Abstract
The Mre11-Rad50-Nbs1 (MRN) complex plays a key role in the DNA damage response, presenting challenges for DNA viruses and retroviruses. To inactivate this complex, adenovirus (Ad) makes use of the E1B-55K and E4-open reading frame 6 (ORF6) proteins for ubiquitin (Ub)-mediated, proteasome-dependent degradation of MRN and the E4-ORF3 protein for relocalization and sequestration of MRN within infected-cell nuclei. Here, we report that Mre11 is modified by the Ub-related modifier SUMO-2 and Nbs1 is modified by both SUMO-1 and SUMO-2. We found that Mre11 and Nbs1 are sumoylated during Ad5 infection and that the E4-ORF3 protein is necessary and sufficient to induce SUMO conjugation. Relocalization of Mre11 and Nbs1 into E4-ORF3 nuclear tracks is required for this modification to occur. E4-ORF3-mediated SUMO-1 conjugation to Nbs1 and SUMO-2 conjugation to Mre11 and Nbs1 are transient during wild-type Ad type 5 (Ad5) infection. In contrast, SUMO-1 conjugation to Nbs1 is stable in cells infected with E1B-55K or E4-ORF6 mutant viruses, suggesting that Ad regulates paralog-specific desumoylation of Nbs1. Inhibition of viral DNA replication blocks deconjugation of SUMO-2 from Mre11 and Nbs1, indicating that a late-phase process is involved in Mre11 and Nbs1 desumoylation. Our results provide direct evidence of Mre11 and Nbs1 sumoylation induced by the Ad5 E4-ORF3 protein and an important example showing that modification of a single substrate by both SUMO-1 and SUMO-2 is regulated through distinct mechanisms. Our findings suggest how E4-ORF3-mediated relocalization of the MRN complex influences the cellular DNA damage response.
INTRODUCTION
The Mre11-Rad50-Nbs1 (MRN) complex is a sensor and effector of the DNA damage response (DDR) and plays an important role in DNA repair pathways (reviewed in reference 31). It is composed of meiotic recombination 11 (Mre11), radiation-sensitive 50 (Rad50), and Nijmegen breakage syndrome 1 (Nbs1) proteins. Mre11 binds DNA and has endo- and exonuclease activities, Rad50 contains coiled-coil domains that tether DNA termini, and Nbs1 mediates protein-protein interactions at the DNA damage sites through the forkhead-associated (FHA) and BRCA1 carboxyl-terminal (BRCT) domains (31). Nbs1 is phosphorylated by kinase ataxia-telangiectasia mutated (ATM), and the MRN complex is required for full activation of ATM- and ATM-Rad3-related (ATR) signaling in response to DNA damage (31). The ends of the adenovirus (Ad) linear double-stranded DNA (dsDNA) genome are recognized by cellular sensors as DNA damage, initiating a DDR (51). If unabated, the DDR will result in ligation of Ad genomes in an end-to-end manner and the formation of concatemers (51). The DDR severely inhibits viral DNA replication. Ad has evolved two mechanisms to inhibit this process. The Ad type 5 (Ad5) E1B-55K and E4-open reading frame 6 (ORF6) proteins form an E3 ubiquitin (Ub) ligase complex with cellular proteins cullin 5 (CUL5), Rbx1, and elongins B and C (24, 42) and inactivate the MRN complex by directing Ub-mediated, proteasome-dependent degradation (47). The Ad5 E4-ORF3 protein sequesters MRN in nuclear track structures within infected-cell nuclei to inhibit MRN activity (18, 47). E4-ORF3 recruits numerous nuclear proteins into these structures, including promyelocytic leukemia (PML) and other PML-nuclear body (PML-NB) associated proteins, such as Sp100 and Daxx, to inactivate cellular antiviral defense mechanisms induced by interferon and a DDR (51).
Ubiquitination and sumoylation have emerged as important posttranslational modifications that regulate DDRs and DNA repair (reviewed in references 5 and 15). Proliferating cell nuclear antigen (PCNA) is a well-known example and is modified by either Ub or SUMO at the same Lys residue (K164) (20). Monoubiquitination of PCNA promotes DNA repair by recruitment of translesion synthesis DNA polymerases to sites of DNA damage. PCNA residue K164 may be polyubiquitinated, which promotes DNA damage repair by a template-switching mechanism. PCNA is sumoylated at residue K164 during S phase, which recruits the DNA helicase Srs2 via a SUMO interaction motif (SIM) to restrict DNA recombination. The importance of the role of protein sumoylation in the regulation of a DDR is becoming increasingly apparent (reviewed in references 5 and 15). The SUMOs (SUMO-1, SUMO-2, and SUMO-3), as well as components of the SUMO machinery, accumulate at sites of DNA damage to direct the sumoylation of proteins involved in DNA repair, such as BRCA1 (21, 35). Sumoylation increases BRCA1 Ub ligase activity. The E3 SUMO ligases, protein inhibitor of activated STAT-1 (PIAS1) and PIAS4, localize at sites of DNA damage and are required to recruit other effectors involved in a DDR and for efficient DNA repair to occur (21, 35). The exact role(s) that SUMOs play during a DDR remains to be elucidated.
In mammals, at least four SUMO isoforms have been identified (reviewed in reference 22). SUMO-2 and SUMO-3 share 95% amino acid homology in precursor forms and 97% homology in mature forms; thus, they are often termed SUMO-2/3. SUMO-1 and SUMO-2/3 have only 50% homology and modify different substrates. It is thought that SUMO-2/3 modification is regulated more dynamically in response to various stimuli, such as heat shock, oxidative stress, and pathogens, since the unconjugated, free SUMO-2/3 population is larger than SUMO-1 in mammalian cells (22). It remains mostly unclear how the sumoylation system confers substrate specificity and SUMO paralog specificity on a wide range of substrates using the only E2 SUMO enzyme, Ub-like conjugase 9 (Ubc9), and a limited number of E3 SUMO ligases (22).
Viruses activate, exploit, or inhibit the host sumoylation system (reviewed in references 52 and 53). Several studies have shown that sumoylation of cellular proteins is enhanced by viral proteins. For example, Daxx sumoylation is stimulated by human cytomegalovirus (HCMV) protein pp71 (26), and PML sumoylation is stimulated by Kaposi's sarcoma-associated herpesvirus (KSHV) K-bZIP and latency-associated nuclear antigen 2 (LANA2) proteins (12, 34). KSHV K-bZIP is a SUMO-2/3-specific SUMO ligase that stimulates sumoylation of cellular binding partners, such as p53 and the retinoblastoma tumor suppressor (pRb) (12). The Ad E1B-55K protein has intrinsic E3 SUMO-1 ligase activity and directs the sumoylation of p53 (37, 39). Sumoylation may be exploited by viral infection. With herpes simplex virus type 1 (HSV-1), the viral protein ICP0 induces the global proteasomal degradation of sumoylated host cell proteins, including PML (9). With Epstein-Barr virus (EBV), the viral protein LMP1 targets Ubc9 and causes increased global protein sumoylation (6). Finally, viral infection may interfere with sumoylation pathways. With human Ad, the E1A proteins function as negative regulators of sumoylation. E1A interaction with pRb represses SUMO conjugation to pRb (32), and interaction with the E2 SUMO conjugase Ubc9 interferes with the polysumoylation process (56). The avian Ad CELO expresses a protein, GAM1, that promotes degradation of the SUMO E1 and E2 enzymes and inhibits global protein sumoylation (8). The human papillomavirus type 16 (HPV16) E6 protein binds PIAS4 and inhibits sumoylation of PIAS4 substrates (7). There also are numerous examples of viral proteins that are sumoylated. With Ad, the human Ad5 E1B-55K protein is sumoylated at Lys104, and this modification is required for transformation of primary baby rat kidney cells (17). Sumoylation of E1B-55K blocks its own nuclear export (30) but promotes p53 export to the cytoplasmic aggresome (39). E1B-55K sumoylation also regulates its binding to cellular PML isoforms, where E1B-55K binds to PML isoform IV (PML-IV) in a sumoylation-dependent manner but binds to PML-V in a sumoylation-independent manner (54).
