Abstract
A new method for measuring the hydrogen exchange of macromolecules in solution is described. The method uses tritium to trace the movement of hydrogen, and utilizes Sephadex columns to effect, in about 2 minutes, a separation between tritiated macromolecule and tritiated solvent great enough to allow the measurement of bound tritium. High sensitivity and freedom from artifact is demonstrated and the possible value of the technique for investigation of other kinds of colloid-small molecule interaction is indicated. Competition experiments involving tritium, hydrogen, and deuterium indicate the absence of any equilibrium isotope effect in the ribonuclease-hydrogen isotope system, though a secondary kinetic isotope effect is apparent when ribonuclease is largely deuterated. Ribonuclease shows four clearly distinguishable kinetic classes of exchangeable hydrogens. Evidence is marshaled to suggest the independently measurable classes II, III, and IV (in order of decreasing rate of exchange) to represent “random-chain” peptides, peptides involved in α-helix, and otherwise shielded side-chain and peptide hydrogens, respectively.
Protons attached to nitrogen, oxygen, and sulfur of molecules dispersed in liquid solution are, in general, able to exchange with mobile protons in the solvent. The rate of exchange of protons occupying specific molecular sites may be modified, or even largely controlled, by the details of molecular architecture in their immediate vicinity. Thus measurement of the kinetics of the hydrogen exchange of macromolecules can provide a fine probe for identifying and quantitating details of conformation and of changes in conformation of macromolecules.
A practicable method for the measurement of hydrogen-exchange kinetics was first developed and used in the study of structure in proteins and polypeptides by Linderstrøm-Lang and his co-workers (Linderstrøm-Lang, 1955). Typically, in this method, deuterated protein is separated from deuterated solvent by freeze-drying, the protein is redissolved in water, and the kinetics of the loss of deuterium by the protein is followed by analyzing samples of solvent, after various times of protein incubation, for deuterium content. Deuterium content is determined by density measurement of tiny samples of solvent isolated from the protein by a second freeze-drying step. The method, though ingenious in conception, seems unusually demanding in execution and makes use of procedures which might be considered questionable. While the method is capable of considerable internal precision, a number of the operations involved, freezing, drying, heating (60°), redissolution, and massive replacement of exchangeable hydrogen by deuterium, might cause changes in the very structures one is attempting to measure.
A number of other methods have been used. For example, Leach and Springell (1962) have explored the use of tritium together with modifications of the drying procedures, in the classical freeze-drying methods. Investigations of hydrogen-deuterium exchange using the techniques of nuclear magnetic resonance (Wishnia and Saunders, 1962) and of infrared spectroscopy both on protein films (Haggis, 1957) and solutions (Nielsen, et al., 1960; Blout, et al., 1961) have been reported. Though they suffer from some of the same drawbacks indicated previously and have not yet reached the degree of precision attained by the Linderstrøm-Lang method, the inherent capability of the infrared and nuclear magnetic resonance methods of distinguishing various chemical groups within the exchanging molecule and of performing continuous measurements without problems of separation may make them very useful in studies of this kind.
It is the purpose of this paper to describe and illustrate the use of a new method which was designed to measure the hydrogen exchange of macromolecules. The method uses tritium as a tracer to follow the movement of hydrogen between macromolecule and solvent, and utilizes columns of Sephadex gel to effect rapid separation between tritiated macromolecule and tritiated solvent SCI that tritium carried by the macromolecule can be measured.
The technique of gel filtration in Sephadex columns has been used by a number of workers to obtain separations between molecular species on the basis of size.1 In this method, one takes advantage of the differential permeability of the porous Sephadex grains to molecules of disparate size. Molecules small enough to penetrate into the grains wash through the column more slowly than larger, more excluded ones. The speed of the separation is limited by the rate of diffusion of the smaller molecule into the grains. It is intended, in this paper, to show that in favorable cases, as in the separation of macromolecules from labeled water, extensive separations can be attained in minutes, perhaps even in seconds, and to demonstrate the use of this technique for investigations of macromolecular structure by the measurement of hydrogen exchange between macromolecules and solvent. The method shows high precision and an apparent freedom from artifacts introduced by handling procedures. It gives promise of being able to identify and quantitate various classes of structure within macromolecules through their distinctive kinetic hydrogen-exchange character.
Discussion of the method will be directed to the study of the hydrogen exchange of proteins. It is clear, however, that the same techniques could prove useful for a more general range of investigation concerned with interactions between small molecules and colloidal particles. One thinks immediately of enzymes and small substrates, haptenes and their specific antibodies, and macromolecules and salts. Another area of potentially fruitful utilization is suggested by the observation that colloids as large as human erythrocytes (7 mμ) pass unattenuated through the columns (Englander, unpublished observations). Thus one might hope to study transport through membranes in suspensions of cells and cell organelles when transport times are of the order of 1 minute or more.
Materials and Measurements
Coarse grade G-25 Sephadex was supplied by A. B. Pharmacia, Uppsala. Tritiated water was obtained from New England Nuclear Corporation at a concentration of 1c/g and used at dilutions of approximately 1000-fold. D2O was used as supplied (Bio-Rad, 99.8%). C14-labeled polyethyleneglycol was prepared from a sample of 20 × 103 mw (Dow Chemical Corp.) polymer kindly supplied by W. Stockmayer. One-ml aqueous radioisotope samples were counted in a dioxane based scintillating fluid described by Bray (1960). A Nuclear-Chicago Model 703 automatic liquid scintillation counter was used. Standard dilutions of initial sample were always counted along with the samples of column effluent, though reproducibility from day to day was outstanding. Preparations of bovine pancreatic ribonuclease used were 5 × recrystallized Worthington lot number 593 and “chromatographically homogeneous” Gallard-Schlesinger, lot A1633. The sample of oxidized ribonuclease was obtained through the courtesy of C. B. Anfinsen and J. Cooke. Ribonuclease concentration was measured in a Zeisa PMQ II spectrophotometer using the molar extinctions of 10.6 × 106/cm at 278 mμ for native and 8.40 × 106/cm at 275 mμ for oxidized (Harrington and Schellman, 1956).2 Relative polyproline (Yeda Research and Development-Schwan Bioresearch) concentrations were estimated by optical rotation in a Rudolph photoelectric spectropolarimeter.
Experimental
Column Preparation
Though other materials have also been used as molecular sieves, Sephadex, because of its ready availability and generally highly satisfactory characteristics, was used in this work.3 Sephadex is a cross-linked dextran preparation in granular form. The pore size of the graim varies inversely with the degree of cross linking. Since it was desirable to have small protein molecules completely excluded from the grains, only the smallest pore size of Sephadex, grade G-25, which completely excludes dextran fractions of 4500 mw, has been examined. To give the columns the possibility of very rapid flow rates, only the coarse grain size was used, though at flow rates so large that the diffusion rate of the small molecule becomes limiting, the separation may benefit by the presence of finer grains.
