Abstract
We have discovered a novel polyomavirus present in multiple human stool samples. The virus was initially identified by shotgun pyrosequencing of DNA purified from virus-like particles isolated from a stool sample collected from a healthy child from Malawi. We subsequently sequenced the virus' 4,927-bp genome, which has been provisionally named MW polyomavirus (MWPyV). The virus has genomic features characteristic of the family Polyomaviridae but is highly divergent from other members of this family. It is predicted to encode the large T antigen and small T antigen early proteins and the VP1, VP2, and VP3 structural proteins. A real-time PCR assay was designed and used to screen 514 stool samples from children with diarrhea in St. Louis, MO; 12 specimens were positive for MWPyV. Comparison of the whole-genome sequences of the index Malawi case and one St. Louis case demonstrated that the two strains of MWPyV varied by 5.3% at the nucleotide level. The number of polyomaviruses found in the human body continues to grow, raising the question of how many more species have yet to be identified and what roles they play in humans with and without manifest disease.
INTRODUCTION
Over the past 5 years, seven novel polyomaviruses have been discovered in humans, including KI polyomavirus (KIPyV) (2), WU polyomavirus (WUPyV) (13), Merkel cell polyomavirus (MCPyV) (11), human polyomavirus 6 (HPyV6) (40), human polyomavirus 7 (HPyV7) (40), trichodysplasia spinulosa-associated polyomavirus (TSPyV) (45), and human polyomavirus 9 (HPyV9) (42). Polyomaviruses also infect a wide variety of mammalian and avian hosts, including the recently described novel polyomaviruses of bats (Myotis species) (32), sea lions (Zalophus californianus) (8, 47), multimammate mice (Mastomys species) (33), canaries (Serinus canaria) (19), orangutans (Pongo species) (17), squirrel monkeys (Saimiri species) (46), chimpanzees (Pan troglodytes subsp. verus) (28), and gorillas (Gorilla gorilla) (28).
Viruses in the Polyomaviridae family typically possess ∼5,000-bp circular, double-stranded DNA genomes. The genome can be divided into three parts—the regulatory region, the early region, and the late region. The regulatory region, also called the noncoding control region (NCCR), contains the origin of replication and promoters for the early and late regions. Transcription occurs bidirectionally from the regulatory region. The early region is expressed from a common primary transcript and is alternatively spliced to produce the large T antigen (LTAg) and small T antigen (STAg) prior to viral replication. LTAg and STAg typically share the first ∼80 amino acids. The late region is expressed after viral replication has begun and encodes the structural proteins VP1, VP2, and VP3. VP1, the major structural protein, typically comprises over 70% of the viral particle and is the antigenic portion of the virus to which most natural antibodies are made (20).
Disease associations have been established for some of the human polyomaviruses. The two well-studied human polyomaviruses BK polyomavirus (BKPyV) and JC polyomavirus (JCPyV) are important human pathogens. BKPyV is known to cause BK nephropathy, which can lead to renal allograft failure, and hemorrhagic cystitis, while JCPyV is the etiological agent of progressive multifocal leukoencephalopathy (PML). Both viruses are ubiquitous worldwide, with seroprevalence rates of 55 to 85% for BKPyV and 44 to 77% for JCPyV (25). Following primary infection in childhood, BKPyV and JCPyV establish persistent latent infections that can periodically reactivate, leading to shedding of infectious virus in the urine (36). Primary infection and periodic reactivation are typically asymptomatic unless the host is immunocompromised, in which case life-threatening illness can occur (36). MCPyV is associated with Merkel cell carcinoma (MCC), a rare but aggressive skin cancer. MCPyV DNA is found in ∼80% of MCC tumors and is clonally integrated into a subset of these (14). TSPyV has been linked to trichodysplasia spinulosa, a very rare skin condition associated with immunosuppression following organ transplantation (24). It is unclear if the other human polyomaviruses play a role in disease.
The recently discovered human polyomaviruses have all been identified through the use of molecular methods for detection of viral nucleic acids. WUPyV and KIPyV were discovered using high-throughput Sanger sequencing (2, 13). MCPyV was identified using digital transcriptome subtraction, which entails pyrosequencing of a cDNA library followed by subtraction of human reads to identify novel viral sequences (11). HPyV6 and TSPyV were discovered using rolling circle amplification (RCA) (40, 45), and consensus PCR primers were utilized to find HPyV7 and HPyV9 (40, 42).
We used shotgun pyrosequencing of purified virus-like particles (VLPs) recovered from a fecal sample to discover a novel polyomavirus in the stool of a healthy child from Malawi. The virus was also detected in 12 additional stool samples from the United States, indicating it has a wide geographic distribution. As stool is not a sterile site, it is currently unknown whether this polyomavirus actively infects humans. Finally, we compared the whole genome nucleotide sequences of the index Malawi case and a case from St. Louis and found these two strains to have 5.3% nucleotide variation.
MATERIALS AND METHODS
Human studies.
This study was approved by the College of Medicine Research and Ethics Committee of the University of Malawi and the Human Research Protection Office of Washington University in St. Louis. The index stool specimen was obtained from a healthy, breast-fed, 15-month-old female living in Mayaka, Malawi, in September 2008 as part of a global gut microbiome survey (48).