We found that Ad5 infection relocalizes SUMO proteins into nuclear tracks containing the E4-ORF3 protein. Since Ad infection has been shown to induce Ub-mediated degradation of the MRN complex, we examined whether Ad infection modulates sumoylation of this complex. Here, we show that the MRN complex components, Mre11 and Nbs1, are sumoylated following Ad5 infection and that E4-ORF3-mediated relocalization of the MRN complex into nuclear tracks is critical for this process. Upon wild-type Ad5 infection, Mre11 and Nbs1 sumoylation reaches a peak at early times after infection and declines thereafter. SUMO-1 deconjugation from Nbs1 depends on degradation of Mre11 by the viral E3 ligase complex containing E1B-55K and E4-ORF6, whereas SUMO-2 deconjugation is independent of viral early-gene products. Ad-induced sumoylation of Nbs1 and Mre11 is serotype specific, since only subgroup C Ad5 E4-ORF3 directs this process, whereas E4-ORF3 proteins from other Ad subgroups do not. Our findings suggest a molecular mechanism of Ad-mediated SUMO conjugation and deconjugation of components of the MRN complex and a clue to study consequences of MRN protein sumoylation during a DDR.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Three HeLa cell lines stably expressing hexahistidine (6His)-tagged SUMOs (49), generously provided by R. T. Hay (University of Dundee), were maintained in the same medium containing 1 μg/ml of puromycin. The following viruses were used in these studies: dl309 (phenotypically wild-type Ad5) (27); inORF3 (ΔE4-ORF3) (25); dl355 (ΔE4-ORF6) (25); dl355/inORF3 (ΔE4-ORF3/ΔE4-ORF6) (25); dl355/pmD105A/L106A (18), dl355/pmL103A (19), and dl355/pmN82A (19) (ΔE4-ORF6/point mutations in E4-ORF3); dl338 (ΔE1B-55K) (41); dl1520 (ΔE1B-55K) (4); dl367 (ΔE1B-55K/ΔE4-ORF6) (14); and dl366 (ΔE4) (25). E1 replacement viruses that express hemagglutinin (HA)-tagged E4-ORF3 of Ad3, -4, -5, -9, and -12 and mutant Ad5 E4-ORF3 proteins under the control of the CMV promoter were described previously (18, 19). E1B-55K mutant H354 (55) was a gift from A. J. Berk (University of California—Los Angeles). Viral preparation and infection were performed as described previously (18).
Plasmids.
The E4-ORF3 expression plasmid pcDNA3-E4-ORF3-WT was generated by insertion of the PCR fragment corresponding to the Ad5 E4-ORF3 coding region into the pcDNA3 vector. The SENP-1 coding region was amplified by PCR and inserted into the pEGFP-C1 plasmid (Clontech) at the EcoRI site.
Antibodies.
The anti-E4-ORF3 rat monoclonal antibody (6A-11) was provided by T. Dobner (Heinrich-Pette Institute), the anti-DBP (B6-8) and anti-E1B-55K (2A6) mouse monoclonal antibodies were from A. J. Levine (Princeton University), and the rabbit polyclonal anti-DBP antibody was from P. van der Vliet (University of Utrecht). The anti-E1A rabbit polyclonal antibody and anti-RanGAP1 and anti-p53 mouse monoclonal antibodies were purchased from Santa Cruz Biotechnology, the anti-Mrell and anti-Nbs1 mouse monoclonal antibodies were from Genetex, the anti-Mre11 and anti-Nbs1 rabbit polyclonal antibodies were from Novus, the anti-SUMO-1 and anti-SUMO-2/3 rabbit polyclonal antibodies were from Zymed, and the anti-HA rabbit polyclonal antibody was from Rockland.
In vivo sumoylation assay.
HeLa cells (1 × 107) expressing 6His-tagged SUMO-1 or SUMO-2 were infected with the viruses indicated in the text, and SUMO conjugates were prepared as described previously (49). Briefly, cell pellets were resuspended in lysis buffer (6 M guanidinium-HCl, 10 mM Tris, 100 mM sodium phosphate, pH 8.0, 5 mM imidazole, and 5 mM β-mercaptoethanol) and sonicated (10 strokes at 40% duty cycle). The lysates were incubated with Ni2+-nitrilotriacetic acid (NTA) beads (Qiagen) at room temperature for 3 h. The beads were washed once with lysis buffer, once with pH 8.0 wash buffer (8 M urea, 10 mM Tris, 100 mM sodium phosphate buffer, pH 8.0, 0.1% Triton X-100, 5 mM imidazole, and 5 mM β-mercaptoethanol), and three times with pH 6.3 wash buffer (same as pH 8.0 wash buffer except for sodium phosphate buffer, pH 6.3). 6His-SUMOs were eluted from Ni2+-NTA beads in buffer containing 200 mM imidazole, and the eluted proteins were analyzed by Western blotting as described previously (45).
Immunofluorescence.
HeLa cells grown on glass coverslips in a 24-well plate were transfected with 0.2 μg pEGFP or pEGFP-SENP1 per well using polyethylenimine (PEI) (Polysciences). Twenty-four hours later, the cells were infected with 200 particles/cell of dl309. At 8 h postinfection (p.i.), the cells were fixed with 4% formaldehyde and permeabilized with phosphate-buffered saline (PBS) buffer containing 0.5% Triton X-100. After blocking in 10% goat serum, Ad DNA-binding protein (DBP) and Mre11 and Nbs1 were detected with rabbit anti-DBP and mouse anti-Mre11 or anti-Nbs1 antibodies. Ad DBP and SUMO-2/3 were detected using mouse anti-DBP and rabbit anti-SUMO-2/3 antibodies. Alexa 350-conjugated (Molecular Probes) and tetramethyl rhodamine isocyanate (TRITC)-conjugated (Zymed) antibodies were used as secondary antibodies. Cell images were acquired on an Axiovert 200 M digital deconvolution microscope (Zeiss) and analyzed using Axiovision software. For the analysis of E4-ORF3 proteins of different Ad serotypes, HeLa cells were infected with 500 particles/cell of E1 replacement viruses expressing HA-tagged E4-ORF3 proteins of Ad serotypes 3, 4, 5, 9, and 12 for 24 h and immunostained using mouse anti-Nbs1 and rabbit anti-HA antibodies. Immunofluorescence assays of SUMO-1 and -2/3 were performed using HeLa cells infected with dl309 or inORF3 for 8 h, and virus-infected cells were monitored using DBP- and E4-ORF3-specific antibodies, respectively.
Immunoprecipitation.
HeLa cells were infected with 200 particles/cell of dl309 and incubated for 8 h. The cell monolayers were lysed in 0.5 ml SDS lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM N-ethylmaleimide, and 0.2 mM iodoacetamide) per 107 cells, incubated on ice for 5 min, and boiled for 20 min. After centrifugation at 12,000 × g for 10 min, 2.5 ml immunoprecipitation (IP) dilution buffer (16.7 mM Tris, pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100, and 1 mM PMSF) was added to the supernatant. The lysates were precleared with protein A-agarose beads (Roche) for 1 h and incubated with anti-SUMO-2/3 antibody overnight, followed by addition of protein A-agarose for 3 h. The beads were washed five times with IP dilution buffer and analyzed by SDS PAGE and Western blotting.
RESULTS
Ad infection induces SUMO-1 and SUMO-2 conjugation of Nbs1 and SUMO-2 conjugation of Mre11.