The term “separation” in this paper refers to the degree of contamination of either of the eluted species by the other. Since in this sense separations of the order of 10−6 or more were desirable in these experiments, the effort was made to combat possible sources of non-ideality of flow in the columns. The glass tubing of the columns was siliconed (Siliclad, Clay-Adams, N. Y.) to minimize flow down the walls. To prevent the formation of continuous channels during packing, columns were packed in ½-cm to 1-cm layers. Each layer was consolidated by a pressured flow of water through it before the next layer was added. A disk of sharkskin filter paper, cut to fit the column closely, covered the top surface of the gel. The ability of the surface to resist being disrupted upon addition of samples and solvent was improved by allowing solvent to drain below the surface from time to time. To prevent unnecessary smearing of the elution patterns, the gel rested on a freely draining sintered glass disk, and effluent was not allowed to pool below the disk before flowing into the collection tubes. Sephadex, suspended in water, was prepared for packing by repeated decanting to remove “fines” and by stirring under vacuum to remove adsorbed air. The lifetime of a good column is limited only by a gradual accumulation of dust which tends eventually to reduce flow rates.
The Separation
In a study of the kinetics of the hydrogen exchange of a protein, most general interest attaches to the region of the kinetic curve in which the protein retains between 1 and 0.1 of an original exchangeable hydrogen atom per amino acid residue. In this region, protein in aqueous solution at a concentration of 1%binds between −3 and 10−4 of the mobile hydrogen in the solution and the same fraction of isotope used to tag hydrogen. (Questions of isotope effect are discussed below.) Therefore if, as in this investigation, one attempts to measure protein-bound hydrogen by separating tagged protein from unbound hydrogen isotope, a relative separation of 10−6 or more is desirable. Further, since the bound hydrogens exchange with time constants of the order of an hour, it is desirable to effect the separation in minutes.
Clearly for the study of systems in which the colloid may carry a larger fraction of the small molecular species present, either because of greater specificity of interaction as with specific antibody and haptene, or because of greater colloid concentration as in study of interactions between cells in suspension and small molecule metabolites, a less extensive separation is acceptable.
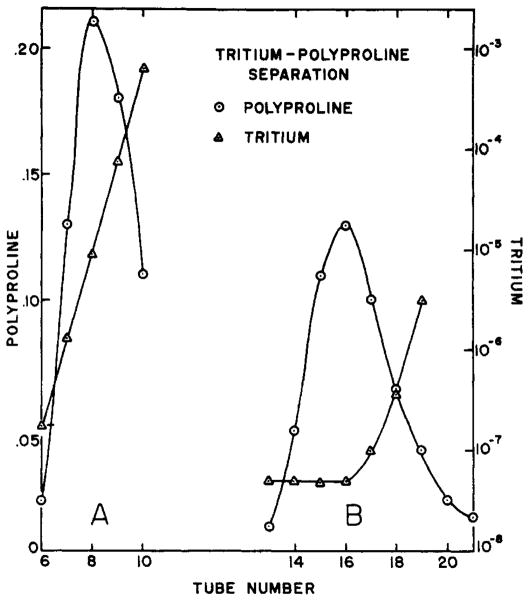
Figure 1B depicts a separation effected in about 2 minutes between polyproline, which has no exchangeable hydrogens, and tritiated water. At zero time a 1-ml sample of polyproline in tritiated water was applied to the top surface of a 6-cm high by 3-cm in diameter column of G-25 Sephadex. As the sample drained through the covering filter paper, a column of solvent was layered above the gel and successive 1.2-ml samples of effluent were collected. The concentration of polymer and tritium in each sample is plotted as the fraction of that in the original. Most of the polymer has been freed of tritium by a relative factor of more than 10−6.
Fig. 1.
Separation of polyproline from THO in Sephadex: columns. (A) 3-cm X 3-cm column. (B) 3-cm x 6-cm high column. Concentration is in terms of fraction of that; in the original sample.
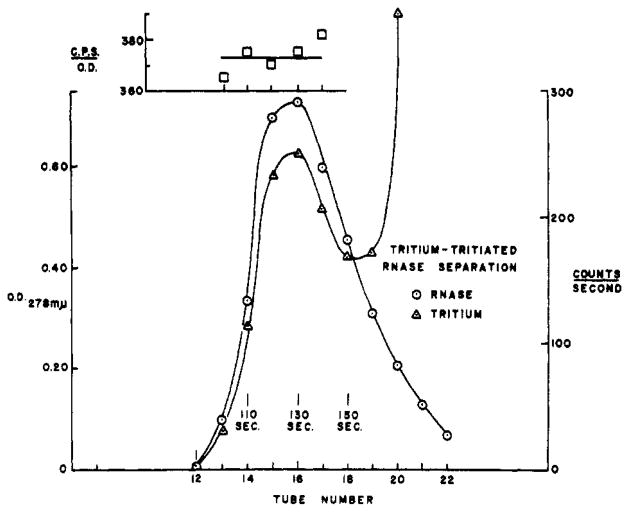
Figure 2 shows an equivalent run performed with native ribonuclease. Ribonuclease at a concentration of 7 mg/ml was initially warmed in tritiated water at 70°, above the temperature of a reversible configurational transition (Hermans and Scheraga, 1959), where all its exchangeable hydrogen is able to come to equilibrium with the tritium in the solvent. After cooling, 1ml was passed through the column. It is evident that a quantity of tritium was carried through the column by the protein. The number of original exchangeable hydrogens per molecule borne by the effluent ribonuclease is directly proportional to the ratio of tritium-to-protein concentration in the effluent samples. This parameter, calculated for each of the samples as tritium activity divided by optical density at 278 mμ, is shown in the inset to Figure 2. Each of the 5 samples yields an independent estimate of this parameter and it is evident that the precision is good.
Fig. 2.
Separation of tritiated ribonuclease from THO in 3-cm × 6-cm Sephadex column. The inset shows, for samples through the peak region, tritium activity divided by optical density.
