Background: The mitochondrial Oxa1 insertase binds ribosomes.
Results: Spacers separating the insertase and ribosome-binding domains of Oxa1 impair biogenesis of cytochrome c oxidase.
Conclusion: Tight ribosome binding of Oxa1 is dispensable for membrane insertion but is required for assembly of translation products.
Significance: In addition to its role as an insertase, Oxa1 promotes the subsequent assembly of respiratory chain complexes.
Keywords: Membrane Biogenesis, Membrane Proteins, Mitochondria, Protein Assembly, Ribosomes, Oxa1
Abstract
The terminal enzyme of the respiratory chain, cytochrome c oxidase, consists of a hydrophobic reaction center formed by three mitochondrially encoded subunits with which 9–10 nuclear encoded subunits are associated. The three core subunits are synthesized on mitochondrial ribosomes and inserted into the inner membrane in a co-translational reaction facilitated by the Oxa1 insertase. Oxa1 consists of an N-terminal insertase domain and a C-terminal ribosome-binding region. Mutants lacking the C-terminal region show specific defects in co-translational insertion, suggesting that the close contact of the ribosome with the insertase promotes co-translational insertion of nascent chains. In this study, we inserted flexible linkers of 100 or 200 amino acid residues between the insertase domain and ribosome-binding region of Oxa1 of Saccharomyces cerevisiae. In the absence of the ribosome receptor Mba1, these linkers caused a length-dependent decrease in mitochondrial respiratory activity caused by diminished levels of cytochrome c oxidase. Interestingly, considerable amounts of mitochondrial translation products were still integrated into the inner membrane in these linker mutants. However, they showed severe defects in later stages of the biogenesis process, presumably during assembly into functional complexes. Our observations suggest that the close proximity of Oxa1 to ribosomes is not only used to improve membrane insertion but is also critical for the productive assembly of the subunits of the cytochrome c oxidase. This points to a role for Oxa1 in the spatial coordination of the ribosome with assembly factors that are critical for enzyme biogenesis.
Introduction
The biogenesis of proteins relies on the synthesis of polypeptides and the subsequent folding into their native structures. Chaperones and assembly factors directly associate with ribosomes to couple synthesis and folding processes kinetically and/or mechanistically (1–3). In the case of the bacterial ribosome, the region around the polypeptide exit tunnel serves as binding platform for the trigger factor as well as for a deformylating enzyme and an aminopeptidase. Similarly, in the eukaryotic cytosol, dedicated chaperones of the Hsp70 family form a ribosome-associated complex that contributes to co-translational protein folding (4, 5).
In addition, during the synthesis of hydrophobic membrane proteins, ribosomes can associate with membrane-bound protein transport channels like the Sec61 translocon of the endoplasmic reticulum or the SecY complex of the bacterial inner membrane. The direct binding to the translocon might have several advantages: it might minimize the time that hydrophobic transmembrane domains are water-exposed, provide an ion-tight seal during the translocation process, or energetically drive the nascent chains across the lipid bilayer (ribosomal pushing) (6, 7). It is experimentally difficult to assess the relevance of translocon binding by the ribosome in particular because the binding presumably relies on several low affinity interactions that cannot be easily dissected (8).
The translation system of mitochondria differs from that of the bacterial or prokaryotic ribosome in several respects: the mitochondrial genome encodes only a handful of proteins, which are almost exclusively hydrophobic membrane proteins. In the yeast Saccharomyces cerevisiae, eight proteins are synthesized in the mitochondrial matrix: cytochrome b of the bc1 complex; subunits 1–3 (Cox1, Cox2, and Cox3) of cytochrome c oxidase; Atp6, Atp8, and Atp9 of the ATPase; and the ribosomal protein Var1. Presumably due to its specialization in the synthesis of hydrophobic membrane proteins, the mitochondrial ribosome is tightly associated with the mitochondrial inner membrane and, at least in yeast, can only be released upon treatment with detergent or urea (9, 10). Moreover, mitochondrial ribosomes are physically bound to the protein insertion machinery of the inner membrane (11–15). Membrane insertion of nascent chains is facilitated by Oxa1, which integrates both mitochondrial translation products and some nuclear encoded proteins into the inner membrane (16–18). Oxa1 comprises two domains: a membrane-embedded core region that is closely related to the core domains of YidC and Alb3 insertases that are present in membranes of bacteria and plastids, respectively (19), and a C-terminal ribosome-binding domain that is absent in its bacterial homologs. This positively charged stretch binds the large subunit of mitochondrial ribosomes in proximity to the polypeptide exit tunnel and is required for the biogenesis of the mitochondrial respiratory chain (15, 20). Oxa1 cooperates with Mba1, a peripheral membrane protein that serves as membrane receptor of mitochondrial ribosomes (9, 21). Like Oxa1, Mba1 binds ribosomes in proximity to the exit tunnel (22). Additional, so far uncharacterized membrane anchors apparently exist, as both ribosomal subunits remain membrane-bound even in the absence of Oxa1 and Mba1. It is conceivable that the binding of the mitochondrial insertion machinery to the proximity of the polypeptide exit tunnel is used to thread nascent chains through the membrane. Alternatively, the physical interaction of Oxa1 might simply ensure that Oxa1 is present when new proteins are synthesized without having any mechanistic relevance for the translocation process per se.
In this study, we inserted flexible linkers of different lengths between the membrane domain of Oxa1 and its ribosome-binding region. In the absence of Mba1, these variants caused defects in the biogenesis of respiratory chain complexes that became more severe with increasing lengths of the linkers. Interestingly, even upon insertion of the longest linkers of 200 amino acid residues, the intermembrane space (IMS)2 domain of the Oxa1 substrate Cox2 was translocated across the membrane, albeit with reduced efficiency. However, the synthesized subunits were unable to assemble into functional cytochrome c oxidase complexes. Our observations imply that, in addition to its role in protein translocation, Oxa1 exhibits a crucial function in the assembly of cytochrome c oxidase that depends on spatial orientation of the Oxa1-ribosome complex.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Media
All yeast strains used in this study are derivatives of W303-1A (MAT a, ade2 ura3 leu2 his3 trp1). The OXA1, MDM38, and MBA1 genes were deleted by LEU2-, KanMX-, and HIS3-containing cassettes, respectively. For construction of the Oxa1 linker mutants, the sequences corresponding to the OXA1 promoter and amino acids 1–317 and 318–402 were separately amplified by PCR and cloned into the SacI and XhoI sites of a pRS424 vector. In the resulting Oxa1 sequence, the insertase domain (amino acids 1–317) and the ribosome-binding domain (amino acids 318–402) were separated by restriction sites for BamHI and PstI. These sites were opened to insert the Nsp1 linker fragment sequences that were amplified from the pSF362-pQE80N plasmid; these sequences corresponded to residues 2–101 and 2–201 of the Nsp1FS mutant protein (23). The Oxa1 variants expressed from the resulting constructs were named Oxa1100 and Oxa1200, respectively. These plasmids or a control plasmid with the wild-type OXA1 gene was transformed into Δoxa1 single mutants or Δoxa1 Δmba1 double mutants.
Yeast cultures were grown at 30 °C in 1% yeast extract and 2% peptone or in minimal synthetic medium supplemented with 2% glycerol, 2% galactose, or 2% glucose (24). Mitochondria were isolated as described previously (24).
Analysis of Mitochondrial Translation Products
Mitochondrial translation products were radiolabeled in whole cells (in vivo) or in isolated mitochondria (in organello) as described previously (25). To assess the accessibility of cysteine residues, mitochondria were incubated in 0.6 m sorbitol, 0.5 m NaCl, and 20 mm Tris (pH 7.4) in the presence of 1 mm mPEG-12 or mPEG-24 for 15 min at 4 °C. Prolonged incubation at temperatures above 20 °C should be avoided to prevent modification of cysteine residues in the matrix. Modification of cysteine residues was stopped by the addition of 2 mm DTT.