A total of 514 stool specimens from St. Louis were tested for MWPyV. Stool samples were from children, age 0 to 18 years, with diarrhea and were submitted to the St. Louis Children's Hospital, St. Louis, MO, microbiology laboratory for bacterial culture from July 2009 to June 2010.
Sample preparation and 454 pyrosequencing.
VLPs were purified as described earlier (37) with minor modifications. In brief, 50 mg of a frozen fecal sample was resuspended in 400 μl of SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris [pH 7.5], and 0.002% gelatin [wt/vol]). Following centrifugation (2,500 × g for 10 min at 4°C) and filtration through 0.45- and 0.22-μm-pore-size Millex filters (Millipore) to remove bacterial cells and large particles, the sample was treated with chloroform (0.2 volumes) for 10 min and centrifuged for 5 min at 2,500 × g. The aqueous phase was treated with Baseline-Zero DNase (2.5 U/ml) (Epicentre) for 1 h at 37°C to remove free DNA, followed by an incubation at 65°C for 15 min to inactivate the enzyme. To extract VLP-associated DNA, the solution was treated with 10 μl 10% SDS and 3 μl proteinase K (20 mg/ml) for 20 min at 56°C. Subsequently, 35 μl of 5 M NaCl and 28 μl of a solution of 10% cetyltrimethylammonium bromide-0.7 M NaCl were introduced. After a 10-min incubation at 65°C, an equal volume of chloroform was added, and the mixture was centrifuged for 5 min at 8,000 × g at room temperature. The supernatant was transferred to a new tube, and an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added, followed by centrifugation for 5 min at 8,000 × g at room temperature. The supernatant was collected, and the DNA was purified using Qiagen MiniElute columns by following the manufacturer's instructions, with a final elution volume of 35 μl.
Purified VLP-derived DNA (1 μl) was used as an input in a 20-μl RCA reaction mixture using the Illustra GenomiPhi V2 kit (GE Healthcare) as recommended by the manufacturer (n = four independent reactions). After 90 min of amplification, the four reactions were pooled and purified using the Qiagen DNeasy kit. DNA (500 ng) was subjected to 454 FLX Titanium pyrosequencing.
Analysis of pyrosequencing reads.
The individual 454 reads were analyzed using a custom bioinformatics pipeline as previously described (7). In brief, unique, high-quality reads were aligned against the reference human genome and the GenBank nucleotide database using BLASTn. Reads with no hits or hits with an E-value greater than e−5 were then aligned using BLASTx to the GenBank nr (nonredundant) database, and reads aligning to viral sequences with the lowest E value were identified.
Complete genome sequencing.
PCR primers were designed to span the gaps between the six reads showing significant similarity to polyomaviruses generated by pyrosequencing to obtain an initial whole-genome sequence. The sequences for these primers are available upon request. The complete MWPyV genome derived from the index Malawi case (designated strain MA095, GenBank JQ898291) was sequenced to greater than 3× coverage using four sets of overlapping PCR primers. They were (listed 5′ to 3′) ACTTAAACCATGTTCCTGACTCTGT (ES087) and ACAGAGATTACAGCACCCATATACT (ES091), GCATCTGCCCTGGTACAAACA (ES088) and CAGACAACTCAGAAGTTTCCACCTC (ES092), GAAGTAGAAGGAGAGGAAAATGCCG (ES089) and TGCTGTTGAGGATACACAACAAGAC (ES093), and AGGCTGCTTAAAGGCCTATGAATG (ES090) and CTGAAACACCAGTTGCTCCAGC (ES094). Amplicons from independent PCRs were cloned into pCR4 (Invitrogen) and bidirectionally sequenced. The complete genome from St. Louis sample WD976 (strain WD976; GenBank accession no. JQ898292) was amplified and sequenced to greater than 3× coverage in the same manner, using the same primer pairs.
Genome annotation.
Open reading frames (ORFs) were predicted using NCBI ORF Finder. The LTAg and STAg ORFs were manually scanned for conserved splice donor and acceptor sites. Conserved motifs in the TAgs and in the NCCR were identified using NCBI CD-Search software (30) and by manual identification. Prediction of putative binding sites for transcription factors was performed using AliBaba software, version 2.1 (15). The NCCR region was scanned for palindrome patterns using the EMBOSS palindrome software (38).
Phylogenetic analysis.