The kinetics of the loss of tritium by the protein can be gauged by a modification of the above sequence. In a procedure which will be referred to as a one column separation the sample is allowed to drain about halfway through the column and then the flow is stopped. The sample incubates in solution within the column with the free, solvent tritium reduced to less than 1% of its concentration in the original mixture, as is indicated by the elution pattern of a 3-cm by 3-cm column shown in Figure 1A. Bound tritium moving out of the protein at a rate characteristic of the ambient temperature and solvent conditions is replaced by hydrogen. At any given time, flow through the column is resumed. The protein, in traversing the lower half of the column, leaves behind the tritium it has lost during its in-column incubation and emerges bearing a residuum of tritium inversely related to the time it has spent in the column. A series of such points in time outline the exchange-out (back-exchange) kinetics of the protein. The duration of a one-column exchange-out experiment is limited by the loss of separation caused by the diffusion of trailing tritium into the protein region.
The kinetics of exchange-in (forward exchange) may be similarly measured. At zero time tritium is added to a protein solution. At various times thereafter samples are passed through a Sephadex column. The protein emerges bearing the quantity of tritium that has exchanged-in during the period of incubation minus the quantity lost during the time of passage through the column.
A two-column separation provides a rather more versatile technique for study of the kinetics of exchange reactions. The tritiated sample is passed through a fist column to reduce the free tritium by a factor of 100 or more. Effluent protein is pooled and incubated under any given conditions for any given time. Aliquotas can then be passed through a second column to remove unbound tritium. This procedure affords the experimenter a number of advantages not provided by the simpler one-column separation. The in-test-tube incubation makes possible the analysis of exchange kinetics at long times and the easy variation of experimental conditions throughout the incubation period. Further the two-column procedure can produce extreme separations though with some sacrifice of protein recovery.
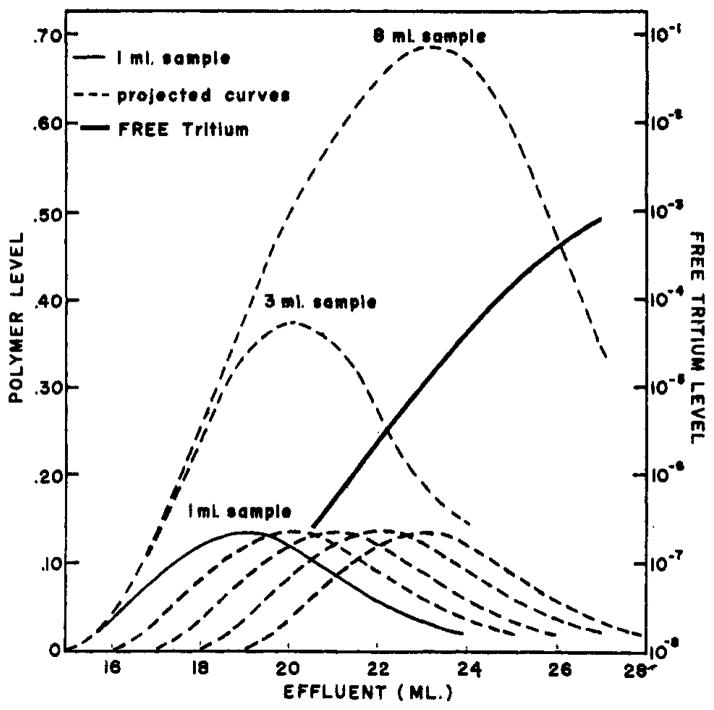
Several practical aspects of manipulation of this procedure may be illustrated by a conceptual experiment. Suppose that more than 1 ml of polymer-tritium sample is applied to the column of Figure 1B. Given the data for a 1-ml sample, the elution pattern of a larger sample can be constructed by recourse to a principle of independence; that is, since there is no question of saturation of the capacity of Sephadex columns so that each molecule of polymer and tritium moves through the column independently of the fate of others, the content of each ml of added sample will also move, spread, and dilute independently of every other ml. Thus the elution pattern arising from the contents of each ml of a larger sample will duplicate that of the ml preceding it, but will be displaced backward by 1ml. The situation is diagramed in Figure 3, The successive distributions of polymer originating in successive ml of added sample are shown individually as replicas of each other. The total elution pattern of polymer resulting from N ml of original sample is just the sum of the first N patterns. The summation is shown for N = 3 ml and 8 ml. The same rule obtains for the tritium pattern. However, because the tritium level increases so sharply just behind the polymer peak, the major fraction of tritium at any point in the polymer region of the effluent comes from the first ml of added sample.
Fig. 3.
Construction of elution patterns that would result from a multiple-ml sample by invoking a principle of independence of project from data for a 1-ml sample.
Conclusions of use in designing experiments can be drawn from summation curves of the type shown in Figure 3. For example, an 8-ml sample will yield 10 ml of effluent with macromolecule and tritium concentration reduced by factors of 2 and 104, respectively (ml No. 17 to 26). This gives the experimenter a stock of sample aliquots which may be manipulated at will and then passed through a second column to determine the effect of the manipulation on the release of tritium. As has been shown by Flodin (1960, Fig. 4B), a sufficiently large sample will yield effluent diluted hardly at all but with free tritium reduced by more than 100-fold. A 3-ml sample will yield 3 ml of effluent sample with polymer and tritium diluted by factors of 3 and 107, respectively (ml No. 19 to 21). Upon passage of this volume through a second equivalent column the 3 peak ml will contain 10% of the original polymer now separated from initially free tritium by a factor of over 1012!
Fig. 4.
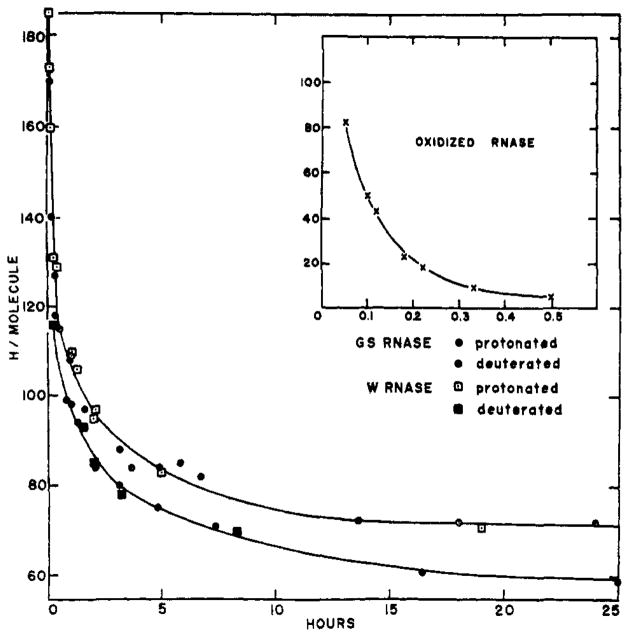
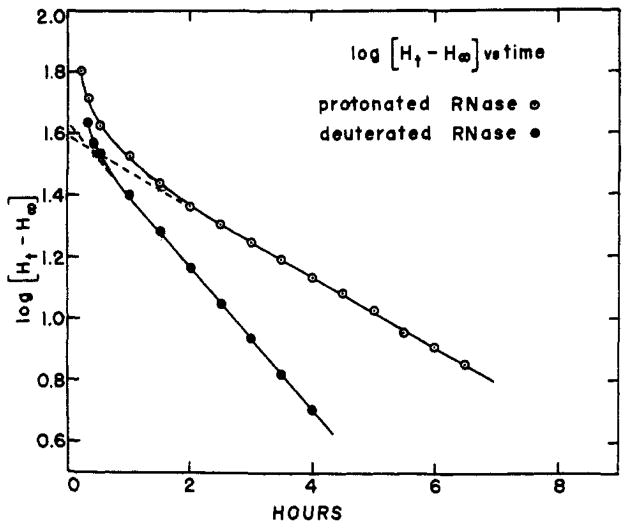
Exchange-out curves (by loss of tritium) of protonated and deuterated ribonuclease and of oxidized ribonuclease against water at 4°, pH 4.7, 0.1 M NaC1. W and GS refer to Worthington and Gallard-Schlesinger preparations, respectively.