Immunoprecipitation and Co-immunoprecipitation
To isolate the Cox20-containing protein complex, translation products were radiolabeled in 120 μg of isolated mitochondria. Mitochondria were washed and lysed in ice-cold lysis buffer (1% digitonin, 20 mm Tris (pH 7.4), 150 mm NaCl, and 1 mm EDTA). The extract (200 μl) was cleared by centrifugation (10 min, 16,000 × g, 4 °C) before protein A-Sepharose beads and 5 μl of anti-Cox20 serum were added. After incubation for 1 h at 4 °C, the beads were extensively washed and analyzed by SDS-PAGE. For immunoprecipitation of Cox2, mitochondria were lysed under denaturing conditions in 20 μl of 1% SDS that was diluted 25-fold with 0.1% Triton X-100, 20 mm Tris (pH 7.4), 150 mm NaCl, 2 mm PMSF, and 5 mm EDTA.
Blue Native (BN) PAGE and Cytochrome c Oxidase Assembly Assay
To follow assembly of cytochrome c oxidase, radiolabeled precursor proteins were imported into mitochondria. Cox5a and Cox13 were synthesized in the presence of [35S]methionine in reticulocyte lysate (26). Import into isolated yeast mitochondria was performed in import buffer (250 mm sucrose, 10 mm MOPS/KOH (pH 7.2), 80 mm KCl, 2 mm KH2PO4, 5 mm MgCl2, 5 mm methionine, 3% BSA, 2 mm NADH, 2 mm ATP, 5 mm creatine phosphate, and 0.1 mg/ml creatine kinase) at 25 °C. Import was stopped by the addition of a 1% mixture of 2.5 mm antimycin A, 0.25 mm valinomycin, and 5 mm oligomycin in ethanol. After in vitro import, mitochondria were washed with 250 mm sucrose, 20 mm MOPS/KOH (pH 7.2), and 1 mm EDTA and processed for SDS- or BN-PAGE analysis, followed by detection of radiolabeled proteins by digital autoradiography.
For BN-PAGE, mitochondria were solubilized in 1% digitonin, 20 mm Tris-HCl (pH 7.4), 5 mm EDTA, 100 mm NaCl, 10% (w/v) glycerol, and 2 mm PMSF for 30 min at 4 °C. The lysate was cleared at 20,000 × g for 15 min at 4 °C. After the addition of 10× loading dye (5% Coomassie G-250, 500 mm 6-aminohexanoic acid, and 100 mm BisTris (pH 7.0)), the supernatant was separated on a 4–13% polyacrylamide gradient gel.
RESULTS
Insertion of Linkers Causes Growth Defects on Non-fermentable Carbon Sources
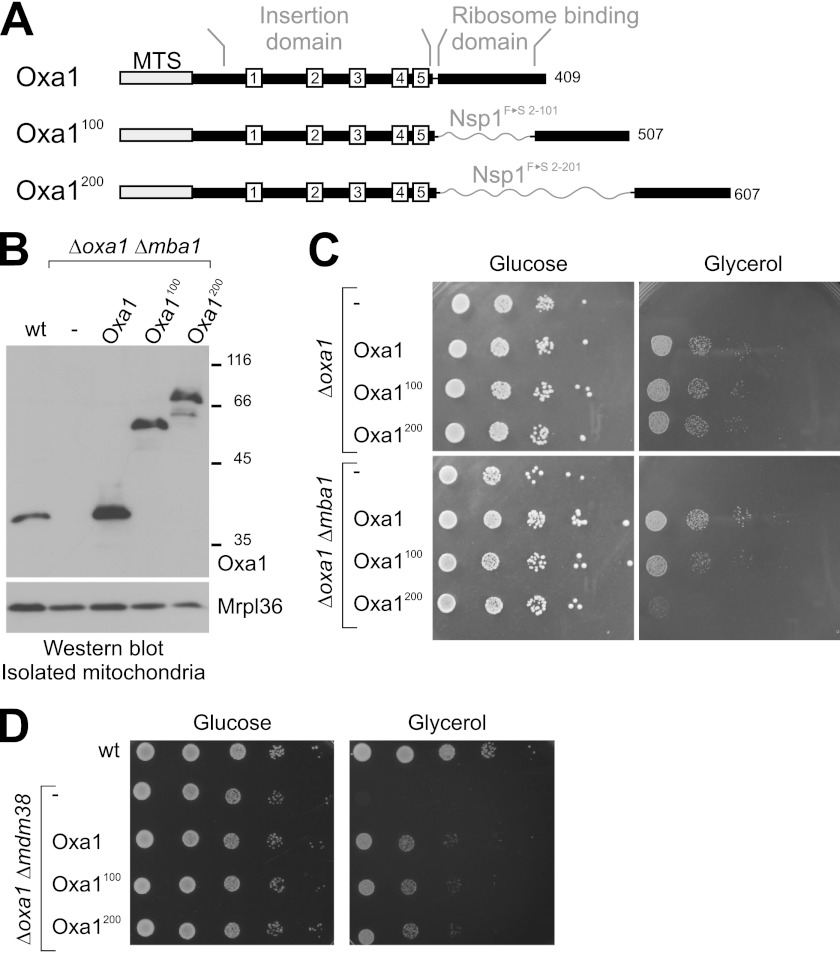
Oxa1 shares a hydrophobic core region with bacterial YidC proteins that comprises five transmembrane regions. In contrast to its bacterial homologs, Oxa1 contains a C-terminal stretch region of ∼100 amino acid residues that protrudes into the matrix. This region is of α-helical amphipathic structure and is rich in positively charged residues (11–15). It was speculated that the C-terminal region of Oxa1 tethers the ribosome to the insertion complex so that nascent chains are threaded efficiently across the membrane. To characterize the relevance of the ribosome-binding domain of Oxa1 for membrane biogenesis in mitochondria, we constructed Oxa1 expression plasmids in which we introduced a pair of restriction sites into the OXA1 sequence after codon 317 (Fig. 1A). These sites were used to construct Oxa1 variants into which spacers of 100 and 200 residues were inserted directly C-terminal of the fifth transmembrane domain (Oxa1100 and Oxa1200). For these linker regions, we chose sequences corresponding to positions 2–101 and 2–201, respectively, of the Phe-to-Ser variant of the nucleofilament Nsp1 (23). In the corresponding Nsp1 sequence, phenylalanine residues were exchanged for serine residues resulting in an intrinsically unstructured nature of this sequence. The reasoning for using the Phe-to-Ser variant of the Nsp1 sequence was to physically separate the membrane domain from the ribosome-binding region of Oxa1 without adding a novel structured domain to the Oxa1 protein.
FIGURE 1.
In the Oxa1 linker mutants, the ribosome-binding and insertase domains of Oxa1 are separated. A, structure of the Oxa1 linker mutants. The mitochondrial targeting sequence (MTS) and the five transmembrane domains are shown as boxes. Flexible regions derived from residues 2–101 or 2–201 of an Nsp1FS mutant (23) were inserted between the membrane-embedded insertase domain and the ribosome-binding domain of Oxa1. B, the linker mutants of Oxa1 are expressed to similar levels as wild-type Oxa1. Oxa1 and the mitochondrial protein MrpL36 were detected by Western blotting in mitochondria isolated from the wild-type and mutant strains. C and D, cells of the mutants indicated were grown overnight to log phase. Serial dilutions (10-fold) were spotted on 1% yeast extract and 2% peptone plates containing glucose or galactose as the carbon source and incubated at 30 °C for 2 and 3 days, respectively.