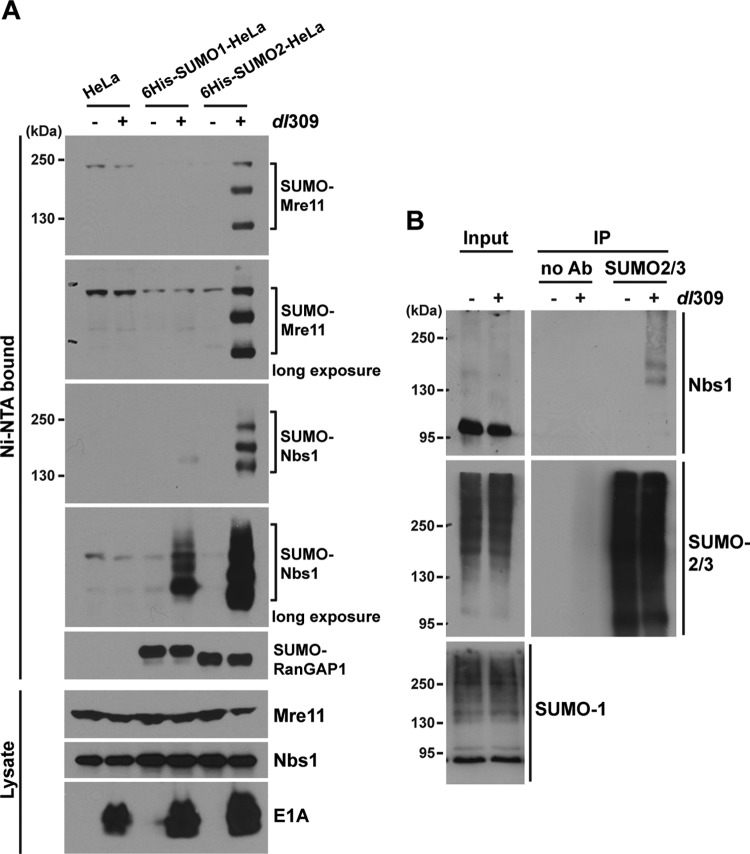
Previous studies showed that the MRN complex is degraded through a Ub-proteasome pathway during Ad infection (47). To investigate whether the MRN complex is also a substrate of sumoylation, we examined sumoylation using HeLa cells stably expressing 6His-tagged SUMO-1, SUMO-2, or SUMO-3 (49). These cell lines express the mature forms of the SUMO proteins. The SUMO-2 and SUMO-3 proteins share 97% amino acid sequence homology and are often termed SUMO-2/3. The parental HeLa and 6His–SUMO-1- and 6His–SUMO-2-expressing HeLa cells were mock infected or infected with wild-type Ad5 dl309 for 8 h. SUMO conjugates were isolated with Ni2+-NTA beads under denaturing conditions, and Mre11 and Nbs1 were analyzed by Western blotting. We detected at least three high-molecular-weight bands corresponding to sumoylated Mre11 and Nbs1 from lysates of Ad5-infected SUMO-2-expressing HeLa cells (Fig. 1A). We also observed SUMO-1 conjugation to Nbs1, but it was weaker than that of SUMO-2; SUMO-1 conjugation to Mre11 was undetectable in these assays. This is the first observation of SUMO modification of Mre11 and Nbs1 in mammalian cells. Sumoylation of Rad50, another component of the MRN complex, was not detected using this assay system. When we examined SUMO-2 and SUMO-3 separately, identical sumoylation patterns were observed (data not shown). No change in the sumoylation pattern of RanGAP1 was observed during Ad infection (Fig. 1A). Unmodified forms of Nbs1 and Mre11 migrate at ∼95 kDa and ∼80 kDa in SDS-PAGE, respectively. Since each SUMO conjugate reduces the mobility of a modified protein by ∼20 kDa, the reductions in Nbs1 and Mre11 mobilities due to Ad-induced sumoylation indicate that two SUMO conjugations occurred with the fastest-migrating SUMO-modified forms and that slower-migrating proteins reflect additional SUMO conjugates on these proteins. Only a small percentage of the total levels of Nbs1 and Mre11 were sumoylated during Ad infection. This is a common observation with sumoylated proteins (49). Sumoylation of Nbs1 was confirmed by immunoprecipitating endogenous SUMO-2/3 from Ad-infected HeLa cell extracts, followed by Western blot analysis (Fig. 1B, IP). We note that the total levels of SUMO-1 and SUMO-2/3 conjugates were not changed in Ad-infected cells (Fig. 1B, input). These results demonstrate that components of the MRN complex, Mre11 and Nbs1, are sumoylated during Ad5 infection.
Fig 1.
Ad infection induces sumoylation of Mre11 and Nbs1. (A) HeLa and 6His-tagged-SUMO-1- and SUMO-2-expressing HeLa cells were uninfected or infected with 200 particles/cell using wild-type Ad5 dl309 for 8 h. SUMO conjugates were prepared using Ni2+-NTA beads under denaturing condition and analyzed by Western blotting using anti-Mre11 and anti-Nbs1 antibodies (Ni-NTA bound). RanGAP1 was analyzed as a sumoylation control. Total cell lysates (Lysate) were analyzed by Western blotting using anti-Mre11 and anti-Nbs1 antibodies; E1A served as a marker of Ad infection. (B) Total cell lysates from dl309-infected HeLa cells were prepared (Input), and anti-SUMO-2/3 antibody (Ab) was used for immunoprecipitation, followed by Western blot analysis using anti-Nbs1 antibody (IP).
Ad5 E4-ORF3 is necessary and sufficient to induce sumoylation of Mre11 and Nbs1.
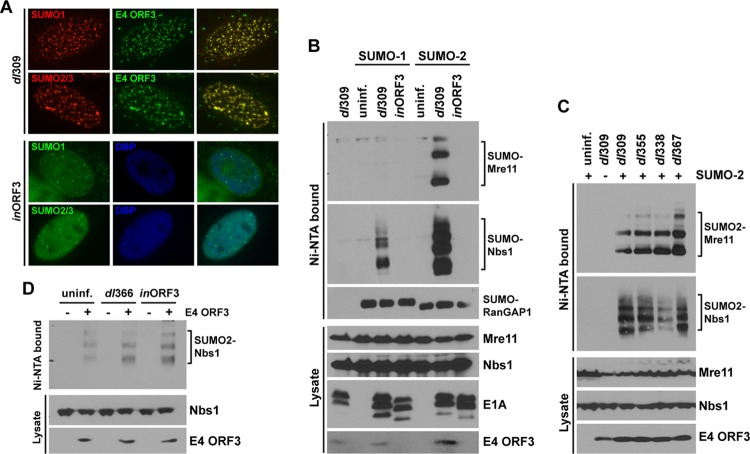
During the early stage of Ad infection, the E4-ORF3 protein reorganizes the MRN complex into filamentous nuclear structures referred to as tracks (11, 16). A previous report showed that ectopically expressed SUMO-1, but not endogenous SUMO-1, is relocalized into nuclear dot- or track-like structures in rat kidney cells stably expressing Ad5 E1B-55K (17). Since we observed Ad-mediated sumoylation of the MRN complex at 8 h p.i. (Fig. 1), we decided to examine the subcellular localization of endogenous SUMO proteins in Ad-infected cells before E1B-55K expression is detectable. The SUMO-1 and SUMO-2/3 proteins colocalized with E4-ORF3 in nuclear tracks in Ad5-infected HeLa cells at 8 h p.i., while infection with an E4-ORF3-deficient mutant virus, inORF3, did not change SUMO localization (Fig. 2A) and displayed a punctate pattern observed in uninfected cells (data not shown).
Fig 2.