Data Analysis
Data from the exchange experiments in following sections were used to calculate the parameter hydrogen atoms/molecule of protein, as follows.
where: Co, is the tritium activity (normally ~2 × 106 cps/ml) in the protein-tritium equilibration mixture; C is the tritium activity carried by protein, as counted in column effluent; D is the optical density of protein in column effluent; εp is the molar extinction coefficient; and 111 is the atom concentration of H in H2O and very nearly of D in D2O. The ratio C / D for a given run is taken as the average of the several estimates of this value from the different samples through the effluent protein peak (inset, Fig. 2). In the ribonuclease experiments, it has proved satisfactory to measure for each run only one background sample and four samples in the peak region. Brief experience with the columns shows where the protein peak is to be expected and which samples it is desirable to measure.
In a current tritium-exchange study of structure and interaction with gelatin and collagen, which show very low absorption in the 280-mμ range, Dr. von Hippel and I have resorted to a different approach to the measurement of protein concentration in Sephadex column effluent. The method is based on the facts that sufficiently large polymers (in the absence of adsorption effects) move through Sephadex columns exactly together and that radioactivity of C14 and of tritium can be independently counted by liquid scintillation techniques when those substances are present in the same simple. We have been able to use C14-labeled polyethyleneglycol, added to the initial sample at known activity, to trace the movement of protein in the columns. Details of this technique will be reported elsewhere.
Studies with Ribonuclease
Application of the methods described to the problems of protein hydrogen exchange have been worked out using ribonuclease as a test protein. The results obtained are presented below. These results not only illustrate the use of the method but also have general implications for the hydrogen-exchange approach to protein structure.
(1) Oxidized ribonuclease
Oxidized ribonuclease, by physical chemical criteria, exists as an un-aggregated, random chain in aqueous solution (Harrington and Schellman, 1956). The exchange-out curve for oxidized ribonuclease at pH 4.7 and 4° in 0.1 M NaCl is shown in Figure 4. The curve falls rapidly to 4 hydrogens/molecule, a residuum which may be due to experimental artifact. The exchanging hydrogens describe a good first-order plot with a half-time (t½) of 4.7 minutes and involving a total of 115 hydrogens.
Of the calculated (from amino acid composition) 245 exchangeable hydrogens of ribonuclease at this pH, intrinsically rapid rates are expected for those bound to oxygen (Weinberg and Zimmerman, 1955) and many of those bound to nitrogen (Nielsen et al., 1960; Bradbury et al., 1962). The 119 peptide protons are intrinsically slower. Infrared studies of Bryan and Nielsen (1960) on the hydrogen-deuterium exchange of poly-D,L-alanine show all the peptide protons to exchange in a single first-order reaction, which, together with other considerations, led these authors to suggest a random-chain conformation for this polypeptide in aqueous solution. (See also Elliott, 1962; but also Rosenheck and Doty, 1961). Their tabulated pH 4.7 rates at 22° and 10° predict at t½; at 4° close to the rate found for the peptide nitrogen protons of oxidized ribonuclease. Thus, by the criterion of hydrogen exchange, both poly-D,L-alanine and oxidized ribonuclease, in solution under these conditions, possess equivalent, uniform, therefore presumably random-chain conformations, and we may expect the 5-minute half-time to be characteristic for peptide protons in random-chain sections of proteins. Further the agreement found between the theoretical number of slow hydrogens in oxidized ribonuclease and the number measured by tritium exchange suggests that sizable equilibrium isotope effects do not occur in these reactions.
(2) Native ribonuclease
Figure 4 shows the exchange-out pattern of native ribonuclease under these same conditions. Worthington (W) and Gallard-Schlesinger (GS) preparations were used. The W points fell 6 H/molecule higher than corresponding GS data. The difference may be due to some contaminant protein in the W preparation or to some aging effect on it, since the GS sample was both the newer and the purer of the two. (Weight average molecular weights, measured by short-column equilibrium sedimentation, were 14,500 for GS [measured by K. Y. Wong] and 15,500 for W. The accepted ribonuclease weight is 13,700.) To bring the data for the two preparations into consonance, 6 H/molecule were subtracted from all W results. Most of the very short-time points plus the W points at 1.25 hours and 2 hours, and the GS points at 1 hour, 1.5 hours, and 3 hours represent one-column runs. The agreement between two-column runs, in which protein was in contact with the column for less than 5 minutes, and one- column runs speaks against the introduction of artifact through contact with the columns.
The exchange-out kinetics of native ribonuclease show a more complex behavior than do the random- chain data. As has long been realized, the structure of the protein modifies the exchange rates of its protons. Visual inspection shows the curve to be composed of at least four kinetically distinguishable classes of exchangeable hydrogen. The different kinetic classes presumably mirror different aspects of structure within the protein. An “instantaneous” class (I) was lost before the earliest measurement was made. A second class (II), exchanging fairly rapidly with a rate similar to that of the oxidized protein, is clearly different from a third group (III)having an apparent half-time of some hours. A fourth group (IV) is lost very slowly if at all.
The accuracy of the curves seems sufficiently great as to invite mathematical analysis. It is of paramount interest to inquire whether the kinetic exchange curve can show us the different kinds of structure present in the protein. If the exchange rate of any proton is determined largely by the immediate structure holding it, we may hope by analysis to identify distinct kinetic classes of protons with exchange rates characteristic of specific classes of structure, e.g., α-helix, random coil. If, on the other hand, the exchange rate of a particular proton is effectively modified by details of protein structure outside its immediate locale, we may expect to find a wide continuum of exchange rates, and this would greatly complicate the effort to recognize specific aspects of structure.