These Oxa1 variants were introduced into yeast strains lacking either the endogenous OXA1 gene or lacking both OXA1 and MBA1. When mitochondria were isolated from the resulting mutants, the Oxa1 variants could be detected by Western blotting at the expected sizes and at levels that were comparable with the Oxa1 levels in wild-type mitochondria (Fig. 1B).
To test the functionality of these variants, we spotted cultures of these strains on plates containing fermentable (glucose) or non-fermentable (glycerol) carbon sources (Fig. 1C). Whereas all strains grew well on glucose, they differed considerably in their ability to grow under respiratory conditions: All Oxa1 variants complemented the Δoxa1 mutant, indicating that, in the presence of Mba1, the spacers did not compromise mitochondrial protein biogenesis. In contrast, in the mutants lacking Mba1, the spacers in Oxa1 caused a strong growth defect that correlated with the lengths of the insertion: growth of the Oxa1100 mutant was ∼10-fold reduced, and the Oxa1200 mutant was even more severely affected (Fig. 1C).
Mdm38 is an inner membrane protein that, in addition to Oxa1 and Mba1, binds to mitochondrial ribosomes (51, 52). Expression of Oxa1 in Δoxa1 Δmdm38 double mutants partially repressed the phenotype of the double mutant, confirming earlier studies demonstrating that Δmdm38 cells show reduced respiratory activity. In this case, as in the Δoxa1 single mutant, the spacer variants did not further reduce the ability to grow on glycerol. This points to a rather specific cooperative role for Mba1 and the C-terminal region of Oxa1 in mitochondrial membrane protein biogenesis.
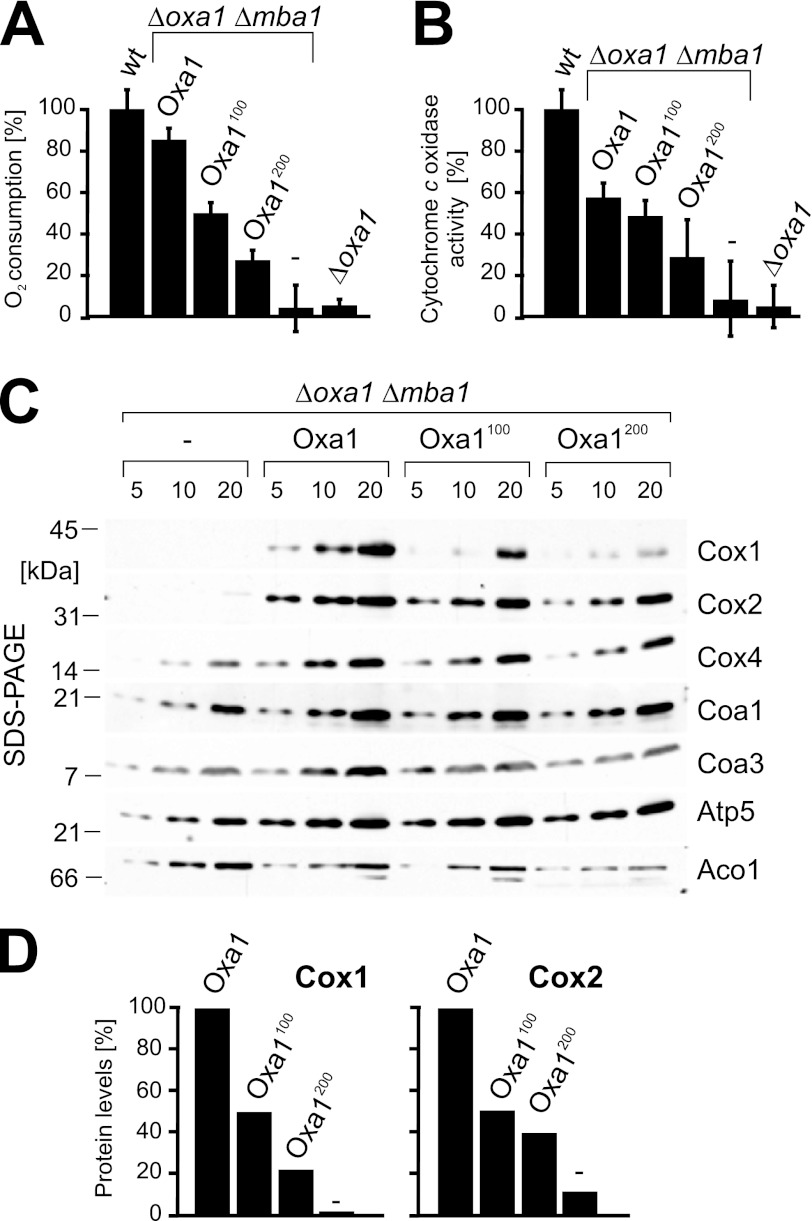
Insertion of Linkers into Oxa1 Leads to Reduced Levels of Respiratory Chain Complexes
The reduced growth of the linker-containing strains pointed to a defect in respiratory activity. To test this more directly, the different strains were grown on galactose medium. Mitochondrial fractions were then isolated, and the oxygen consumption was measured using a Clark electrode (Fig. 2A). The results correlated well with the growth phenotype, showing decreased oxygen consumption with increasing lengths of the linker segments. Oxa1 plays a crucial role in the biogenesis of cytochrome c oxidase, and direct measurements of the cytochrome c oxidase activity indeed revealed strong defects in the mutants. In the Δmba1 mutants expressing Oxa1, the cytochrome c oxidase activity levels were reduced to ∼50%, which correlated well with previous results published for Δmba1 strains (21). In the presence of the linker stretches, the cytochrome c oxidase was further reduced and dropped to ∼28% in the Oxa1200 strain.
FIGURE 2.
Insertion of linkers into Oxa1 leads to reduced levels of respiratory chain complexes. A, mitochondria (10 μg) from the indicated strains were incubated in the presence of 7 mm NADH. The oxygen consumption per time was measured in three independent measurements. B, the activity of cytochrome c oxidase was analyzed in 500 μg of mitochondria after lysis with Triton X-100. C, steady-state levels of mitochondrial proteins were assessed by Western blotting. The positions of molecular mass markers are indicated. D, quantification of Cox1 and Cox2 levels in Δoxa1 Δmba1 strains expressing the indicated proteins from plasmids.
Next, we analyzed the steady-state levels of cytochrome c oxidase subunits by Western blotting (Fig. 2, C and D). The insertion mutants showed considerably reduced levels of Cox1 and, to a slightly lesser degree, Cox2, whereas nuclear encoded proteins of the inner membrane (Cox4, Coa1, Coa3, and Atp5) or the matrix (Aco1) were not affected. It should be noted that the Cox2 protein accumulated in the mature form, whereas in Δoxa1 mutants, the maturation of Cox2 is blocked (16, 17). Due to the proteolytic instability of the precursor form, no Cox2 was detectable in Δoxa1 Δmba1 mitochondria (Fig. 2C). From this, we conclude that the insertion of a flexible linker between the membrane domain and the ribosome-binding domain of Oxa1 strongly interferes with the biogenesis or stability of cytochrome c oxidase.