E4-ORF3 is necessary and sufficient to induce sumoylation of Mre11 and Nbs1. (A) Subcellular localization of endogenous SUMO-1 and -2/3 was determined in dl309- or inORF3-infected HeLa cells by immunofluorescence (IF) at 8 h p.i. Rabbit anti-SUMO-1 and anti-SUMO-2/3, rat anti-E4-ORF3, and mouse anti-DBP antibodies were used for immunostaining. TRITC-conjugated anti-rabbit secondary antibodies were used with dl309-infected cells to detect SUMO proteins, and fluorescein isothiocyanate (FITC)-conjugated anti-rat secondary antibody was used to detect E4-ORF3. FITC-conjugated anti-rabbit secondary antibodies were used with inORF3-infected cells to detect SUMO proteins, and Alexa 350-conjugated anti-mouse secondary antibody was used to detect Ad DBP as a marker of infected cells. (Right) Merged images. (B) 6His-SUMO-expressing HeLa cells were left uninfected (uninf.) or infected with dl309 or inORF3 (ΔE4-ORF3) for 8 h. SUMO conjugates were analyzed as described for Fig. 1. (C) 6His–SUMO-2-expressing HeLa cells (+SUMO-2) were left uninfected or infected with dl309, dl355 (ΔE4-ORF6), dl338 (ΔE1B-55K), or dl367 (ΔE1B-55K/ΔE4-ORF6) for 8 h. SUMO conjugates were analyzed as described for Fig. 1. Parent HeLa cells (−SUMO-2) infected with dl309 were used as a negative control. E4-ORF3 expression levels were analyzed using whole-cell lysates. (D) SUMO-2-expressing HeLa cells were transfected with an empty vector (−E4-ORF3) or an Ad5 E4-ORF3 expression vector (+E4-ORF3). Twenty-four hours later, the cells were left uninfected or infected with dl366 (ΔE4) or inORF3 (ΔE4-ORF3) for 8 h, followed by analysis of SUMO conjugates as described for Fig. 1. E4-ORF3 expression levels were analyzed using whole-cell lysates.
We examined the effect of the E4-ORF3 protein on Ad-induced sumoylation of the MRN complex. HeLa cells expressing 6His-SUMO proteins were infected with wild-type Ad5 (dl309) or the E4 ORF3-deficient mutant virus (inORF3) for 8 h, and Mre11/Nbs1 sumoylation was analyzed as described above. Interestingly, SUMO-1 and SUMO-2 conjugation of Mre11 and Nbs1 was completely ablated in inORF3-infected cells (Fig. 2B), indicating that the E4-ORF3 protein is essential to induce sumoylation of Mre11 and Nbs1. Since the Ad E1B-55K and E4-ORF6 proteins function to recruit a Ub ligase complex to the MRN complex (24, 42) and E1B-55K itself is a known substrate (17) and ligase (37, 39) of sumoylation, we tested whether E1B-55K and E4-ORF6 are required for E4-ORF3-induced sumoylation of Mre11 and Nbs1. 6His–SUMO-2–HeLa cells infected with mutant viruses dl338 (ΔE1B-55K), dl355 (ΔE4-ORF6), and dl367 (ΔE1B-55K/ΔE4-ORF6) showed levels of Mre11 and Nbs1 sumoylation similar to those in wild-type Ad5 infection (Fig. 2C), demonstrating that E1B-55K and E4-ORF6 do not affect SUMO-2 conjugation of Mre11 and Nbs1.
To further investigate whether E4-ORF3 is sufficient to induce Mre11 and Nbs1 sumoylation, 6His–SUMO-2–HeLa cells were transfected with an empty or an E4-ORF3-containing expression vector, followed by mock infection or infection with dl366 (ΔE4) or inORF3 (ΔE4-ORF3) virus. SUMO-conjugated proteins were analyzed as described above. Interestingly, E4-ORF3 expression alone was sufficient to induce Nbs1 sumoylation; there were no significant differences between uninfected and ΔE4- or ΔE4-ORF3-infected cells (Fig. 2D). These data demonstrate that E4-ORF3 expression induces sumoylation of the Nbs1 and Mre11 proteins and that this process does not require any other Ad gene products.
E4-ORF3-mediated sumoylation of Mre11 and Nbs1 requires relocalization into nuclear track structures.
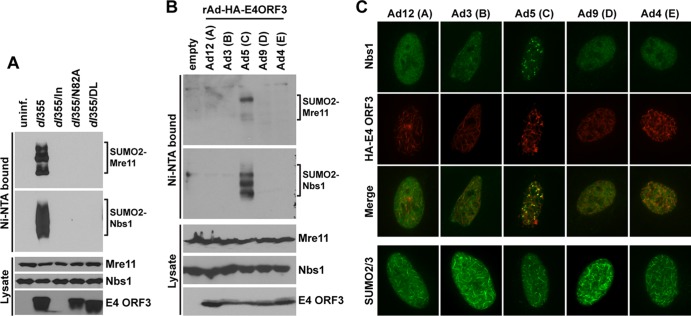
As the major strategy employed by E4-ORF3 to inactivate components of the cellular DNA damage response is reorganization into nuclear tracks (19, 47), we examined whether relocalization of the MRN complex is critical for E4-ORF3-induced sumoylation of Mre11 and Nbs1. We used several well-characterized E4-ORF3 point mutants, including N82A, which is completely nonfunctional (19, 47), and D105A/L106A, which forms nuclear track structures and relocalizes PML, but not the MRN complex (19). The D105A/L106A mutant is able to relocalize SUMO proteins into nuclear track structures (data not shown). Mre11 and Nbs1 sumoylation was not observed in cells infected with viruses harboring either of these E4-ORF3 point mutations (Fig. 3A), indicating that relocalization of the MRN complex by E4-ORF3 may be required for Mre11 and Nbs1 sumoylation.
Fig 3.
Relocalization of Mre11 and Nbs1 into nuclear tracks is required for E4-ORF3-induced sumoylation. (A) 6His–SUMO-2-expressing HeLa cells were left uninfected or infected with dl355 (ΔE4-ORF6), dl355/in (ΔE4-ORF6/ΔE4-ORF3), dl355/N82A (ΔE4-ORF6/E4-ORF3-N82A point mutation), or dl355/DL (ΔE4-ORF6/E4ORF3-D105A/L106A point mutations) for 8 h. SUMO conjugates were analyzed as described for Fig. 1. E4-ORF3 expression levels were analyzed using whole-cell lysates. (B) E1 replacement Ad vectors expressing HA-tagged E4-ORF3 proteins of Ad12, -3, -5, -9, and -4, representing subgroups A, B, C, D, and E, respectively, were used to infect 6His–SUMO-2–HeLa cells. After 24 h, SUMO conjugates were analyzed as described for Fig. 1. E4-ORF3 levels were determined in whole-cell lysates using anti-HA antibody. rAd, recombinant Ad. (C) HeLa cells were infected with E1 replacement Ad vectors as described for panel B. Nbs1 localization (FITC-conjugated secondary antibody) and E4-ORF3 localization (TRITC-conjugated secondary antibody) were analyzed by IF using anti-Nbs1 and anti-HA antibodies, respectively. Merged images are shown in the third row from the top. SUMO-2/3 localization in cells infected with these viruses is shown in the bottom row.