A nonarbitrary way of identifying a sizable class of hydrogens exchanging with first-order kinetics among all the hydrogens generating the curves of Figure 4 is to construct a plot of slope of the curve (dH/dt) vs. the ordinate (Ht). Such a plot, constructed for the protonated ribonuclease curve of Figure 4, is straight over a large fraction of the visually identifiable class III and a short extrapolation to dH/dt = 0 shows this class to end at H∞ = 73. The total size of the class and its rate constant can be found by using the smoothed curve of Figure 4 to construct a plot of log (Ht – 73) vs. time. This curve is shown in Figure 5. Extrapolation to zero time shows the upper limit of this class to be H0 = 113. Thus class III contains 40 hydrogens which exchange with a half-time of 2.6 hours. The same procedure applied to the lower curve of Figure 4 showing the loss of tritium by 99+ % deuterated ribonuclease indicates a presumably equivalent class III between the ordinates 72 H/molecule and 115H/molecule, but with a half-time of exchange of 1.3 hours. These results also set the size of class IV at about 70 H/molecule in both. If it is assumed that class II is a single first-order group its size can be estimated by using the upper limit of class III (H0 = 113) and the early-time protonated ribonuclease data to construct a plot of log (Ht - 113) vs. time. The somewhat scattered points can be fit with a straight-line plot showing a size for this class of 79±10 H/molecule and with a slope indicating a half-time of exchange of 8 ± 1.5 minutes. These results are summarized in Table I.2
Fig. 5.
Semilog plots of Figure 4 curves showing class III loss of tritium by protonated and by deuterated ribonuclease to be first-order reactions, equivalent in size but differing in rate.
Table I.
The Size2 and Time Constant Of the Kinetic Classes Composing the Exchange-out Curves of Figure 4
| Class | Protonated Ribonuclease
|
Deuterated Ribonuclease
|
||
|---|---|---|---|---|
| Size (H/molecule) | t½(min) | Size (H/molecule) | t½ (min) | |
| II | 80 | 8 | ||
| III | 40 | 160 | 43 | 80 |
| IV | 73 | Slow | 72 | Slow |
Kinetic class I is evidently generated by those groups in the protein which are intrinsically fast exchangers not sufficiently inhibited by local structure to render them measurable by this method. It seems reasonably secure, from the results with oxidized ribonuclease, to identify class II with peptide hydrogens in more or less random-chain conformation. Classes III and IV must arise from protons involved in hydrogen bonds and/or sterically protected from easy contact with water. It is tempting to suppose that class III, composed of hydrogens having a uniform exchange rate and therefore, perhaps, arising from one, uniform structural class, represents α-helix. This picture would ascribe to class IV “masked” and/or bonded hydrogens of amino acid side chains and possibly also “masked” α-helical hydrogens. We might then expect class IV, a potpourri of various structural features to be decomposable into measurable subgroups by various stratagems. Some measurements of ribonuclease exchange at higher pH indicate that this is indeed the case (see also Hvidt, 1955).
Before proceeding further with discussion of the measurement of structural classes, it is necessary to consider the possible operation of isotope effects in these measurements. Indeed, a search for isotope effects seems doubly desirable in view of the evident disagreement between results shown in Figure 4 and those previously reported for ribonuclease by the use of the Linderstrøm-Lang deuterium-exchange method.
Isotope Effects
By observing the behavior of tritium we wish to deduce certain facets of the interaction between protein and hydrogen. A differential selection by the protein of one of the isotopes at the expense of the other would yield misleading results. It is therefore necessary to inquire whether tritium provides a proper index for the behavior of hydrogen, that is, whether isotope effects seriously affect these measurements.
In designing their now classical methods for measurement of the hydrogen exchange of proteins, Linderstrøm-Lang and his co-workers (1955) avoided complications due to possible equilibrium isotope effects by resorting to complete substitution of all hydrogen-exchanging sites with deuterium so that no question of selection between the two alternative isotopes could arise. The use of tritium in trace amounts eliminates this safeguard but makes possible an internal assay for possible isotope effects. This is so because the isotope present in bulk quantity can be chosen to be either protium, deuterium, or mixtures of the two, either can be used as the solvent or “in” the protein, and the kinetics of the exchange of tritium between protein and solvent, in either direction, can be studied. The various possibilities allow for a thoroughgoing investigation of a number of different competitive situations. Some experiments of this nature have been carried out with ribonuclease. The results at hand indicate the absence of any equilibrium isotope effect in this system.
We wish to ask whether there exists an unequal competition among the hydrogen isotopes for sites on the protein; that is, whether, at equilibrium, the ratio of T to H within the protein is equal to the T–H ratio in the solvent. Stated formally in terms of the exchange formula
| (1) |
with respect to the ith site on the protein, we are asking if the equilibrium constant for equation (1), KT, H = ki/k−i, is equal to unity.
(1) (T/H)equilibrium vs. (T/D) equilibrium
If there is an unequal competition between T and H, we may expect that an unequal competition will also obtain between T and D and, since the selective factors among the three isotopes will very likely differ, that the two inequalities will be different. That is, if KT,H = 1, then, with respect to formula (2)
| (2) |
we expect KT,D = k′i/k′−i = 1, and that KT,H = KT,D.
The upper curve of Figure 4 shows the exchange-out kinetics of ribonuclease which was initially brought to equilibrium with T in H2O. The lower curve was obtained with ribonuclease initially equilibrated with T in D2O. For both cases data were taken only after separation of tritiated protein from tritiated solvent in H2O-filled columns, so that the exchange measured is that of T within the protein against H in the solvent. Thus differences between the two kinetic curves can arise only in the equilibration procedure.
A perceptible, albeit small, difference is evident; the close agreement between the two exchange-out curves indicates that KT,H and KT,D are not very different. The kinetic analysis summarized in Table I reveals the curves to be essentially identical in terms of the size of the slower kinetic classes, differing only in the exchange rates of the slower hydrogens. Thus, at least with respect to those sites of ribonuclease involved in structure, KT,H = KT,D. It is unlikely that both are equal to any number other than unity.