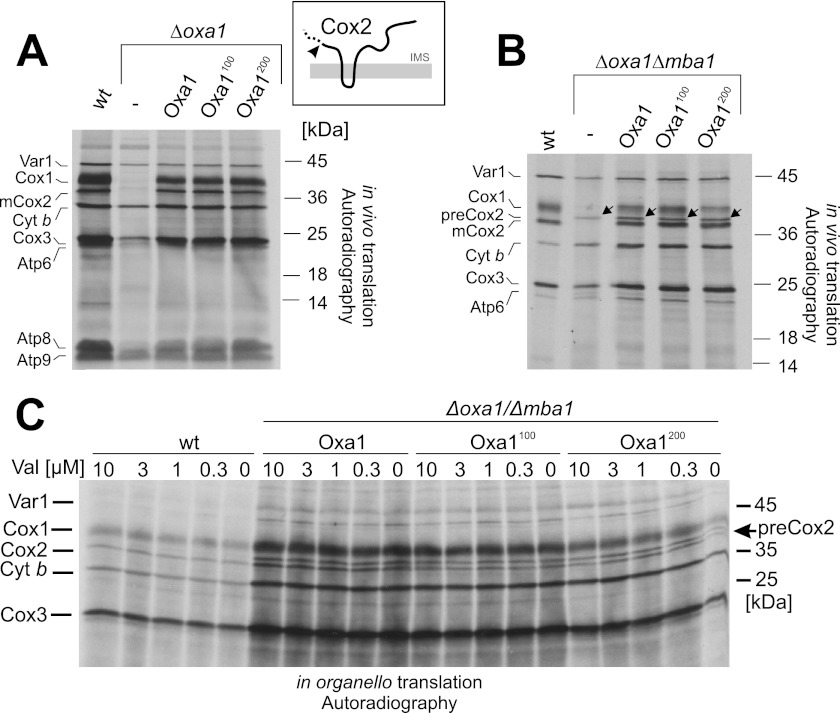
Insertion of Linkers into Oxa1 Does Not Impair Processing of Cox2
The Oxa1 insertase was shown to be required for membrane insertion of Cox2 (16). In yeast, Cox2 is synthesized with an N-terminal leader peptide that is removed by the Imp1 protease after translocation of the N terminus into the IMS (27, 28). Mutants lacking Oxa1 fail to insert Cox2 into the inner membrane, which therefore accumulates in the matrix in its precursor form and is rapidly degraded (16, 29). To follow the maturation of newly synthesized Cox2, we labeled mitochondrial translation products with [35S]methionine in whole yeast cells (in vivo) after blocking cytosolic translation with cycloheximide (Fig. 3A). In cells lacking Oxa1, the levels of Cox1, Cox2, and Cox3 and, to some degree, also those of other translation products, were strongly diminished (Fig. 3A). The small amounts of Cox2 that were synthesized in Δoxa1 cells were present in the precursor form. In cells carrying the linker-containing variants of Oxa1, no changes in the expression patterns were found, showing that even the longer linker does not prevent maturation and hence insertion of Cox2.
FIGURE 3.
Insertion of linkers into Oxa1 does not impair processing of Cox2. A, mitochondrial translation products were radiolabeled with [35S]methionine in yeast cells of the indicated strains after inhibition of cytosolic protein synthesis with cycloheximide. The inset shows the topology of Cox2. The arrowhead depicts the low levels of Cox2 precursor that accumulate in Δoxa1 mutants. B, translation products were radiolabeled as described for A in Mba1-deficient strains. Note that the levels of mature Cox2 are similar between the linkerless and linker-containing Oxa1 variants. C, mitochondria were isolated from the indicated strains and incubated in the presence of different concentrations of valinomycin and [35S]methionine. After incubation for 15 min at 30 °C, radiolabeling was stopped by the addition of unlabeled methionine, and the samples were analyzed by SDS-PAGE and autoradiography. Cyt b, cytochrome b.
Next, we analyzed mitochondrial protein synthesis in the strains lacking Mba1. Δmba1 mutants have been shown to exhibit insertion defects for Cox2, although here still considerable amounts of Cox2 are mature (21). As shown in Fig. 3B, mutants lacking Oxa1 and Mba1 did not process newly synthesized Cox2. However, the expression of linker-containing Oxa1 variants strongly relieved this block, and about half of the produced Cox2 protein was found in the mature form. Interestingly, the block in Cox2 processing was similar for all three Oxa1 variants. From this, we conclude that the defect in the biogenesis of cytochrome c oxidase in the linker-containing Oxa1 mutants is not due to defects in processing of the Cox2 precursor.
Next, we asked whether insertion of the linkers into Oxa1 renders the insertion of Cox2 more dependent on the membrane potential. To test this, we incubated mitochondria isolated from different yeast mutants in the presence of increasing concentrations of valinomycin, which dissipates the proton gradient across the inner membrane. We added [35S]methionine to radiolabel translation products (“in organello translation”). This again showed that about half of the produced Cox2 proteins accumulated in the precursor form in the Δmba1 mutants expressing the Oxa1 variants (Fig. 3C). The degree of Cox2 processing was not further diminished upon the addition of valinomycin. In summary, these data indicate that, in the presence of Mba1, even the 200-residue spacer did not impair the insertion of the N terminus of Cox2 by Oxa1. In the absence of Mba1, some of the Cox2 protein accumulated in the precursor form; however, this species was not considerably increased in the presence of the linker mutants of Oxa1. This suggests that the defect in the biogenesis of cytochrome c oxidase is not caused by impaired processing of newly synthesized Cox2.
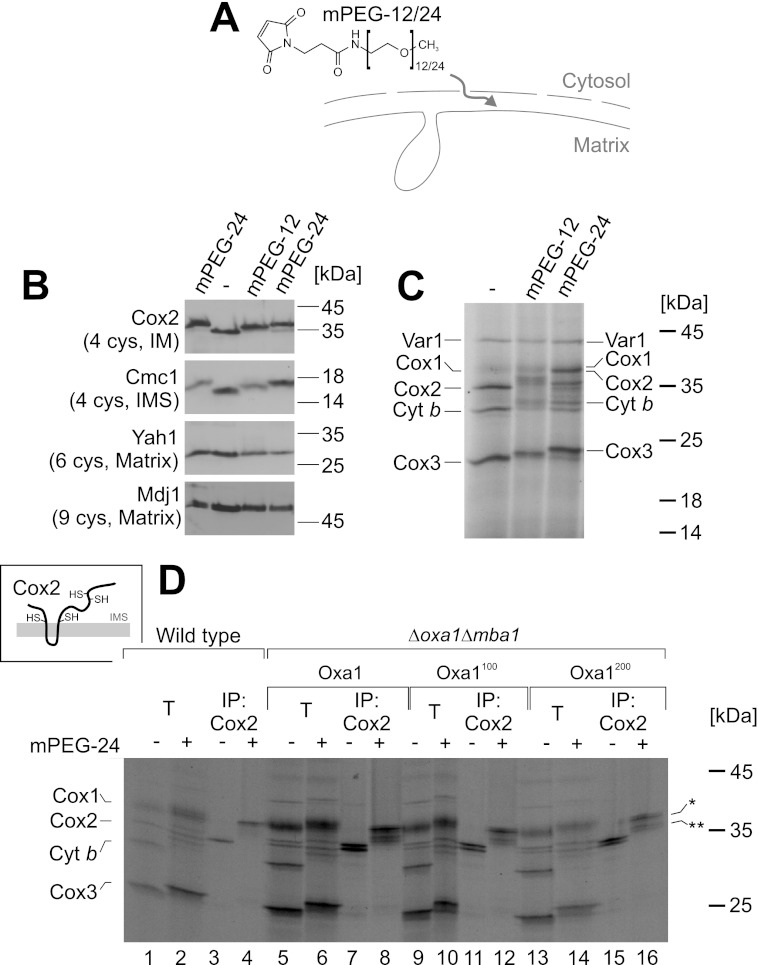
Insertion of Linkers into Oxa1 Does Not Prevent Protein Insertion into the Inner Membrane
The observed maturation of the Cox2 precursor is a good indication that the insertion of the N terminus of Cox2 is not impaired in the Oxa1 linker mutants. However, this does not rule out defects in the translocation of the large C-terminal IMS domain of Cox2. In earlier studies, protein translocation was assessed by protease accessibility assays, which can be problematic due to the inherent inefficiency to rupture the outer membrane of respiratory-deficient mutants by hypotonic swelling. We therefore set out to develop a swelling-independent assay to monitor inner membrane protein topology. In bacteria, a topology assay has been established that uses the modification of cysteine residues by membrane-impermeable alkylating reagents (30, 31). To assess the accessibility of protein thiols in mitochondria, we employed the alkylating maleimide derivatives mPEG-12 and mPEG-24 (Fig. 4A), which bind irreversibly and specifically to cysteine residues, thereby adding molecular masses of 0.6 and 1.2 kDa per moiety, respectively. Both molecules are not membrane-permeable. Therefore, we tested whether they are able to enter the IMS, e.g. through the pores of β-barrel proteins in the outer membrane that facilitate the diffusion of molecules of up to 3–5 kDa. Mitochondria were incubated with mPEG-12 and mPEG-24, and the migration of cysteine-containing proteins was analyzed by Western blotting (Fig. 4B). Although we did not observe size shifts for matrix proteins (Yah1 and Mdj1), proteins of the IMS (Cmc1) or inner membrane proteins containing IMS-exposed domains (Cox2) shifted after exposure to these alkylating reagents. With this, we could show that mPEG-12 and mPEG-24 are able to pass the outer mitochondria membrane but cannot cross the inner membrane. Hence, these alkylating reactions specifically modify regions of mitochondrial proteins that are exposed to the IMS.