It has been shown that only Ad5 E4-ORF3, but not E4-ORF3 from Ad4 and Ad12, relocalizes the MRN complex into nuclear tracks, although the E4-ORF3 proteins of all three Ad serotypes disrupt PML-NB (48). To test the E4-ORF3 serotype specificity for sumoylation of Mre11 and Nbs1 and to examine the correlation between relocalization and sumoylation, 6His–SUMO-2–HeLa cells were infected with E1-deleted recombinant Ad vectors expressing HA-tagged E4-ORF3 proteins of Ad3, -4, -5, -9, and -12 (Ad subgroups B, E, C, D, and A, respectively. Consistent with the results using the E4-ORF3 point mutant D105A/L106A (Fig. 3A), only Ad5 E4-ORF3, but not the E4-ORF3 proteins from any other Ad subgroups, induced sumoylation of Mre11 and Nbs1 (Fig. 3B). As previously reported for Ad4, -5, and -12 (48), only subgroup C Ad5 relocalized Nbs1 into E4-ORF3-containing tracks in infected HeLa cells, while Ad3, -4, -9, and -12 did not (Fig. 3C, top three rows). In contrast, the E4-ORF3 proteins of all of these serotypes were able to redirect SUMO-2/3 from punctate structures observed in uninfected and inORF3-infected cells (Fig. 2A) into nuclear tracks (Fig. 3C, bottom row). Taken together, our findings indicate that relocalization of MRN into nuclear tracks is required for E4-ORF3-induced sumoylation of Mre11 and Nbs1.
These results do not reveal if Mre11 and Nbs1 sumoylation is the result or the cause of MRN relocalization into E4-ORF3-containing nuclear tracks. If inhibition of sumoylation results in a defect in MRN complex relocalization, it would indicate that sumoylation is required for E4-ORF3-mediated MRN relocalization. To test this, the cellular sumoylation system was suppressed by ectopically expressing SUMO-specific protease 1 (SENP1) to direct SUMO deconjugation (2) (Fig. 4). In uninfected cells, SUMO-2/3 expression levels and localization did not change upon transfection of a control enhanced green fluorescent protein (EGFP) expression vector in comparison to nontransfected cells (Fig. 4, uninf. SUMO-2/3, EGFP). In contrast, a significant reduction in nuclear SUMO-2/3 staining was observed in cells transfected with an EGFP-SENP1 expression vector (Fig. 4, uninf. SUMO-2/3, EGFP-SENP1); such a decrease was previously reported with SUMO-1 and ectopic expression of SENP1 (2). In dl309-infected cells, SUMO-2/3 relocalization into nuclear tracks was not altered by EGFP expression (Fig. 4, dl309 SUMO-2/3, EGFP), but SUMO-2/3 expression was significantly ablated by EGFP-SENP1 expression (Fig. 4, dl309 SUMO-2/3, EGFP-SENP1). The same results were obtained with SUMO-1 and EGFP-SENP1 expression (data not shown). Mre11 and Nbs1 localization was examined at 8 h postinfection, as described above. Both Mre11 and Nbs1 were effectively relocalized by Ad5 infection in EGFP-SENP1-expressing cells (Fig. 4, dl309 Mre11/Nbs1, EGFP-SENP1). Since SUMO levels were effectively suppressed by EGFP-SENP1 expression but Mre11 and Nbs1 were still relocalized by E4-ORF3, these results suggest that Mre11 and Nbs1 sumoylation occurs after relocalization into nuclear tracks rather than as a prerequisite for MRN relocalization.
Fig 4.
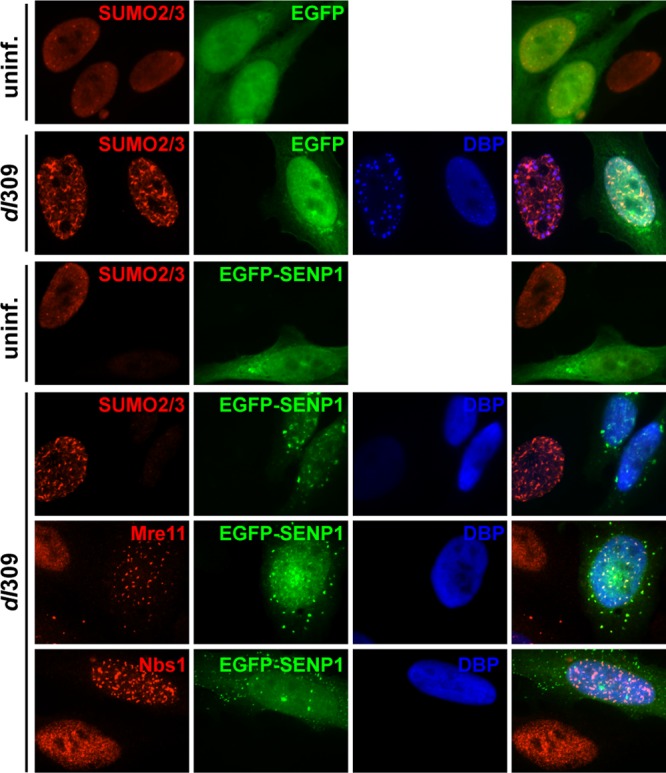
Mre11 and Nbs1 are relocalized into nuclear tracks by Ad infection in SENP1-expressing cells. EGFP or EGFP-SENP1 expression plasmids were transfected into HeLa cells. After 24 h, the cells were left uninfected (uninf.) or infected with dl309 (dl309) for 8 h and analyzed by IF using rabbit anti-SUMO-2/3 antibody or mouse anti-Mre11 and anti-Nbs1 antibodies and TRITC-conjugated secondary antibodies. EGFP and EGFP-SENP1 were visualized using EGFP tags. Ad DBP was used to identify Ad-infected cells using a mouse antibody for SUMO-2/3 IF and a rabbit antibody for Mre11/Nbs1 IF and Alexa 350-conjugated secondary antibodies. Merged images are shown on the right.
Mre11- and Nbs1-SUMO conjugates are transient during wild-type Ad5 infection.
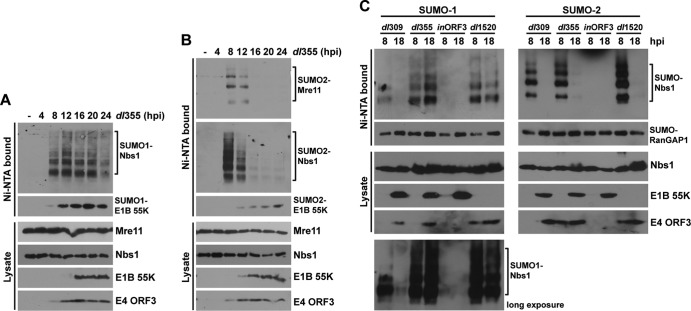
During the late phase of Ad infection, E4-ORF3 nuclear tracks are maintained (18), but MRN components are translocated into viral replication centers (19) and/or cytoplasmic aggresomes (1, 33). This led us to question if Mre11- and Nbs1-SUMO conjugates were stable into the late phase of Ad infection. 6His–SUMO-1- and -2-expressing HeLa cells were infected with wild-type Ad5 for 4, 8, 12, 16, and 24 h, and Mre11 and Nbs1 SUMO conjugation levels were determined as described above. Mre11 and Nbs1 sumoylation reached a peak at 8 h p.i., when E4-ORF3 expression was evident, and then all Mre11- and Nbs1-SUMO conjugates significantly decreased by 12 h p.i. and became undetectable later than 16 h p.i. (Fig. 5). This was not surprising, since the E1B-55K/E4-ORF6 Ub-ligase complex mediates proteasomal degradation of the MRN complex at late times after infection. Consistent with the previous reports that Mre11 is degraded faster than Nbs1 and Rad50 during Ad infection (29, 47), Mre11 protein levels were greatly diminished by 12 to 16 h p.i., whereas Nbs1 protein levels were constant until 16 h p.i. (Fig. 5). These results are consistent with the observation that E4-ORF3 cannot relocalize Nbs1 in the absence of Mre11 (1) and suggest that sustained Nbs1 sumoylation requires continued colocalization with E4-ORF3 in nuclear tracks. These results also demonstrate that the conjugation of SUMO-1 and SUMO-2 to Nbs1 and SUMO-2 to Mre11 is transient during wild-type Ad5 infection. No changes in the pattern of RanGAP1 sumoylation by SUMO-1 or SUMO-2 were evident throughout the time course of Ad infection (Fig. 5).
Fig 5.