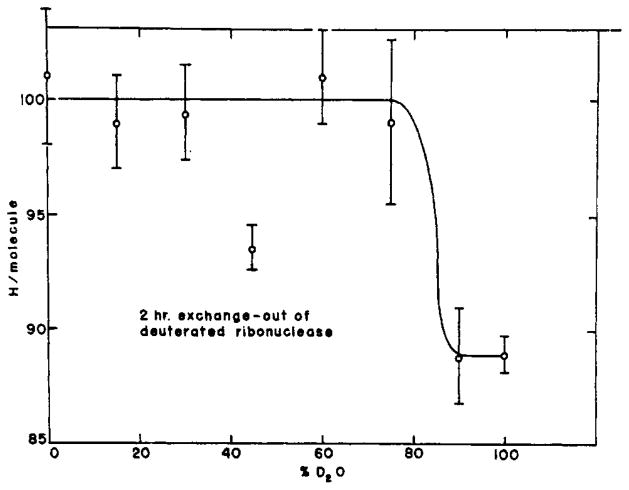
It is interesting to note that, since both curves of Figure 4 measure the exchange of T within the protein against H in the 100% H2O solvent, the effect on the exchange rate of deuterating the protein must be a secondary one operating through some modification of the protein itself. A similar effect has been noted by Benson (1959) for protonated and deuterated myoglobin. Presumably this phenomenon is explicable in terms of some general effect on the protein resulting from the small modification in hydrogen-bonding parameters upon deuteration (e.g., Tomita et al., 1961). The present method may be able to give information pertinent to this effect. We might for instance inquire whether the effect increases continuously as the ratio of D to H in the initial equilibration solvent is increased, or whether it appears suddenly at some sufficiently high concentration of deuterium. Several experiments directed toward this question have yielded data generally resembling those shown in Figure 6. The figure depicts the number of original hydrogens per molecule still remaining after 2 hours of exchanging out vs. the deuterium content of the equilibration solvent. These experiments suggest. that the effect appears cooperatively above some critical level of deuterium, though the error of measurement has been sufficiently great as to preclude firm conclusions.
Fig. 6.
The change-over of ribonuclease from the slower to the more rapidly exchanging form as a function of initial degree of deuteration.
(2) Exchange-in vs. exchange-out
An experiment different in design from those so far discussed involves the measurement of the exchange of solvent tritium into the protein. If ki = k−i (formula 1), the results of exchange-in experiments are predictable from the exchange-out data at hand. Conversely, exchange-in data can match the values predicted from exchange-out results only if no isotope effects are operating.
Some data of this kind for ribonuclease are shown in Table II. In these experiments the addition of a trace of THO to the protein solution marks the zero time. At the times specified an aliquot of the incubation mixture was applied to a Sephadex column. During the separation of protein from tritiated solvent in the column, some of the fast tritium is unavoidably lost. It seems advisable to allow the protein to exchange-out in the column for a time calculated to remove most of the fast T and very little of the slow. The 16-minute exchange-out time chosen for these experiments removes all but 16 of the fast hydrogens and only 3 (calculated) of the slow ones, while bringing the measurement to a part of the exchange curve where only about 2 hydrogens are lost per minute. Thus for the data in Table II, the earliest time points depend largely upon the faster hydrogens while the later times measure largely the slower ones. For an exchange-in time t, the number of hydrogens per molecule predicted on the assumption of no isotope effect is, closely, the difference between the ordinates on the exchange-out curve corresponding to the times 16 minutes and t. A small correction, 3 H/molecule at most, must be added to this value. The fraction of 3 H to be added is the same as the fraction of class III not yet labeled at the end of the exchange-in period.
Table II.
Comparison of Exchange-in Data With Values Predicted From Exchange-out Data And The Assumption Of No Isotope Effects
| Exchange-in Time(hr) | H/molecule Predicted | H/molecule Found |
|---|---|---|
| 0.8 | 22 | 24 |
| 1.0 | 24 | 28 |
| 1.6 | 31 | 32 |
| 2.1 | 35 | 37 |
| 3.8 | 43 | 43 |
| 6.1 | 49 | 52 |
| 21.5 | 58 | 62 |
| 26.0 | 58 | 64 |
The data of Table II further support the previous conclusion as to the absence of an equilibrium isotope effect.
DISCUSSION
The Method
The original development of protein hydrogen-exchange techniques, following closely the recognition of the role of hydrogen-bonded helical structure in proteins, was based on the stated assumption that slowly exchanging hydrogens would identify α-helix. The early demonstrations of “instantaneous” exchange in structureless polypeptides and of classes of slower hydrogens in native proteins (Lenormant and Blout, 1953; Linderstrøm-Lang, 1958) tended to support this idea, and hydrogen-exchange data, even today, are still being interpreted on this basis. It has been variously pointed out that this approach may be oversimplified. Hydrogen in side-chain hydrogen bonds and those simply shielded by neighboring structure might also be expected to show inhibited exchange. On the other side, the rapidity of exchange of the hydrogens buried in DNA structure4 and the failure to observe enough slow hydrogens in myoglobin (Benson, 1959; Beychok et al.,1962) to account for its helical content as determined by X-ray diffraction (Kendrew, et al.,1960) further challenge the basic assumption.
Nevertheless, the general recognition of the potential power of this approach to macromolecular structure has led to a proliferation of methods designed to measure hydrogen exchange. The present method is one of this list. It possesses several desirable features; chief among these are high precision (Figs. 2 and 4) and an evident freedom from artifacts. The use of tritium in trace quantities and of Sephadex columns to separate labeled protein from labeled solvent avoids the freezing, drying, and dissolution steps, and the massive replacement of H by D employed by other methods. The method can work with low protein concentrations. (Dr. von Hippel and I have routinely used gelatin and collagen at 1 mg/ml.) It is rapid, easy to handle, and versatile in application. The use of a trace isotope, it is true, raises questions of equilibrium isotope effect, but the method is internally capable of evaluating these effects.
Ribonuclease Data
Analysis of the ribonuclease-exchange data shows four distinct kinetic classes, the sizes of which, in terms of H/molecule, are given in Table I.
The hydrogen exchange of ribonuclease has previously been extensively investigated by several methods. Published data can be described in terms of three kinetic classes: an “instantaneous” class corresponding to class I plus class II of this study and often plus part of classes III and IV; an intermediate class; an exceedingly slow class. With the classical deuterium exchange techniques under the same conditions as used here, Schildkraut and Scheraga (1960) found 25 H/ molecule in the intermediate class and 45 very slow, while Ottesen and Stracher (1960) found 50 intermediate and 33 very slow H/molecule. Hvidt (1955) in earlier work found about 15 and 45 hydrogens, respectively, in the two classes. The intermediate class exchanged with an apparent half-time of 1 hour to 1.5 hours so that it corresponds to class III (t½; = 1.3 hours for deuterated ribonuclease) of this communication, while the very slow class is the obvious mate of class IV. Leach and Springell (1962) studying the use of tritium and milder drying procedures with the Linderstrøm-Lang method found, with their most cautious procedure, 45 H/molecule in class III and 70 H/molecule in class IV, in good agreement with the present results.