Protein sequences associated with the reference genomes for 27 polyomaviruses were obtained from GenBank; these included baboon polyomavirus (NC_007611; SA12) (6), bat polyomavirus (NC_011310; BatPyV) (32), B-lymphotropic polyomavirus (NC_004763; LPyV) (34), BKPyV (NC_001538) (43), Bornean orangutan polyomavirus (NC_013439; OraV1) (17), bovine polyomavirus (NC_001442; BPyV) (41), California sea lion polyomavirus (NC_013796; SLPyV) (8), hamster polyomavirus (NC_001663; HaPyV) (9), JCPyV (NC_001699) (12), MCPyV (HM011557) (40), murine pneumotropic virus (NC_001505; MPtV) (31), murine polyomavirus (NC_001515; MPyV) (16), simian virus 40 (NC_001669; SV40) (27), squirrel monkey polyomavirus (NC_009951; SqPyV) (46), Sumatran orangutan polyomavirus (FN356901; OraV2) (17), TSPyV (NC_014361) (45), HPyV6 (NC_014406) (40), HPyV7 (NC_014407) (40), KIPyV (NC_009238) (2), WUPyV (NC_009539) (13), avian polyomavirus (NC_004764; APyV) (39), canary polyomavirus (GU345044; CaPyV) (19), crow polyomavirus (NC_007922; CPyV) (23), finch polyomavirus (NC_007923; FPyV) (23), goose hemorrhagic polyomavirus (NC_004800; GHV) (22), chimpanzee polyomavirus (NC_014743; ChPyV) (10), and HPyV9 (NC_015150) (42). The predicted open reading frames for MWPyV LTAg, VP1, and VP2 were aligned with the corresponding proteins from the 27 known polyomaviruses using Fast Statistical Alignment (FSA) software, version 1.15.2 (5). For the LTAg analysis, unalignable regions were removed, and the remainder of the alignment was concatenated. Maximum likelihood trees were generated using PhyML, version 3.0 (18), with 1,000 bootstrap replicates and the best model as determined by Prot Test software, version 2.4 (1); these were RtRev for VP1 and LG for VP2 and LTAg.
Nucleic acid extraction.
Stools which had been frozen at −80°C were diluted approximately 1:6 in phosphate-buffered saline (PBS) and filtered through 0.45-μm-pore-size membranes prior to extraction. Total nucleic acids were extracted using an Ampliprep Cobas automated extractor (Roche) and eluted in a volume of 75 μl. The samples were arrayed in a 96-well plate for storage at −80°C.
Real-time PCR screening of the St. Louis cohort.
A TaqMan real-time PCR assay was designed to target the MWPyV LTAg using Primer Express software (Applied Biosystems). Primers and probe used for this assay were 5′-TGAGAAGGCCCCGGTTCT-3′ (ES105), 5′-GAGGATGGGATGAAGATTTAAGTTG-3′ (ES106), and 5′-FAM-CCTCATCACTGGGAGC-MGBNFQ-3′ (ES107) (where FAM is 6-carboxyfluorescein). The resulting amplicon was 73 bp. Standard curves were generated using serial 10-fold dilutions ranging from 5 × 106 to 5 copies of a positive-control plasmid (plasmid K-p31) per reaction. The 25-μl PCR mixtures consisted of 5 μl of extracted sample, 1× universal TaqMan real-time PCR master mix (Applied Biosystems), 12.5 pmol of each primer, and 4 pmol of the probe. Samples were tested in a 96-well-plate format, with eight water negative controls (one per row) and one positive control containing 50 copies of plasmid per plate. The cycling conditions were 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 s followed by 60°C for 1 min. Reactions were run on an ABI 7500 real-time thermocycler (Applied Biosystems). The threshold of all plates was set at a standard value, and the data were analyzed using the ABI software. Samples were counted as positive if their threshold cycle (CT) value was <35.
Nucleotide sequence accession numbers.
The sequences reported here were deposited in GenBank under the accession numbers JQ898291 (index case, strain MA095) and JQ898292 (St. Louis case, strain WD976).
RESULTS
Discovery of a novel polyomavirus by pyrosequencing.
MW polyomavirus was discovered in a stool sample from a child from Malawi; the sample was collected in September 2008 as part of a global gut microbiome survey project (48). Following purification of VLPs by passage through 0.45- and 0.22-μm-pore-sized filters and subsequent DNase treatment, DNA was extracted from the VLPs and amplified using the highly processive phi29 polymerase. The resulting material was subjected to 454 pyrosequencing. Six reads were identified with limited similarity to known polyomaviruses. Three of the initial six reads could be assembled into one 959-bp contig, with the highest scoring BLASTx hit possessing 36% amino acid identity to LPyV STAg. The other three reads all aligned to the VP1 protein of known polyomaviruses by BLASTx and shared 64%, 48%, and 59% amino acid identity to JCPyV VP1, TSPyV VP1, and JCPyV VP1, respectively.
Complete genome sequencing and genome analysis.