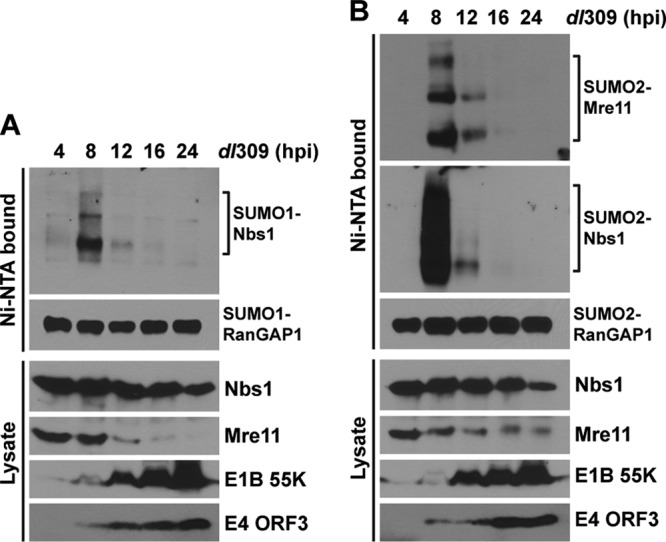
Sumoylation of Mre11 and Nbs1 during a time course of wild-type Ad5 infection. 6His–SUMO-1-expressing (A) and SUMO-2-expressing (B) HeLa cells were infected with dl309 for different periods (4, 8, 12, 16, and 24 h). SUMO conjugates were analyzed as described for Fig. 1. Total cell lysates were examined by Western blotting using the indicated antibodies.
Deconjugation of SUMO-1 and SUMO-2 from Nbs1 displays different kinetics during infection with E1B-55K and E4-ORF6 mutant viruses.
To further study desumoylation mechanisms at late times after Ad infection, E1B-55K/E4-ORF6-mediated MRN degradation was prevented by infecting cells with the E4-ORF6 mutant virus dl355. Mre11 and Nbs1 sumoylation levels were determined at 4, 8, 12, 16, 20, and 24 h postinfection. A more complex pattern of Nbs1 SUMO-1 and SUMO-2 conjugation was observed in dl355-infected cells (Fig. 6A and B); we do not know the basis of this effect. Deconjugation of SUMO-2 from Mre11 and Nbs1 showed the same pattern observed during wild-type Ad5 infection (Fig. 6B). In contrast to results observed with wild-type Ad5, E4-ORF3-induced SUMO-1 conjugation to Nbs1 was constant even at late times after infection with the E4-ORF6 mutant virus (Fig. 6A), implying that conjugation/deconjugation of SUMO-1 and SUMO-2 to Nbs1 is regulated by distinct mechanisms during the late phase of Ad infection. We next examined SUMO-1 and SUMO-2 conjugation to Nbs1 at 8 and 18 h p.i. with wild-type Ad5 or E1B-55K and E4-ORF6 mutant viruses. Conjugation of both SUMO-1 and SUMO-2 to Nbs1 was significantly decreased during the late phase of wild-type Ad5 (dl309) infection, whereas only SUMO-2 conjugation was reduced during infection with E4-ORF6 (dl355) or E1B-55K (dl1520) mutant virus infection (Fig. 6C). SUMO-1 conjugation to Nbs1 was not reduced at late times after infection with either mutant virus (Fig. 6C), suggesting that E4-ORF6 and E1B-55K play an important role in deconjugation of SUMO-1 from Nbs1.
Fig 6.
SUMO-1 and SUMO-2 deconjugation from Nbs1 shows different kinetics during infection with E1B-55K- or E4-ORF6-deficient viruses than with wild-type Ad5. (A) 6His–SUMO-1 conjugation to Nbs1 during a time course of infection with dl355 (ΔE4-ORF6) infection, analyzed as described for Fig. 1. (B) 6His–SUMO-2 conjugation to Mre11 and Nbs1 during a time course of dl355 (ΔE4-ORF6) infection, analyzed as described for Fig. 1. (C) 6His–SUMO-1- and -2-expressing HeLa cells were infected with dl309, dl355 (ΔE4-ORF6), inORF3 (ΔE4-ORF3), or dl1520 (ΔE1B-55K) for 8 or 18 h. 6His–SUMO conjugation to Nbs1 was analyzed as described for Fig. 1.
To ascertain if E1B-55K/E4-ORF6-mediated proteasomal degradation of Mre11 results in deconjugation of SUMO-1 from Nbs1 during Ad infection, we analyzed Nbs1 sumoylation in cells infected with the E1B-55K insertion mutant H354, that interferes with MRN degradation but still allows the degradation of p53. At 20 h p.i., SUMO-1 conjugation to Nbs1 was retained in cells infected with dl1520 (E1B-55K-null) and H354, while wild-type Ad5 infection abrogated Nbs1 sumoylation (Fig. 7A). Taken together, these results show that E4-ORF3-induced SUMO-1 conjugation to Nbs1 is stable at late times of Ad infection, when the E1B-55K/E4-ORF6 E3 Ub-ligase complex is inactive for Mre11 degradation.
Fig 7.
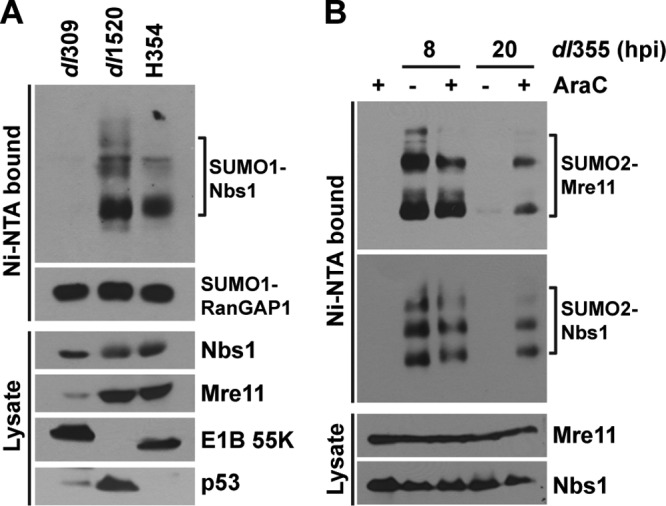
Distinct mechanisms of SUMO-1 and -2 deconjugation from Mre11 and Nbs1 are employed during Ad infection. (A) 6His–SUMO-1-expressing HeLa cells were infected with dl309, dl1520 (ΔE1B-55K-null mutant), and the E1B-55K point mutant H354 for 20 h, and Nbs1 sumoylation was analyzed as described for Fig. 1. p53 was used to monitor the substrate-specific degradation defect of the E1B-55K H354 mutant virus. (B) 6His–SUMO-2-expressing HeLa cells were left uninfected or infected with dl355 (ΔE4-ORF6) for 8 or 20 h in the absence (−) or presence (+) of 20 μg/ml AraC. 6His–SUMO-2 conjugation was analyzed as described for Fig. 1.
Inhibition of viral DNA replication and late-gene expression restores SUMO-2 conjugation of Nbs1.
To determine whether any early Ad gene products other than E1B-55K and E4-ORF6 play a role in deconjugation of SUMO-2 from Nbs1, we analyzed Nbs1 sumoylation in the presence of AraC, a nucleoside analog that inhibits DNA synthesis and consequently late gene expression. 6His–SUMO-2 HeLa cells were infected with the E4-ORF6 mutant virus dl355, and culture medium containing AraC was added for 8 and 20 h p.i. AraC treatment did not affect Nbs1 sumoylation in uninfected cells (Fig. 7B), although the drug is known to induce stalled replication forks and a DNA damage response. AraC-treated and untreated dl355-infected cells showed similar levels of Nbs1 sumoylation at 8 h p.i., but SUMO-2 conjugation to Nbs1 was retained until 20 h p.i. only in AraC-treated cells (Fig. 7B). This result suggests that viral DNA replication, an Ad late-gene product(s), or a process induced during the late phase of infection is required for deconjugation of SUMO-2 from Nbs1.