In view of the accuracy and freedom from artifact of the Sephadex method a search of the previous methods for the source of the discrepancies seems justified. The failure of the Linderstrøm-Lang method to detect class II is obvious. The supposition that class II deuterons were lost in the cryosublimation step is supported by a number of observations in the literature. Hydrogen exchange between dry protein and vapor-phase water is well known. In the cryosublimation step of the Linderstrøm-Lang method, largely protonated ice is removed from deuterated protein by sublimation. Protein-vapor (or protein-ice) exchange would spuriously increase the measured loss of D by the protein. In fact, infrared studies of the exchange of poly-D,L-alanine (Bryan and Nielsen, 1960) and some di- and tri-peptides (Nielsen et al., 1960) did show less early- time exchange than was found by Linderstrøm-Lang (1958) and his co-workers, who in using the cryosublimation step lost some of the polyalanine deuterium and all of the small-peptide deuterium. The “instantaneous” exchange reported for oxidized ribonuclease and other random-chain polypeptides, when compared with the present data (t½; = 5) is also revealing. This effect of loss of loosely held deuterium during cryosublimation constitutes an extension of the artifact, discovered by Hvidt and Linderstrøm-Lang (1955), involving exchange between dry protein and a damp P2O5-trap.
In view of the time relationships involved the same artifact could act to decrease the measured size of class III but would not noticeably affect class IV deuterons unless some other effect also entered. That other effects might enter is suggested by consideration of the factors that stabilize protein molecules and the modification of these factors accomplished by removing proteins from water into vacuum. Hydrogen bonds which stabilize helical regions of proteins are more stable in vacuum than in water. The stability of hydrophobic bonds, which depends partly on the lyophobic nature of water, would on the other hand be reduced by drying. Thus drying and heating of proteins could bring about significant conformational changes and render previously protected hydrogens more vulnerable to exchange with vapor-phase hydrogen. The classical techniques, then, are capable of distorting the hydrogen exchange picture of macromolecules, and the differences between the present results and previously reported data very probably stem from artifacts arising in those techniques.
Isotope Effects
Equilibrium isotope effects have been reported for the biphasic exchange systems dry protein—gaseous ammonia (Lobunez and Karush, 1959) and cellulose-THO (Lang and Mason, 1960). Nielsen (1960) noted an effect in the acid-catalysis region of N-methylacetamide. The general literature on isotope effects in proton-transfer reactions is extensive (Bell, 1959).
Evidently the system at hand is free of such effects. Leach and Springell (1962)) using a modified Linderstrøm-Lang drying technique in tritium exchange studies of native ribonuclease, found very nearly the theoretical number of exchangeable hydrogens. The present data for oxidized ribonuclease measure closely the theoretical number of exchangeable hydrogens. Further, when exchange-out data of ribonuclease equilibrated with T in H2O and D2O are compared, the individually measurable classes for the two cases are found to be of the same size (Table I). That is, for classes III and IV, KT,H = KT,D. The obvious physical argument makes it seem very unlikely that both equilibrium constants could have any value other than unity. Independently, comparison of rate of exchange of T against H into and out of ribonuclease shows ki = K−i (formula 1))so that again KT,H = 1. The weight of evidence indicates the lack of any isotope effects in the THO-H2O-ribonuclease exchange system. In view of the demonstrated lack of such effects for both random-chain and structured regions of ribonuclease and the presumed similarities in the exchange mechanisms for all proteins, the generality of this conclusion for proteins in aqueous solution seems probable.5
The present data show that though there is on kinetic isotope effect within the T-H (ribonuclease) system, there is a difference between the T-H and the T-D systems. It will be remembered that, for class III, deuterated ribonuclease lost T twice as rapidly as did protonated ribonuclease (t½; = 1.3 hours and 2.6 hours, respectively). The experiment shows this to be a secondary kinetic isotope effect caused by isotope atoms not participating in the reaction itself but exerting an effect through their very presence in the protein. (It is of interest to note again the agreement between the ½ for class III found here for deuterated ribonuclease in T-H exchange with that shown in published work on deuterated ribonuclease in D-H exchange.)
Consideration of this phenomenon leads to an interesting observation. As the 99+% deuterated protein (in H2O) of Figure 4 loses its T, it also loses its D at the same rate. Yet the loss of T retains its first-order character throughout the class III region (Fig. 5). We conclude that the D in ribonuclease which is effective in increasing the class III rate by a factor of 2 is that involved in class IV structure.
The accuracy of the analysis of the exchange-out curve of deuterated ribonuclease may be considered questionable in view of the relatively short segment of the curve included in the apparently first-order region. An alternative explanation for the small difference in the exchange-out curves of Figure 4 would be the loss, upon deuteration, by the protein of an amount of structure responsible for the maintenance of 10 H/molecule in the class IV region.
Identification of Structure
Hydrogen exchange methods rely on kinetic analysis to identify classes of structure and measure the number of hydrogens involved in each. This is as true of infrared and nuclear magnetic resonance methods, which can monitor particular chemical groups, as it is of the “separate and measure” methods, including the Linderstrøm-Lang and the Sephadex methods, which do not share this ability. Little specific information, however, connecting rate of exchange with kind of structure has been available. The present results when considered together with some data in the literature allow some progress in this direction.
Especially pertinent to the following discussion is the work of Blout et al. (1961). These workers used the amide II band to follow the loss of peptide hydrogens by protonated ribonuclease dissolved (at time zero) in D2O at room temperature. They detected the exchange of about 30 backbone protons between 10 minutes and 24 hours and measured a long-lived, 24-hour residuum of 14 peptide hydrogens. The remaining 75 peptides, then, exchanged rapidly.
The present data for oxidized ribonuclease show two kinetic classes of exchangeable hydrogens. One is too fast to measure by the Sephadex method. The second, evidently involving the peptide hydrogens, has a t½ of 5 minutes. Classes I and II of native ribonuclease are, in rate, analogous to these. It seems evident that kinetic class I of native ribonuclease represents intrinsically very rapidly exchanging side-chain groups not sufficiently slowed by their local structure as to render their exchange measurable by the present method. The good agreement between the exchange times of class II hydrogens (8 minutes) and the peptide hydrogens of oxidized ribonuclease (5 minutes), and between the size of class II (80 H) and the number of fast. peptides found by Blout et al. (1961) (75 H ) indicates that kinetic class II represents relatively unhindered peptide groups. (Blout’s somewhat faster class II. rate is presumably a result of the higher temperature used and perhaps also of events occurring upon solution of the dry protein.) It may be assumed, especially in view of the following discussion, that class II peptides are not involved in α-helix. The size of class II then limits the amount of helix possible in the protein.