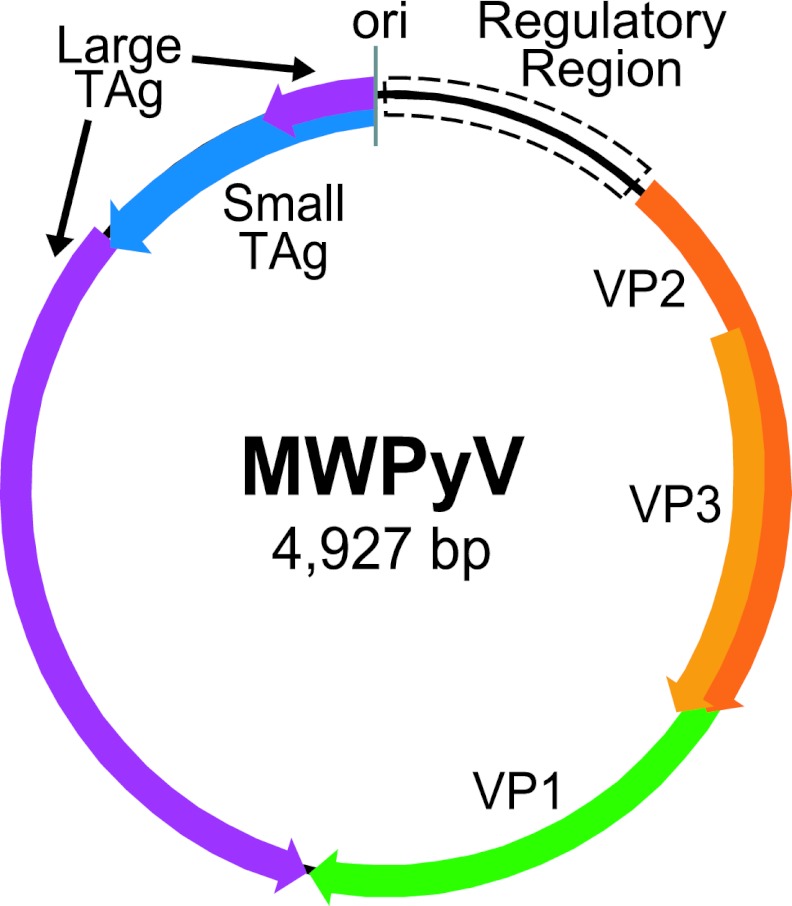
A series of PCR primers was designed based on the initial six reads. Sequencing of the resulting amplicons yielded a complete genome of 4,927 bp (Fig. 1). ICTV has set the demarcation criteria for proposed new polyomaviruses at 81% nucleotide identity over the whole genome (21). Based on the limited sequence similarity to any known polyomaviruses, we named the novel virus MW polyomavirus (MWPyV) after its discovery in Malawi. The overall GC content of MWPyV was 37%, which is very similar to those of WUPyV (39%), BKPyV (39%), and JCPyV (40%). The MWPyV genome organization was characteristic of the known polyomaviruses and included an early region coding on one strand for LTAg and STAg and a late region coding on the opposite strand for the structural proteins VP1, VP2, and VP3. The sizes of the predicted ORFs were comparable to those of known polyomaviruses (Table 1).
Fig 1.
Genome organization of MWPyV. ori, origin of replication.
Table 1.
Putative proteins encoded by MWPyV (strain MA095)
| Protein | Putative coding region(s) | Predicted size (aa) | Calculated mass (kDa) | Range (aa) in other polyomaviruses |
|---|---|---|---|---|
| STAg | 4927–4328 | 199 | 23.4 | 124–198 |
| LTAg | 4927–4688, 4332–2566 | 668 | 77.0 | 599–817 |
| VP1 | 1353–2564 | 403 | 43.6 | 343–497 |
| VP2 | 431–1363 | 310 | 34.2 | 241–415 |
| VP3 | 761–1363 | 200 | 22.8 | 190–272 |
The TAgs and VP2 were separated by a regulatory region, which had an A/T-rich tract on the late side of the putative replication origin. The core origin of replication contained three repeats of the consensus pentanucleotide LTAg binding site, G(A/G)GGC (35) (two GAGGC and one GGGGC), and one nonconsensus binding site, TAGGC. Several polyomaviruses (BKPyV, JCPyV, WUPyV, KIPyV, and SV40) contain an imperfect palindrome sequence followed by additional LTAg binding sites to the early side of the four binding sites. Palindrome patterns were identified in MWPyV, but no additional LTAg binding sites were detected in this area. The regulatory region contained several predicted transcription factor binding sites, including multiple binding sites for four factors known to play a role in BKPyV viral transcription and regulation: Sp1, nuclear factor I (NFI), AP1, and C/EBP (29). Multiple binding sites were also identified for HNF-3, USF-2, and Oct-1. Many other transcription factors were predicted to bind to only one site.
Analysis of the MWPyV LTAg ORF revealed a conserved splice donor site immediately after amino acid 80; the position of this site was similar to that found in WUPyV, BKPyV, and JCPyV, which occur after amino acids 84, 81, and 81, respectively. Two consensus splice acceptor sites were identified, which would yield introns of 355 or 463 bp and proteins of 668 or 632 amino acids, respectively. Examination of the protein sequence of the 632-amino-acid form showed that it lacked the Rb-binding motif, which was contained in the excised intron. In contrast, the predicted 668-amino-acid protein included the conserved Rb-binding motif. Based on this analysis, we predicted the LTAg to be 668 amino acids.