FIGURE 4.
Insertion of linkers into Oxa1 does not prevent protein insertion into the inner membrane. A, structure of the maleimide derivatives mPEG-12 and mPEG-24. B, wild-type mitochondria were incubated in the presence of 1 mm mPEG-12 or mPEG-24 or mock-treated. After quenching the alkylating reagents by the addition of 2 mm DTT, mitochondria were reisolated, washed, and analyzed by Western blotting. The total numbers of cysteine residues in the proteins are indicated. The mPEG-24-treated sample was loaded twice to see the migration differences more clearly. C, mitochondrial translation products were radiolabeled in isolated mitochondria before mitochondria were incubated with mPEG-12 and mPEG-24 as described for B. D, following translation and treatment with mPEG-24, mitochondria were lysed and either directly applied to the gel (10% total, T) or used for immunoprecipitation (IP) with Cox2-specific antibodies. As shown in the inset, completely inserted Cox2 exposes four cysteine residues to the IMS. Fully or partially modified Cox2 species are indicated by one or two asterisks, respectively. Cyt b, cytochrome b.
We next radiolabeled translation products in isolated mitochondria before we added mPEG-12 and mPEG-24 (Fig. 4C). Incubation with the alkylating maleimide compounds reduced the mobility of translation products except for the matrix protein Var1 (which contains one cysteine residue), consistent with the inaccessibility of matrix proteins. The observed size shifts of most hydrophobic translation products indicated that they expose cysteine residues to the IMS. We tested the accessibility of newly synthesized translation products in the different Oxa1 mutants (Fig. 4D). To identify the nature of the different shifted bands, we performed immunoprecipitation experiments with antibodies against Cox2. Cox2 is the translation product that comprises the largest IMS domains; it contains four cysteine residues, two of which are directly adjacent to the transmembrane domains (Fig. 4D, inset). In wild-type mitochondria, mPEG-24 treatment caused a significant shift of Cox2 from 34 to 39 kDa (Fig. 4, C and D, lanes 3 and 4), consistent with a modification of all four cysteine residues. A size shift was also observed with endogenous Cox2 as detected by Western blotting (Fig. 4B). The same fully modified Cox2 species was observed in the Δmba1 strains, regardless which Oxa1 variant was present (Fig. 4D, lanes 8, 12, and 16). In addition, lower migrating bands appeared that presumably were derived from the precursor form of Cox2, in which only one or two cysteine residues were alkylated (indicated by asterisks). From this, we conclude that the Oxa1100- and Oxa1200-expressing strains contain fractions of Cox2 in which both IMS domains are translocated across the inner membrane, confirming that a defect in membrane insertion is not the cause of the cytochrome c oxidase deficiency of these strains. Moreover, also Cox1 is alkylated in mitochondria of all four strains, indicating that also this protein is inserted into the inner membrane.
Insertion of Linkers into Oxa1 Does Not Prevent Binding of Cox2 to Cox20
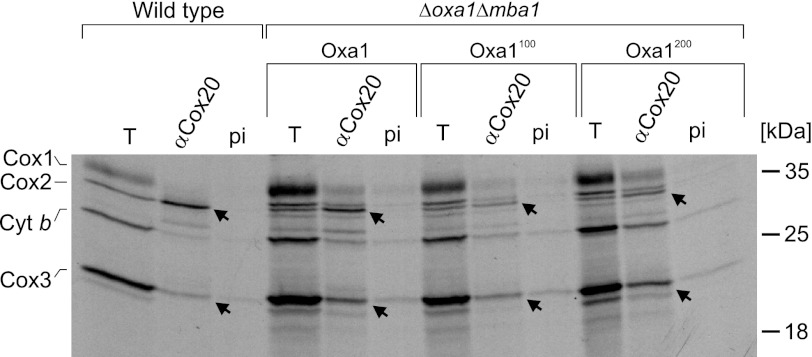
Because the reduced levels of cytochrome c oxidase were apparently not caused by defects in the synthesis or membrane insertion of their subunits, we asked whether steps farther downstream in their biogenesis were affected in these mutants. In the biogenesis pathway of Cox2, membrane insertion of the protein is followed by binding to the Cox2-specific chaperone Cox20 (32, 33). To assess the binding of newly synthesized Cox2 to Cox20, we radiolabeled translation products in mitochondria of the different strains, lysed the mitochondria, and performed co-immunoprecipitation experiments with Cox20-specific antibodies or preimmune serum as a control (Fig. 5). In extracts of wild-type mitochondria, the newly synthesized Cox2 and, to a lesser degree, Cox3 were co-isolated with anti-Cox20 antibodies. Similarly, the mature forms of Cox2 and Cox3 were pulled down with Cox20 from extracts of the linker mutants (Fig. 5, arrows). From this, we conclude that insertion of the linkers into the Oxa1 sequence does not block the interaction of Cox2 and Cox3 with Cox20, pointing to a role for Oxa1 in a later step of cytochrome c oxidase assembly.
FIGURE 5.
Insertion of linkers into Oxa1 does not prevent binding of Cox2 to Cox20. Translation products were radiolabeled in isolated mitochondria. Mitochondria were lysed with Triton X-100 and used for co-immunoprecipitation with serum against Cox20 (αCox20) or with preimmune (pi) serum as a control. The total lanes (T) show 10% of the material used for the co-immunoprecipitations. Arrows depict the Cox2 and Cox3 proteins that were co-immunoprecipitated with anti-Cox20 antibodies. Cyt b, cytochrome b.