DISCUSSION
Here, we provide the first evidence that Mre11 and Nbs1 are SUMO substrates in mammalian cells and that the Ad5 E4-ORF3 protein modulates the cellular sumoylation system. We observed at least three SUMO conjugates of Nbs1 and Mre11 (Fig. 1A); the mobilities of the fastest-migrating sumoylated Nbs1 and Mre11 species are consistent with the conjugation of two SUMO moieties. We expect that these species represent independent sumoylation sites rather than polysumoylation at a single lysine residue, since the numbers of bands are the same with SUMO-1 and SUMO-2 and SUMO-1 is not reported to form multichain linkages (22). We do not know if the E4-ORF3 protein induces SUMO-1 and SUMO-2 conjugation at the same or different lysine residues in Nbs1. Experiments are in progress to identify the sites of Mre11 and Nbs1 sumoylation induced by E4-ORF3 protein expression. Mapping the SUMO conjugation site(s) induced by E4-ORF3 in Mre11 and Nbs1 will be important to analyze the functional consequences of the MRN complex sumoylation in the DDR. Sumoylation of the MRX components Mre11, Rad50, and Xrs2 (Nbs1) was observed in Saccharomyces cerevisiae following the induction of a DDR by replicative stress, and protein sumoylation induced by a DDR is widespread in the organism (13). The MRX complex serves as a positive regulator of sumoylation in the organism, and sumoylation significantly promotes the DDR in this system (13). It seems counterintuitive that E4-ORF3-induced sumoylation of Mre11 and Nbs1 relates to an analogous process in mammalian cells, since the function of the Ad5 E4-ORF3 protein is to inhibit the MRN complex and a DDR in Ad-infected cells (51). We did not observe the induction of MRN sumoylation following the induction of replicative stress or gamma irradiation in HeLa cells (data not shown). E4-ORF3 inhibits ATR, but not ATM, signaling through relocalization of the MRN complex (10), and it will be interesting to explore whether sumoylation of the MRN complex affects ATR signaling.
While we do not yet know the mechanism(s) by which the E4-ORF3 protein regulates Mre11 and Nbs1 sumoylation, our results indicate that E4-ORF3 may affect different aspects of the SUMO conjugation and deconjugation system. This supposition is based on the observation that E4-ORF3 induces SUMO-1 and SUMO-2 conjugation to Nbs1 but only SUMO-2 conjugation to Mre11 (Fig. 1A), suggesting that different SUMO ligases may be involved in these SUMO-1 and SUMO-2 conjugation processes. This idea is consistent with the observation that Ad5 infection did not change total SUMO-1 or SUMO-2/3 conjugate levels (Fig. 1B), suggesting that specific components of the SUMO conjugation/deconjugation system are impacted by the E4-ORF3 protein. In addition, the patterns of Nbs1–SUMO-1 and -SUMO-2 desumoylation differed over a time course in cells infected with the E4-ORF6 mutant virus (Fig. 6A and B), suggesting that different SUMO ligases or deconjugation enzymes may be involved in this process. The fact that deconjugation of SUMO-1 from Nbs1 differed in cells infected with wild-type Ad5 (Fig. 5) and cells infected with the E1B-55K and E4-ORF6 mutant viruses (Fig. 6C) demonstrates that the E1B-55K and E4-ORF6 proteins regulate E4-ORF3-induced Nbs1 desumoylation. The E1B-55K protein is itself a SUMO ligase, and it induces sumoylation of p53 (37, 39). This activity, however, does not contribute to the induction of Nbs1 sumoylation by E4-ORF3, since the E1B-55K mutant virus induced Nbs1 sumoylation normally (Fig. 6C). It is well established that the E1B-55K/E4-ORF6 complex directs ubiquitin-dependent proteasomal degradation of cellular substrates, such as p53 and MRN, and sustained levels of MRN with the E4-ORF6 mutant virus (Fig. 6A) may allow sustained SUMO-1 conjugation. The degradation of Mre11 in wild-type Ad5-infected cells correlates precisely with the loss of Nbs1–SUMO-1 conjugates (Fig. 5), and we speculate that the integrity of the MRN complex is required for SUMO-1 conjugation of its components. In contrast, this is not the case with deconjugation of SUMO-2 from either Mre11 or Nbs1, where the E1B-55K/E4-ORF6 complex did not influence this process (Fig. 6C).
The functional consequence(s) of the sumoylation of Mre11 and Nbs1 induced by E4-ORF3 is not known. With both cellular proteins, recruitment into E4-ORF3-containing nuclear tracks is required for sumoylation. Ad5 E4-ORF3 mutant proteins that are defective in this process (N82A, L103A, and D015A/L106A), as well as E4-ORF3 proteins from Ad subgroups other than subgroup C (Ad3, -4, -9, and -12) that also are defective in this process, were unable to induce sumoylation of Mre11 or Nbs1 (Fig. 3). This could reflect the need for sumoylation of Mre11 and Nbs1 to enter E4-ORF3-containing tracks or that these cellular proteins are sumoylated once they enter these nuclear structures. The fact that ectopic expression of the SUMO protease SENP1 did not interfere with recruitment of Mre11 and Nbs1 into E4-ORF3 nuclear tracks but significantly ablated SUMO expression (Fig. 4 and data not shown) favors the latter possibility. During Ad5 infection, MRN components are transported into cytoplasmic aggressomes, which accelerates their degradation by the E1B-55K and E4-ORF6 proteins (1, 33). These aggresomes also contain the E4-ORF3 protein, and the E4-ORF3 protein increases the formation of E1B-55K-containing aggresomes and Mre11 recruitment in the absence of the E4-ORF6 protein (33). We speculate that sumoylation of Mre11 and Nbs1 induced by E4-ORF3 may facilitate this process to promote MRN degradation. It has been postulated that sumoylation and transport through the nuclear pore may be coupled (40), and there are a number of examples where sumoylation regulates this process to promote either nuclear or cytoplasmic accumulation of a gene product (50). For example, p53 sumoylation is implicated in its nuclear export (50). Further, the import and export of the Ad E1B-55K protein is regulated by sumoylation, and E1B-55K regulates p53 sumoylation and promotes the nuclear export of p53 (30, 39).
Identifying the mechanism of SUMO-2 deconjugation from Nbs1 is also of interest. It mostly remains an enigma how SUMO substrate specificity and paralog selectivity are conferred by a limited number of E2 and E3 enzymes. SIM-mediated interactions may answer part of this question. The SUMO proteases, SENPs, also provide a clue to this puzzle. In humans, six SUMO proteases (SENP-1, -2, -3, -5, -6, and -7) have been identified, and they display different subnuclear localizations, enzyme activities, and SUMO paralog preferences (reviewed in references 34 and 36). Since SUMO-2 conjugation to RanGAP1 was sustained into the late phase of infection with wild-type Ad5 and the E4-ORF6 mutant virus (Fig. 5 and 6C), we assume that SUMO-2 deconjugation is regulated by an Ad-specific and substrate-specific mechanism during the late phase of viral infection. Interestingly, a previous study showed that the Ad protease (AVP) deconjugates tetra-Ub and cleaves pro-ISG15 in the presence of pVIc activating peptide using an in vitro system (3). Future studies are needed to determine if AVP regulates Nbs1 desumoylation.