MWPyV LTAg possessed conserved features common to other polyomavirus LTAgs, including a DNaJ domain containing the conserved region 1 (cr1) sequence and the highly conserved hexapeptide motif HPDKGG. These domains were followed by conserved region 2 (cr2), which contained the Rb-binding motif LxCxE (LSCNE in MWPyV), a putative nuclear localization signal (NLS), a canonical DNA binding domain, and a zinc finger region. Closer inspection of the zinc finger region revealed a conserved C2H2 zinc finger motif with the sequence C324, C327, H334, H339. There are typically three highly conserved amino acids N terminal to the first cysteine (C324) in this motif, including a tyrosine 10 amino acids away, an aspartic acid located 18 amino acids away, and an alanine present 25 amino acids away (35). In MWPyV, the aspartic acid and alanine residues were conserved, while the tyrosine was not and was replaced by a leucine. A conserved leucine-rich hydrophobic region C terminal to the aspartic acid was also present. Following the zinc finger region, the MWPyV LTAg contained the highly conserved ATPase-p53 binding domain, including the two conserved motifs GPXXXGKT and GXXXVNLE. There was no sequence corresponding to the host range domain present in SV40, BKPyV, SA12, and JCPyV (35).
In most polyomaviruses, STAg is encoded by a single unspliced ORF. In HaPyV and MPyV, the STAg transcript is spliced. Analysis of the MWPyV early region did not reveal an obvious splice donor site, so the STAg was predicted to be 199 amino acids. As LTAg and STAg share the first 80 amino acids, the STAg also contained the DNaJ domain. In the unique C-terminal part of STAg, there was a conserved cysteine-rich motif, CX5CX7–8CXCX2CX21–22CSCX2CX3WFG. This motif was conserved in MWPyV with the exception of the initial cysteine residue and the serine residue, which were an isoleucine and a phenylalanine, respectively.
MPyV and HaPyV encode a middle T antigen (MTAg) generated by alternative splicing; the MWPyV genome was scanned for splicing motifs similar to those used by MPyV and HaPyV. No obvious splice sites that would generate an appropriately sized third T antigen protein were identified, suggesting that MWPyV likely does not encode an MTAg.
Some polyomaviruses, including JCPyV and BKPyV, also encode an agnoprotein in the late region between the NCCR and the VP2 start codon. Analysis of the MWPyV sequence in this region yielded one 45-amino-acid ORF on the same strand as the structural proteins. However, because this ORF was not conserved in the other completely sequenced MWPyV strain, strain WD976 (described later in this report), we do not believe that MWPyV encodes an agnoprotein.
Phylogenetic analysis.
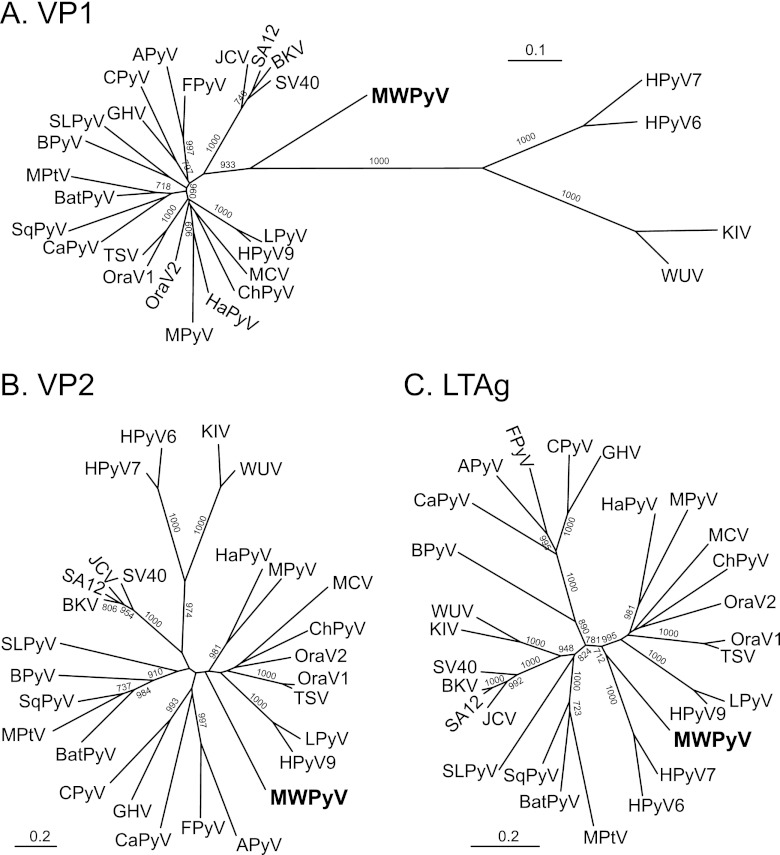
Maximum likelihood analysis of the VP1, VP2, and LTAg proteins demonstrated that MWPyV was highly divergent from all known polyomaviruses (Fig. 2). Analysis of VP1 sequences showed that MWPyV is midway between the Wukipolyomavirus and Orthopolyomavirus genera (Fig. 2A). In contrast, based on VP2 and LTAg sequences, MWPyV clustered with the clade containing HPyV9, LPyV, HaPyV, MPyV, TSPyV, MCV, ChPyV, and the orangutan polyomaviruses (Fig. 2B and C). The discordant phylogenetic relationships suggest that MWPyV might have been derived from an ancestral recombination event.