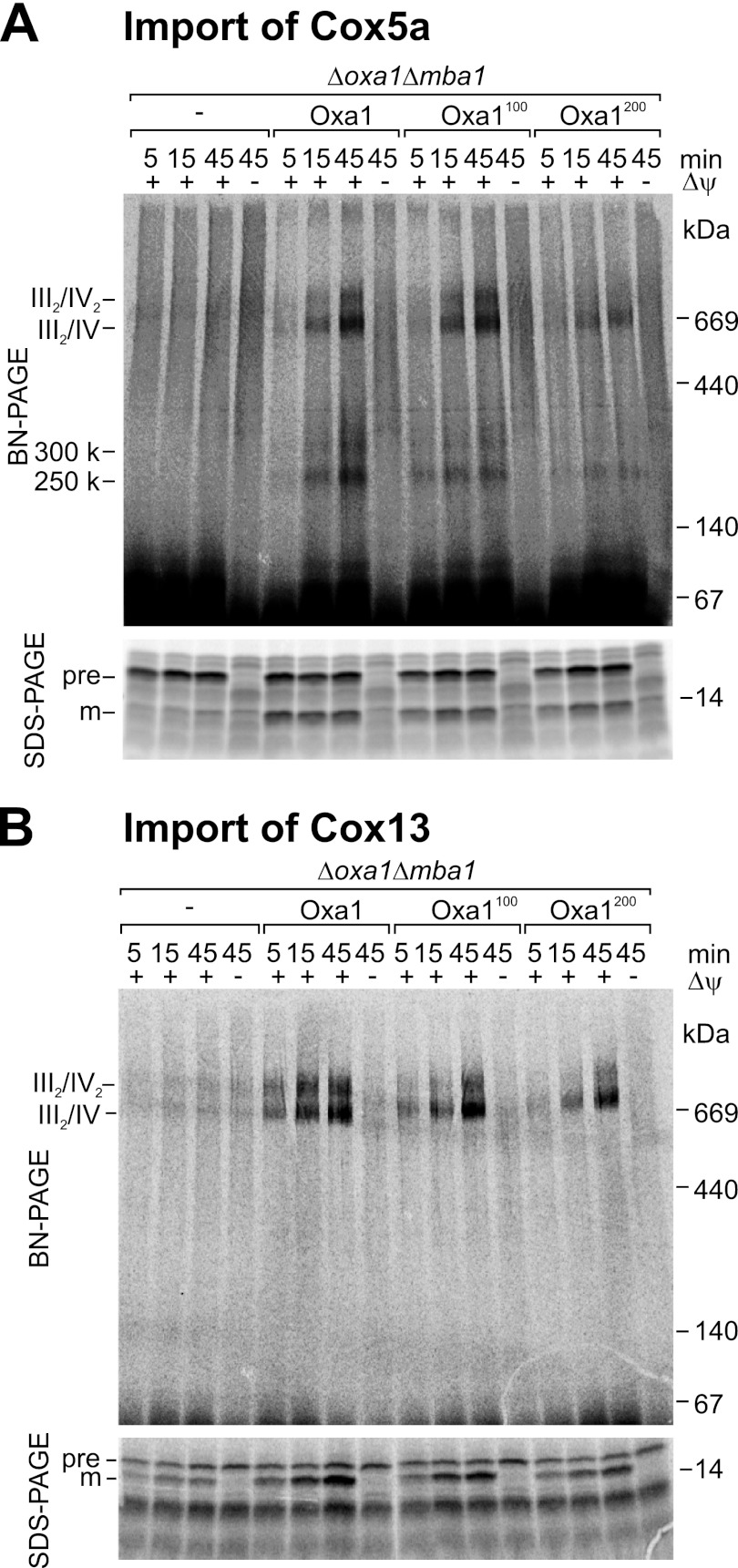
Insertion of Linkers into Oxa1 Impairs Assembly of the Cytochrome c Oxidase Complex
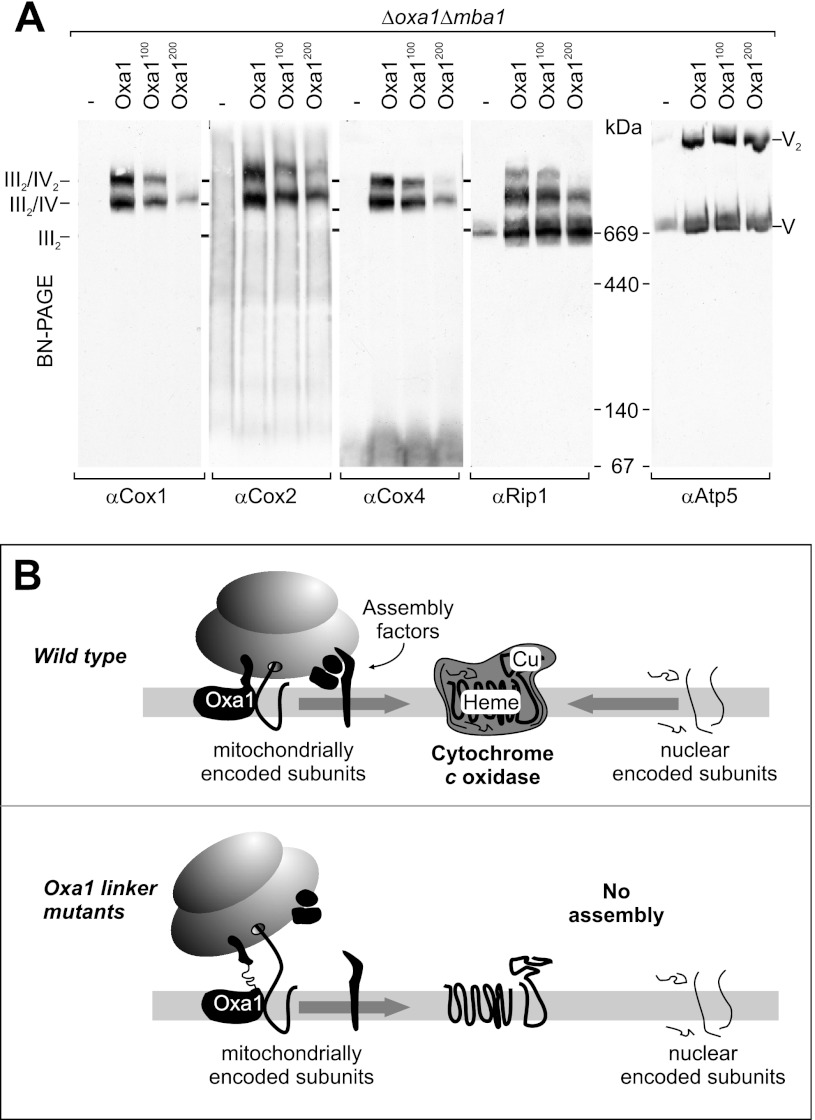
The strong defect in the biogenesis of cytochrome c oxidase in the Oxa1 spacer mutants is apparently not caused by a block of protein insertion. We therefore asked whether these mutants show defect in later stages of cytochrome c oxidase biogenesis, in particular in the complex assembly. The enzymes of the respiratory chain are associated with large supercomplexes presumably to increase the efficiency of channeling electrons from one complex to the next (34). These supercomplexes can be separated by BN-PAGE. To follow the assembly of cytochrome c oxidase experimentally, we radiolabeled the nuclear encoded subunits Cox5a and Cox13 in vitro and followed the assembly after import into isolated mitochondria by BN-PAGE. Cox5a (Fig. 6A) and Cox13 (Fig. 6B) were efficiently imported into the Oxa1 mutant mitochondria, but their assembly into the supercomplexes of the respiratory chain occurred only with considerable reduced kinetics and efficiency. This indicates that the insertion of linker regions into the Oxa1 sequence caused a defect in complex assembly. The strongly reduced levels of these supercomplexes were confirmed by Western blotting of isolated mitochondria. As shown in Fig. 7A, mitochondria lacking Oxa1 and Mba1 contained almost no detectable supercomplexes. Upon expression of wild-type Oxa1, considerable levels of the different supercomplexes III2, III2/IV, III2/IV2, and V2 were present. Whereas the III2 and V2 supercomplexes, which contain two cytochrome bc1 complexes or two ATPase complexes, respectively, were also present in the Oxa1100 and Oxa1200 strains, the levels of the supercomplexes containing cytochrome c oxidase were severely diminished. Thus, spatial separation of the insertion domain and the ribosome-binding region of Oxa1 did not interfere with the insertion of mitochondrial translation products into the inner membrane but rather with a much later stage in the biogenesis of cytochrome c oxidase, indicating that the ribosome binding to Oxa1 has a critical function in coordinating cytochrome c oxidase assembly in the inner membrane of mitochondria.
