biol 12: 520–525 [DOI] [PubMed] [Google Scholar]
- Lim WA, Richards FM, Fox RO 1994. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature 372: 375–379 [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P 2012. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336: 1723–1725 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P 2005. A structural perspective of CTD function. Genes Dev 19: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Qin J, Clore GM, Kennedy WP, Kuszewski J, Gronenborn AM 1996. The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure 4: 613–620 [DOI] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. 2010. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Allen JW, Lee C-J, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P 2011. Cis-proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 286: 5717–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Behav MM, Manley JL, Tong L 2010. Crystal structure of the human symplekin–Ssu72–CTD phosphopeptide complex. Nature 467: 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ 2012. Ssu72 phosphatase dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem 287: 8541–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]