Fig 2.
Phylogenetic analysis of MW polyomavirus. Amino-acid-based trees were generated using the maximum likelihood method with 1,000 bootstrap replicates. Bootstrap values less than 700 are not shown. (A) VP1; (B) VP2; (C) LTAg.
Prevalence of MWPyV.
A TaqMan real-time PCR assay targeting the MWPyV LTAg was designed and validated using a positive-control plasmid; based on the standard curve, the MWPyV assay demonstrated a reliable detection limit of approximately five copies per reaction, yielded a linear regression R2 value of 0.99, and was 93% efficient. This real-time PCR assay was used to screen a cohort consisting of 514 stool samples from children at St. Louis Children's Hospital presenting with diarrhea. Twelve samples (2.3%) from the St. Louis cohort tested positive for MWPyV (Table 2).
Table 2.
Specimens and patients testing positive for MWPyV
| Sample | Patient | Age | Sex | CT | Date (mo/day/yr) | Tested positive | Tested negative |
|---|---|---|---|---|---|---|---|
| WD972 | 1 | 5 yr 0 mo | M | 21.68 | 8/10/09 | E. coli serotype O Rough | Enteric pathogen culturea (except E. coli), Giardia, Cryptosporidium, ova & parasite screen (O&P) |
| WD976 | 1 | 21.85 | 8/11/09 | E. coli serotype O Rough | Enteric pathogen culture (except E. coli), Giardia, Cryptosporidium, C. difficile, O&P | ||
| WD1226 | 1 | 28.21 | 12/22/09 | Enteric pathogen culture, O&P | |||
| WD1239 | 2 | 1 yr 0 mo | M | 31.76 | 12/29/09 | Enteric pathogen culture, C. difficile | |
| WD1314 | 3 | 1 yr 5 mo | F | 30.37 | 2/11/10 | Enteric pathogen culture, rotavirus | |
| WD1300 | 4 | 1 yr 8 mo | F | 34.99 | 2/4/10 | Enteric pathogen culture, rotavirus, viral culture | |
| WD1260 | 5 | 1 yr 4 mo | F | 29.91 | 1/12/10 | Enteric pathogen culture, O&P | |
| WD958 | 6 | 1 yr 8 mo | M | 31.89 | 8/3/09 | Enteric pathogen culture, C. difficile, O&P | |
| WD1039 | 7 | 4 yr 3 mo | F | 31.42 | 9/14/09 | Enteric pathogen culture, C. difficile | |
| WD1233 | 8 | 4 yr 9 mo | M | 32.33 | 12/25/09 | Enteric pathogen culture, rotavirus | |
| WD1055 | 9 | 5 yr 5 mo | M | 30.90 | 9/22/09 | Enteric pathogen culture | |
| WD1442 | 10 | 3 yr 0 mo | M | 32.41 | 5/24/10 | C. jejuni | Enteric pathogen culture (except C. jejuni) |
Enteric pathogen culture includes Salmonella, Shigella, E. coli O157, E. coli Shiga toxins not 0157, Yersinia, Aeromonas, Plesiomonas, and Campylobacter.
Three of the MWPyV-positive samples were obtained from a 5-year-old lung transplant recipient over a period of 4 months from August to December 2009 (patient 1; Table 2). This patient had received a lung transplant 3 years earlier and at the time of sampling presented with persistent, recurrent diarrhea. Two of the samples, WD972 and WD976, were obtained on consecutive days in August 2009, and both samples were positive for Escherichia coli serotype O Rough. The patient again presented with diarrhea in December 2009 (sample WD1226), but this sample had no growth in the enteric pathogen culture (including E. coli) and was negative for ova and parasites (Table 2). The other nine samples came from nine individual patients, ranging in age from 1 to 5 years (Table 2). Eight of the nine patients were negative for all organisms tested except MWPyV. Only patient 10 (sample WD1442) was positive for Campylobacter jejuni.
Strain variation.
To assess the extent of sequence variation between the St. Louis and Malawi isolates, we sequenced the complete genome of MWPyV from St. Louis sample WD976 to greater than 3× coverage. The two whole-genome sequences diverged by 5.3% at the nucleotide level. Strain WD976 had two insertions (11 bp and 1 bp) in the NCCR, which resulted in a genome size of 4,939 bp. The vast majority of the polymorphisms in the coding regions resulted in synonymous mutations. One notable mutation changed the size of the STAg ORF. The predicted TAA stop codon identified in the MA095 strain was mutated to AAA in WD976, resulting in a protein prediction of 206 amino acids, seven amino acids longer than the index genome's STAg.