FIGURE 6.
Insertion of linkers into Oxa1 impairs the assembly of the cytochrome c oxidase complex. 35S-Labeled precursor forms of Cox5a (A) and Cox13 (B) were imported into isolated mitochondria for the indicated times. Non-imported material was removed by treatment with proteinase K. Mitochondria were lysed with digitonin before protein complexes were separated by BN-PAGE (upper panels) or SDS-PAGE (lower panels). pre, precursor; m, mature.
FIGURE 7.
Oxa1 linker mutants contain reduced levels of cytochrome c oxidase-containing supercomplexes. A, digitonin extracts of mitochondria were analyzed by BN-PAGE and Western blotting with antibodies against subunits of the cytochrome c oxidase complex (Cox1, Cox2, and Cox4), the cytochrome bc1 complex (Rip1), or the ATPase (Atp5). B, model of mitochondrial ribosomes serving as a binding platform for assembly factors. The model proposes that the close contact of mitochondrial ribosomes with the inner membrane allows the binding of assembly factors to the sites at which newly synthesized proteins are inserted into the membrane. Accordingly, spatial separation of the membrane-embedded insertase region and the ribosome-binding region of Oxa1 interferes with the coordination of assembly factors and therefore reduces the efficiency of cytochrome c oxidase assembly.
DISCUSSION
The biogenesis of the enzymes of the respiratory chain is a complicated process that is mastered by a plethora of specialized proteins. Only for cytochrome c oxidase, >30 assembly factors were identified, which are involved in translational regulation, membrane integration, heme insertion, copper binding, and the assembly of different subunits (35–39). Although biochemical studies or genetic epistasis analyses attributed specific steps in the biogenesis pathway to most of these factors, the molecular function of most of these components is unknown. This is due in part to the fact that, in contrast to the biogenesis of the ATPase complex (40–42), no reconstituted systems could be developed to follow the complete assembly pathway of cytochrome c oxidase in vitro or in isolated mitochondria.
Oxa1 plays a very early role in cytochrome c oxidase biogenesis, as it catalyzes the membrane integration of its mitochondrially encoded subunits. In this study, we characterized Oxa1 variants in which we inserted flexible linker regions of 100 or 200 residues. In the absence of the ribosome receptor Mba1, these mutants showed defects in the levels of cytochrome c oxidase, which were particularly severe in the Oxa1200 mutant. These defects included reduced growth on non-fermentative carbon sources, reduced respiratory activity of mitochondria, reduced activity of cytochrome c oxidase, reduced steady-state levels of Cox1, and reduced levels of cytochrome c oxidase-containing supercomplexes. However, membrane insertion of mitochondrial translation products, in particular insertion of the Oxa1 substrate Cox2, still occurred in the Oxa1200 mutant with similar efficiency as in the Oxa1 variant that lacked the linker region. Thus, the defects observed in the linker mutants are obviously not caused by a block in membrane insertion but rather by defects farther downstream in the assembly process of cytochrome c oxidase.
How do the linker regions interfere with the assembly of cytochrome c oxidase? The results shown in this study suggest that the spatial proximity of ribosomes to the insertion site in the inner membrane is important for cytochrome c oxidase biogenesis, potentially because this helps to orchestrate the activities of additional assembly factors required for biogenesis of cytochrome c oxidase (Fig. 7B). Loosening the tight contact of ribosomes with the membrane in Δmba1 Oxa1100 and Δmba1 Oxa1200 strains may impair the function of assembly factors, for some of which binding to mitochondrial ribosomes was shown previously. 1) Cox11, the component that inserts copper ions into the newly synthesized Cox1 protein, copurifies with mitochondrial ribosomes (43, 44). 2) Mss51 serves as a chaperone that binds directly to newly synthesized Cox1 and to the assembly factors Cox14, Coa1, Coa3, and Shy1 (45–48). In addition, Mss51 interacts with mitochondrial ribosomes and initiates Cox1 synthesis (49, 50). 3) The inner membrane protein Mdm38 cooperates with Mba1 in ribosome binding and plays a regulatory role in the synthesis and assembly of mitochondrial translation products (51, 52). Thus, the mitochondrial ribosome apparently serves as a complex binding platform for a multitude of assembly factors. The specialization of the mitochondrial translation system in the synthesis of a small number of respiratory chain subunits presumably made a role for the ribosome in the assembly of respiratory chain complexes possible. All proteins synthesized in mitochondria are subunits of complex protein structures. It is not clear why mitochondrial DNA was maintained in evolution at all. The best accepted hypothesis is that mitochondrial translation is important to regulate synthesis and assembly of proteins in a timely and spatially controlled manner (48, 53). Over the last years, studies in yeast identified a number of regulatory feedback loops that make sure that mitochondrial translation products are synthesized only when they can be productively assembled (49, 50, 54–56). This is achieved by a crosstalk of assembly and translation factors on mitochondrial ribosomes. Our results here suggest that the ribosome-binding region of Oxa1 contributes to the coordination of the assembly process. This region might have been added during evolution not only to improve co-translational insertion of membrane proteins (a process that is mastered in bacteria that lack a ribosome-binding domain on YidC) but particularly to position the ribosome on the site of protein insertion to improve its function as an interaction platform for assembly factors. It will be exciting to explore the structure and function of these mitochondrial ribosome assembly complexes in more detail.
Acknowledgments
We thank Markus Hildenbeutel and Linda Janssen for help with some experiments, Steffen Frey and Dirk Görlich (Max Planck Institute) for the mutagenized NSP1 sequence, and Alex Tzagoloff (Columbia University, New York) for the anti-Cox20 antibody.
This work was supported by Deutsche Forschungsgemeinschaft Grant FOR 967 (to J. M. H. and P. R.), the Landesschwerpunkt für Membrantransport in Rheinland-Pfalz (to J. M. H.), the Göttingen Graduate School for Neurosciences and Molecular Biosciences (to B. B.), and the Max-Planck-Gesellschaft (to P. R.).