Ad has evolved a number of mechanisms to inhibit the induction of a DDR from the earliest stages to the late phase of infection. The basic Ad core protein, protein VII, appears to protect the incoming viral genome from recognition by the DDR until the early-gene products E1B-55K, E4-ORF6, and E4-ORF3 are expressed (28). These early Ad gene products inhibit the induction of a DDR as core protein VII is displaced from the viral genome and into the late phase of infection (51). The results of the current study add an additional level of complexity to this picture with the induction of Mre11 and Nbs1 sumoylation by the E4-ORF3 protein. This multifunctional viral protein targets numerous cellular effectors into nuclear tracks to sequester their activities (51), it inhibits p53-induced gene expression by establishing heterochromatin in p53-responsive cellular promoter regions (46), and it plays additional roles in the viral life cycle, including the promotion of cell cycle-independent viral replication (23, 44), the regulation of viral late mRNA splicing and cytoplasmic mRNA accumulation (38), and the regulation of late protein translation (43). Many of these E4-ORF3 functions have been investigated using only subgroup C Ad. An exception is E4-ORF3 relocalization of MRN into nuclear tracks, where Ad5, but not E4-ORF3 proteins of other Ad subgroups, directs this process (48) (Fig. 3C). With the regulation of MRN during Ad infection, subgroup C Ads have evolved redundant mechanisms to inhibit MRN utilizing E1B-55K/E4-ORF6-mediated MRN degradation and E4-ORF3-mediated MRN sequestration. Perhaps the same principle holds true for the process regulated by Ad5 E4-ORF3-induced Mre11 and Nbs1 sumoylation, where another viral product plays a redundant role with non-subgroup C Ads.
ACKNOWLEDGMENTS
We are very grateful to Ronald Hay (University of Dundee) for providing 6His-SUMO cell lines and other reagents. We thank Thomas Dobner (Heinrich Pette Institute) for the anti-E4-ORF3 monoclonal antibody (6A-11), Arnold Levine (Princeton University) for the anti-DBP (B6-8) and anti-E1B-55K (2A6) monoclonal antibodies, Peter van der Vliet (University of Utrecht) for the rabbit polyclonal anti-DBP antibody, and Arnold Berk (UCLA) for the E1B-55K mutant virus H354. We acknowledge the excellent technical assistance of Ilana Shoshani and thank members of our laboratory for informed discussions.
This work was supported by NIH grant CA122677.
Footnotes
Published ahead of print 27 June 2012
REFERENCES
- 1. Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bailey D, O'Hare P. 2004. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 279:692–703 [DOI] [PubMed] [Google Scholar]
- 3. Balakirev MY, Jaquinod M, Haas AL, Chroboczek J. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323–6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barker DD, Berk AJ. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107–121 [DOI] [PubMed] [Google Scholar]
- 5. Bekker-Jensen S, Mailand N. 2011. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 585:2914–2919 [DOI] [PubMed] [Google Scholar]
- 6. Bentz GL, Whitehurst CB, Pagano JS. 2011. Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminal-activating region 3 contributes to LMP1-mediated cellular migration via its interaction with Ubc9. J. Virol. 85:10144–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bischof O, et al. 2006. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol. Cell 22:783–794 [DOI] [PubMed] [Google Scholar]
- 8. Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. 2004. A mechanism for inhibiting the SUMO pathway. Mol. Cell 16:549–561 [DOI] [PubMed] [Google Scholar]
- 9. Boutell C, et al. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 7:e1002245 doi:10.1371/journal.ppat.1002245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carson CT, et al. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 28:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carvalho T, et al. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang PC, et al. 2010. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 285:5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cremona CA, et al. 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45:422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cutt J, Shenk T, Hearing P. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dou H, Huang C, Van Nguyen T, Lu LS, Yeh ET. 2011. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 585:2891–2896 [DOI] [PubMed] [Google Scholar]
- 16. Doucas V, et al. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196–207 [DOI] [PubMed] [Google Scholar]
- 17. Endter C, Kzhyshkowska J, Stauber R, Dobner T. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. U. S. A. 98:11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans JD, Hearing P. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans JD, Hearing P. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fox JT, Lee KY, Myung K. 2011. Dynamic regulation of PCNA ubiquitylation/deubiquitylation. FEBS Lett. 585:2780–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galanty Y, et al. 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462:935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodrum FD, Ornelles DA. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 73:7474–7488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harada JN, Shevchenko A, Pallas DC, Berk AJ. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang M, Hearing P. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hwang J, Kalejta RF. 2009. Human cytomegalovirus protein pp71 induces Daxx SUMOylation. J. Virol. 83:6591–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones N, Shenk T. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683–689 [DOI] [PubMed] [Google Scholar]
- 28. Karen KA, Hearing P. 2011. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 85:4135–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karen KA, Hoey PJ, Young C, Hearing P. 2009. Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection. J. Virol. 83:4565–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kindsmüller K, et al. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U. S. A. 104:6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamarche BJ, Orazio NI, Weitzman MD. 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584:3682–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ledl A, Schmidt D, Müller S. 2005. Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24:3810–3818 [DOI] [PubMed] [Google Scholar]
- 33. Liu Y, Shevchenko A, Berk AJ. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79:14004–14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marcos-Villar L, et al. 2011. Covalent modification by SUMO is required for efficient disruption of PML oncogenic domains by Kaposi's sarcoma-associated herpesvirus latent protein LANA2. J. Gen. Virol. 92:188–194 [DOI] [PubMed] [Google Scholar]
- 35. Morris JR, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462:886–890 [DOI] [PubMed] [Google Scholar]
- 36. Mukhopadhyay D, Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286–295 [DOI] [PubMed] [Google Scholar]
- 37. Muller S, Dobner T. 2008. The adenovirus E1B-55K oncoprotein induces SUMO modification of p53. Cell Cycle 7:754–758 [DOI] [PubMed] [Google Scholar]
- 38. Nordqvist K, Ohman K, Akusjarvi G. 1994. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pennella MA, Liu Y, Woo JL, Kim CA, Berk AJ. 2010. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J. Virol. 84:12210–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pichler A, Melchior F. 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381–387 [DOI] [PubMed] [Google Scholar]
- 41. Pilder S, Moore M, Logan J, Shenk T. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Querido E, et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shepard RN, Ornelles DA. 2004. Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background. J. Virol. 78:9924–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shepard RN, Ornelles DA. 2003. E4orf3 is necessary for enhanced S-phase replication of cell cycle-restricted subgroup C adenoviruses. J. Virol. 77:8593–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sohn SY, Hearing P. 2011. Adenovirus sequesters phosphorylated STAT1 at viral replication centers and inhibits STAT dephosphorylation. J. Virol. 85:7555–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soria C, Estermann FE, Espantman KC, O'Shea CC. 2010. Heterochromatin silencing of p53 target genes by a small viral protein. Nature 466:1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352 [DOI] [PubMed] [Google Scholar]
- 48. Stracker TH, et al. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. 2009. Detection of protein SUMOylation in vivo. Nat. Protoc. 4:1363–1371 [DOI] [PubMed] [Google Scholar]
- 50. Wang YE, Pernet O, Lee B. 2012. Regulation of the nucleocytoplasmic trafficking of viral and cellular proteins by ubiquitin and small ubiquitin-related modifiers. Biol. Cell 104:121–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weitzman MD, Ornelles DA. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686–7696 [DOI] [PubMed] [Google Scholar]
- 52. Wilson VG. 2012. Sumoylation at the host-pathogen interface. Biomolecules 2:203–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wimmer P, Schreiner S, Dobner T. 2012. Human pathogens and the host cell SUMOylation system. J. Virol. 86:642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wimmer P, et al. 2010. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29:5511–5522 [DOI] [PubMed] [Google Scholar]
- 55. Yew PR, Cheng Kao C, Berk AJ. 1990. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology 179:795–805 [DOI] [PubMed] [Google Scholar]
- 56. Yousef A, et al. 2010. Identification of a molecular recognition feature in the E1A oncoprotein that binds the SUMO conjugase UBC9 and likely interferes with polySUMOylation. Oncogene 29:4693–4704 [DOI] [PubMed] [Google Scholar]