DISCUSSION
We used a pyrosequencing strategy to identify a novel polyomavirus present in human stool. The initial discovery was in a stool specimen collected from a healthy child in Malawi. Further screening by real-time PCR demonstrated the presence of the virus in 12 stool samples collected from a cohort of patients in St. Louis, MO. These data demonstrated that MWPyV is geographically widespread in human populations and can be found on two continents. As the ICTV polyomavirus subgroup currently has no systematic naming convention for novel polyomaviruses, we chose to name this new virus using a two-letter convention following the model of BKPyV, JCPyV, KIPyV, and WUPyV; we made this decision for two reasons. First, we did not employ the numerical system used in the naming of HPyV6, HPyV7, and HPyV9 because we have not yet formally demonstrated that this virus infects humans and to avoid potential conflicts in temporal priority in describing novel polyomaviruses. Second, both MCPyV and TSPyV are named based on putative disease associations, but no disease association currently exists for our new virus. Therefore, we chose a two-letter abbreviation reflecting the geographic location of the index case.
The ICTV polyomavirus subgroup recently defined two mammalian genera, Orthopolyomavirus and Wukipolyomavirus, within the family Polyomaviridae based primarily on phylogenetic analysis of the late genes (combined VP1 and VP2) (21). Classification of MWPyV into one of these two genera is confounded by the distinct phylogenetic tree topologies that were generated for the VP1 and VP2 proteins (Fig. 2). The different topologies suggest that MWPyV is derived from an ancestral recombination event. Such recombination among polyomaviruses has been previously suggested (40).
We sequenced two complete genomes of MWPyV, one from the index child in Malawi and one from a child in St. Louis. A high degree of strain variation (5.3%) was observed between these two MWPyV strains, which is comparable to the ∼5% sequence divergence present in strains of BKPyV (26). It contrasts sharply with the very limited variation (<1.2%) seen with WUPyV worldwide (3). The primers and probe used in the aforementioned MWPyV real-time PCR assay were perfectly conserved in both strains and thus detect both stains with equal efficiency. However, it remains to be determined whether even greater variation in MWPyV can be discovered when broader consensus sequence-based assays are used. Others have speculated that sequence variation in BKPyV and JCPyV plays a role in viral pathogenesis and disease severity (4, 44). If MWPyV is ultimately found to be a pathogen, it will be interesting to determine whether there are strain-dependent pathogenic phenotypes. Among the differences we observed were a 7-amino-acid extension of the STAg and an 11-bp insertion in the NCCR in the WD976 strain versus the index Malawi strain. The functional consequences of these alterations remain to be defined.
One critical question is whether MWPyV is a bona fide infectious agent of humans and if so, what disease(s), if any, might be associated with MWPyV infection. The detection of MWPyV in stools of children with diarrhea, many of which have no known infection etiology, raises the possibility that MWPyV might play a role in human diarrhea. Alternatively, it is possible that MWPyV does not cause infection in the gastrointestinal tract but has a tropism for other human organ systems and is shed in stool as a mode of transmission or simply as a by-product. It is also possible that MWPyV is a dietary contaminant and does not actively infect humans. Approaches to answer whether MWPyV is an infectious agent include serological studies to determine whether the host mounts an antibody-based immune response to MWPyV and additional screening of specimens collected from sterile sites, such as serum or cerebrospinal fluid. Further studies will be needed to define whether MWPyV has additional tropisms in the human body and to assess potential associations with human disease.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research and the Bill and Melinda Gates Foundation. E.A.S. is supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program.
Footnotes
Published ahead of print 27 June 2012
REFERENCES
- 1. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 2. Allander T, et al. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bialasiewicz S, et al. 2010. Whole-genome characterization and genotyping of global WU polyomavirus strains. J. Virol. 84:6229–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boldorini R, et al. 2009. Genomic mutations of viral protein 1 and BK virus nephropathy in kidney transplant recipients. J. Med. Virol. 81:1385–1393 [DOI] [PubMed] [Google Scholar]
- 5. Bradley RK, et al. 2009. Fast statistical alignment. PLoS Comput. Biol. 5:e1000392 doi:10.1371/journal.pcbi.1000392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantalupo P, et al. 2005. Complete nucleotide sequence of polyomavirus SA12. J. Virol. 79:13094–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cantalupo PG, et al. 2011. Raw sewage harbors diverse viral populations. mBio 2(5):e00180–12 doi:10.1128/mBio.00180–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Colegrove KM, et al. 2010. Polyomavirus infection in a free-ranging California sea lion (Zalophus californianus) with intestinal T-cell lymphoma. J. Vet. Diagn. Invest. 22:628–632 [DOI] [PubMed] [Google Scholar]