- IMS
- intermembrane space
- BN
- blue native
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 2. Kramer G., Boehringer D., Ban N., Bukau B. (2009) The ribosome as a platform for co-translational processing, folding, and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 [DOI] [PubMed] [Google Scholar]
- 3. Giglione C., Fieulaine S., Meinnel T. (2009) Co-translational processing mechanisms: towards a dynamic 3D model. Trends Biochem. Sci. 34, 417–426 [DOI] [PubMed] [Google Scholar]
- 4. Gautschi M., Lilie H., Fünfschilling U., Mun A., Ross S., Lithgow T., Rücknagel P., Rospert S. (2001) RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. U.S.A. 98, 3762–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jaiswal H., Conz C., Otto H., Wölfle T., Fitzke E., Mayer M. P., Rospert S. (2011) The chaperone network connected to human ribosome-associated complex. Mol. Cell. Biol. 31, 1160–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson A. E., van Waes M. A. (1999) The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15, 799–842 [DOI] [PubMed] [Google Scholar]
- 7. Osborne A. R., Rapoport T. A., van den Berg B. (2005) Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 21, 529–550 [DOI] [PubMed] [Google Scholar]
- 8. Frauenfeld J., Gumbart J., Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., Beckmann R. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ott M., Prestele M., Bauerschmitt H., Funes S., Bonnefoy N., Herrmann J. M. (2006) Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 25, 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watson K. (1972) The organization of ribosomal granules within mitochondrial structures of aerobic and anaerobic cells of Saccharomyces cerevisae. J. Cell Biol. 55, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jia L., Dienhart M., Schramp M., McCauley M., Hell K., Stuart R. A. (2003) Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22, 6438–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J. M. (2003) Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22, 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haque M. E., Spremulli L. L., Fecko C. J. (2010) Identification of protein-protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J. Biol. Chem. 285, 34991–34998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haque M. E., Elmore K. B., Tripathy A., Koc H., Koc E. C., Spremulli L. L. (2010) Properties of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L and its interactions with mammalian mitochondrial ribosomes. J. Biol. Chem. 285, 28353–28362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kohler R., Boehringer D., Greber B., Bingel-Erlenmeyer R., Collinson I., Schaffitzel C., Ban N. (2009) YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol. Cell 34, 344–353 [DOI] [PubMed] [Google Scholar]
- 16. He S., Fox T. D. (1997) Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol. Biol. Cell 8, 1449–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hell K., Neupert W., Stuart R. A. (2001) Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20, 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bohnert M., Rehling P., Guiard B., Herrmann J. M., Pfanner N., van der Laan M. (2010) Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr. Biol. 20, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 19. Dalbey R. E., Wang P., Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 [DOI] [PubMed] [Google Scholar]
- 20. Jia L., Kaur J., Stuart R. A. (2009) Mapping of the Saccharomyces cerevisiae Oxa1-mitochondrial ribosome interface and identification of MrpL40, a ribosomal protein in close proximity to Oxa1 and critical for oxidative phosphorylation complex assembly. Eukaryot. Cell 8, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Preuss M., Leonhard K., Hell K., Stuart R. A., Neupert W., Herrmann J. M. (2001) Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J. Cell Biol. 153, 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gruschke S., Gröne K., Heublein M., Hölz S., Israel L., Imhof A., Herrmann J. M., Ott M. (2010) Proteins at the polypeptide tunnel exit of the yeast mitochondrial ribosome. J. Biol. Chem. 285, 19022–19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frey S., Richter R. P., Görlich D. (2006) FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 [DOI] [PubMed] [Google Scholar]
- 24. Altmann K., Dürr M., Westermann B. (2007) in Mitochondria: Practical Protocols (Leister D., Herrmann J. M. eds) pp. 81–90, Humana Press, Totowa, NJ [Google Scholar]
- 25. Funes S., Herrmann J. M. (2007) Analysis of mitochondrial protein synthesis in yeast. Methods Mol. Biol. 372, 255–263 [DOI] [PubMed] [Google Scholar]
- 26. Brandner K., Rehling P., Truscott K. N. (2005) The carboxyl-terminal third of the dicarboxylate carrier is crucial for productive association with the inner membrane twin-pore translocase. J. Biol. Chem. 280, 6215–6221 [DOI] [PubMed] [Google Scholar]
- 27. Nunnari J., Fox T. D., Walter P. (1993) A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262, 1997–2004 [DOI] [PubMed] [Google Scholar]
- 28. Schneider A., Behrens M., Scherer P., Pratje E., Michaelis G., Schatz G. (1991) Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 10, 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hell K., Herrmann J., Pratje E., Neupert W., Stuart R. A. (1997) Oxa1p mediates the export of the N and C termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 418, 367–370 [DOI] [PubMed] [Google Scholar]
- 30. Facey S. J., Neugebauer S. A., Krauss S., Kuhn A. (2007) The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J. Mol. Biol. 365, 995–1004 [DOI] [PubMed] [Google Scholar]
- 31. Fujihira E., Tamura N., Yamaguchi A. (2002) Membrane topology of a multidrug efflux transporter, AcrB, in Escherichia coli. J. Biochem. 131, 145–151 [DOI] [PubMed] [Google Scholar]
- 32. Hell K., Tzagoloff A., Neupert W., Stuart R. A. (2000) Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 275, 4571–4578 [DOI] [PubMed] [Google Scholar]
- 33. Elliott L. E., Saracco S. A., Fox T. D. (2012) Multiple roles of the Cox20 chaperone in assembly of Saccharomyces cerevisiae cytochrome c oxidase. Genetics 190, 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schägger H., Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tzagoloff A., Dieckmann C. L. (1990) PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., Westermann B., Herrmann J. M., Riemer J. (2009) Systematic analysis of the twin Cx9C protein family. J. Mol. Biol. 393, 356–368 [DOI] [PubMed] [Google Scholar]
- 37. Carr H. S., Winge D. R. (2003) Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36, 309–316 [DOI] [PubMed] [Google Scholar]
- 38. Herrmann J. M., Funes S. (2005) Biogenesis of cytochrome oxidase–sophisticated assembly lines in the mitochondrial inner membrane. Gene 354, 43–52 [DOI] [PubMed] [Google Scholar]
- 39. Fernández-Vizarra E., Tiranti V., Zeviani M. (2009) Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta 1793, 200–211 [DOI] [PubMed] [Google Scholar]
- 40. Rak M., Gokova S., Tzagoloff A. (2011) Modular assembly of yeast mitochondrial ATP synthase. EMBO J. 30, 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wagner K., Perschil I., Fichter C. D., van der Laan M. (2010) Stepwise assembly of dimeric F1Fo-ATP synthase in mitochondria involves the small Fo-subunits k and i. Mol. Biol. Cell 21, 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matthies D., Haberstock S., Joos F., Dötsch V., Vonck J., Bernhard F., Meier T. (2011) Cell-free expression and assembly of ATP synthase. J. Mol. Biol. 413, 593–603 [DOI] [PubMed] [Google Scholar]
- 43. Carr H. S., George G. N., Winge D. R. (2002) Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 277, 31237–31242 [DOI] [PubMed] [Google Scholar]
- 44. Khalimonchuk O., Ostermann K., Rödel G. (2005) Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the CuB site formation of cytochrome c oxidase. Curr. Genet. 47, 223–233 [DOI] [PubMed] [Google Scholar]
- 45. Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., Winge D. R. (2007) Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mick D. U., Vukotic M., Piechura H., Meyer H. E., Warscheid B., Deckers M., Rehling P. (2010) Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 191, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fontanesi F., Clemente P., Barrientos A. (2011) Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 286, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mick D. U., Fox T. D., Rehling P. (2011) Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barrientos A., Zambrano A., Tzagoloff A. (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perez-Martinez X., Broadley S. A., Fox T. D. (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bauerschmitt H., Mick D. U., Deckers M., Vollmer C., Funes S., Kehrein K., Ott M., Rehling P., Herrmann J. M. (2010) Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol. Biol. Cell 21, 1937–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frazier A. E., Taylor R. D., Mick D. U., Warscheid B., Stoepel N., Meyer H. E., Ryan M. T., Guiard B., Rehling P. (2006) Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 172, 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Allen J. F. (2003) The function of genomes in bioenergetic organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 19–37; discussion 37–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gruschke S., Kehrein K., Römpler K., Gröne K., Israel L., Imhof A., Herrmann J. M., Ott M. (2011) Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 193, 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rak M., Tzagoloff A. (2009) F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 106, 18509–18514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herrmann J. M., Woellhaf M. W., Bonnefoy N. (2012) Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]