- 9. Delmas V, Bastien C, Scherneck S, Feunteun J. 1985. A new member of the polyomavirus family: the hamster papovavirus. Complete nucleotide sequence and transformation properties. EMBO J. 4:1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deuzing I, et al. 2010. Detection and characterization of two chimpanzee polyomavirus genotypes from different subspecies. Virol. J. 7:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frisque RJ, Bream GL, Cannella MT. 1984. Human polyomavirus JC virus genome. J. Virol. 51:458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaynor AM, et al. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64 doi:10.1371/journal.ppat.0030064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gjoerup O, Chang Y. 2010. Update on human polyomaviruses and cancer. Adv. Cancer Res. 106:1–51 [DOI] [PubMed] [Google Scholar]
- 15. Grabe N. 2002. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2:S1–S15 [PubMed] [Google Scholar]
- 16. Griffin BE, Fried M, Cowie A. 1974. Polyoma DNA: a physical map. Proc. Natl. Acad. Sci. U. S. A. 71:2077–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groenewoud MJ, et al. 2010. Characterization of novel polyomaviruses from Bornean and Sumatran orang-utans. J. Gen. Virol. 91:653–658 [DOI] [PubMed] [Google Scholar]
- 18. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 19. Halami MY, et al. 2010. Whole-genome characterization of a novel polyomavirus detected in fatally diseased canary birds. J. Gen. Virol. 91:3016–3022 [DOI] [PubMed] [Google Scholar]
- 20. Imperiale MJ, Major EO. 2007. Polyomaviruses. In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21. Johne R, et al. 2011. Taxonomical developments in the family Polyomaviridae. Arch. Virol. 156:1627–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johne R, Muller H. 2003. The genome of goose hemorrhagic polyomavirus, a new member of the proposed subgenus Avipolyomavirus. Virology 308:291–302 [DOI] [PubMed] [Google Scholar]
- 23. Johne R, Wittig W, Fernandez-de-Luco D, Hofle U, Muller H. 2006. Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes. J. Virol. 80:3523–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kazem S, et al. 2012. Trichodysplasia spinulosa is characterized by active polyomavirus infection. J. Clin. Virol. 53:225–230 [DOI] [PubMed] [Google Scholar]
- 25. Knowles WA. 2006. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV). Adv. Exp. Med. Biol. 577:19–45 [DOI] [PubMed] [Google Scholar]
- 26. Krumbholz A, Bininda-Emonds OR, Wutzler P, Zell R. 2009. Phylogenetics, evolution, and medical importance of polyomaviruses. Infect. Genet. Evol. 9:784–799 [DOI] [PubMed] [Google Scholar]
- 27. Lebowitz P, Weissman SM. 1979. Organization and transcription of the simian virus 40 genome. Curr. Top. Microbiol. Immunol. 87:43–172 [DOI] [PubMed] [Google Scholar]
- 28. Leendertz FH, et al. 2011. African great apes are naturally infected with polyomaviruses closely related to Merkel cell polyomavirus. J. Virol. 85:916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang B, Tikhanovich I, Nasheuer HP, Folk WR. 2012. Stimulation of BK virus DNA replication by NFI family transcription factors. J. Virol. 86:3264–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mayer M, Dorries K. 1991. Nucleotide sequence and genome organization of the murine polyomavirus, Kilham strain. Virology 181:469–480 [DOI] [PubMed] [Google Scholar]
- 32. Misra V, et al. 2009. Detection of polyoma and corona viruses in bats of Canada. J. Gen. Virol. 90:2015–2022 [DOI] [PubMed] [Google Scholar]
- 33. Orba Y, et al. 2011. Detection and characterization of a novel polyomavirus in wild rodents. J. Gen. Virol. 92:789–795 [DOI] [PubMed] [Google Scholar]
- 34. Pawlita M, Clad A, zur Hausen H. 1985. Complete DNA sequence of lymphotropic papovavirus: prototype of a new species of the polyomavirus genus. Virology 143:196–211 [DOI] [PubMed] [Google Scholar]
- 35. Pipas JM. 1992. Common and unique features of T antigens encoded by the polyomavirus group. J. Virol. 66:3979–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Randhawa P, Vats A, Shapiro R. 2006. The pathobiology of polyomavirus infection in man. Adv. Exp. Med. Biol. 577:148–159 [DOI] [PubMed] [Google Scholar]
- 37. Reyes A, et al. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 39. Rott O, Kroger M, Muller H, Hobom G. 1988. The genome of budgerigar fledgling disease virus, an avian polyomavirus. Virology 165:74–86 [DOI] [PubMed] [Google Scholar]
- 40. Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schuurman R, Sol C, van der Noordaa J. 1990. The complete nucleotide sequence of bovine polyomavirus. J. Gen. Virol. 71(Pt 8):1723–1735 [DOI] [PubMed] [Google Scholar]
- 42. Scuda N, et al. 2011. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 85:4586–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seif I, Khoury G, Dhar R. 1979. The genome of human papovavirus BKV. Cell 18:963–977 [DOI] [PubMed] [Google Scholar]
- 44. Sunyaev SR, Lugovskoy A, Simon K, Gorelik L. 2009. Adaptive mutations in the JC virus protein capsid are associated with progressive multifocal leukoencephalopathy (PML). PLoS Genet. 5:e1000368 doi:10.1371/journal.pgen.1000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Meijden E, et al. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 6:e1001024 doi:10.1371/journal.ppat.1001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Verschoor EJ, et al. 2008. Molecular characterization of the first polyomavirus from a New World primate: squirrel monkey polyomavirus. J. Gen. Virol. 89:130–137 [DOI] [PubMed] [Google Scholar]
- 47. Wellehan JF, Jr, et al. 2011. Characterization of California sea lion polyomavirus 1: expansion of the known host range of the Polyomaviridae to Carnivora. Infect. Genet. Evol. 11:987–996 [DOI] [PubMed] [Google Scholar]
- 48. Yatsunenko T, et al. 9 May 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]