Since Blout et al. found only 14 very slow peptide protons, the large majority of class IV (70 H ) must represent side-chain hydrogens. (Of these, 20 may be assignable to guanidino groups [Wishnia and Saunders, 1962].) The factors responsible for retaining this large number of hydrogens cannot be detailed, though presumably hydrogen bonding and steric shielding from fruitful contact with water play a role. The 14. peptide hydrogens may be involved in α-helix, but these data clearly show that in general the very slowly exchanging hydrogens of proteins need not relate to helical content.
Blout et al. detected the exchange of 30 peptide hydrogens in the class III region. This, together with the uniform exchange behavior of the 40 class III hydrogens found here, strongly suggests that all of class III represents peptides. (It is noteworthy that the infrared method also underestimated the number of α-helical peptides of hemoglobin and myoglobin which exchange in the class III region [Beychok et al., 1962].)
The isolation of the class III exchange rate is remarkable. We do not see a continuum of rate classes in a transition region between classes II and IV. Rather, the exchange rate of class III is different by more than an order of magnitude from its neighbors. This rate is not very different from the rates measured, under approximately these conditions, for α-helical poly-α,L-glutamic acid (Blout et al., 1961), for the α-helical peptides of myoglobin (Benson, 1959; Beychok et al., 1962), and for the exchange of the slow protons in oxidized ribonuclease which appear and increase in number with increasing LiBr concentration (Stracher, 1960), a treatment which it has been suggested encourages formation of α-helix (Harrington and Schellman, 1957). These similarities in rate suggest that class III represents α-helix.
If class III hydrogens are not involved in α-helix, then their diminished rate of exchange must occur as a result of other structural features of the protein. Participation in individual hydrogen bonds and steric shielding by surrounding structure are the evident alternatives. In either of these two cases, we might expect the maintenance of the class III groups to depend in large measure upon the continued integrity of the extensive superstructure represented by kinetic class IV. We would expect a reasonable degree of interdependence of the hydrogen exchange of both classes. Data in the literature clearly show that the hydrogen exchange of class IV structure can be drastically modified with no observable effect on class III exchange. Ottesen and Stracher (1960) measured the deuterium exchange of ribonuclease at 0° and at 39°. While class IV decreased from 30+H to zero, class III was unchanged (H = 50, t½ 1 hour). Hvidt (1955) studied the deuterium exchange of ribonuclease as a function of pH. Again while class IV fell from 40 H to 10 H as pH increased, class III was not obviously altered. The weight of evidence would seem to identify class III hydrogens with α-helical peptides.
Some of the arguments given are, in themselves, not completely secure. The similarities in rate referred to, in view of differences in experimental conditions among the cases cited, can be taken as only qualitative similarities. It may be that ribonuclease has no α-helix, and that peptides of class III merely approximate the helix-characteristic exchange rate. The postulated effect of LiBr has not yet achieved general acceptance (Bigelow and Geschwind, 1961; Mandelkern and Roberts, 1961). A real question concerns the uniform rate of class III; would one expect all the protons of an α-helix, the ones on the “outer side” and those on the “inner side,” to show the same rate? Though the arguments outlined suggest that kinetic class III does represent α-helix, final resolution of this most interesting question must await further, more direct evidence.
If class III is taken to represent α-helix, the present data show ribonuclease to have no fewer than 40/119 = 34%2 of its peptide hydrogens in α-helix (proline is excluded from the calculation). The estimate rises to 46% if the class IV Blout peptides are added. These values are within the (wide) range estimated by Rosenheck and Doty (1961) from the hypochromic effect in the 200 mμ region, but far higher than the estimate of 17% by optical rotation (see Urnes and Doty, 1961). The difference between these two estimates has been taken as evidence for the existence of a quantity of left-handed helix in ribonuclease. It may be of some interest that the size of left-handed helix indicated by the values given here is close to the size of either of several of the chain segments lying between two disulfide bridges in ribonuclease.
The complete generality of the identification of exchange rates with kinds of structure cannot yet be guaranteed. However, the high precision obtainable, the similarities among rates seen in various proteins and polypeptides, and the fact that rates of different classes differ by more than an order of magnitude make the identification and measurement of structure and changes in structure by hydrogen exchange kinetics seem very hopeful.
Acknowledgments
The author gratefully acknowledges the contribution made by P. H. von Hippel to this work. But for the benefit of his knowledge and insight and the hospitality of his laboratory, this work might not have been done. The author also gratefully acknowledges the efficient and effective help of his wife and laboratory assistant, Joan. Thanks are due to R. C. Fuller and J. Pellerin for making available the facilities of the Department of Microbiology, and to W. Weintraub, a Dartmouth medical student who participated in this work as a summer Fellow of the Dr. Henry R. Viets Student Traineeship in Myasthenia Gravis.
Footnotes
This work was supported by an American Cancer Society postdoctoral grant (PF-86) and by a U. S. Public Health Service grant (A-3412).
Added in Proof
A recent redetermination of the molar extinction coefficient of carefully purified (countercurrent distribution, IRC 50 chromatography) ribonuclease by Eaker (1962) yielded a value of 9.5 × 106. Use of this value alters the numbers of hydrogens in the various kinetic classes as follows: Class II, 71 H; class III, 36 H (30% α-helix); class IV, 65 H.
A complete bibliography is available from A. B. Pharmacia, Uppsala, Sweden.
See note added in proof.
A complete discussion of the preparation, properties, and uses of Sephadex is presented in the dissertation of P. Flodin. 1962.
In agreement with previous reports, the present method indicates the time for loss of hydrogen by DNA to be less than 1 minute under normal solution conditions.
Large enough changes in conditions, e.g., pH, may change the operative exchange-reaction mechanism and lead to an isotope effect (Nielsen, 1960).
References
- Bell RP. The Proton in Chemistry. Ithaca, N. Y: Cornell University Press; 1959. [Google Scholar]
- Benson ES. Compt Rend Trav Lab Carlsberg. 1959;31:235. [PubMed] [Google Scholar]
- Beychok S, de Lozé C, Blout ER. J Mol Biol. 1962;4:421. doi: 10.1016/s0022-2836(62)80099-0. [DOI] [PubMed] [Google Scholar]
- Bigelow CC, Geschwind I. Compt Rend trav Lab Carlsberg. 1961;32:89. [PubMed] [Google Scholar]
- Blout ER, de Lozé C, Asadourian A. J Am Chem soc. 1961;83:1895. [Google Scholar]
- Bradbury EM, Price WC, Wilkinson GR. J Mol Biol. 1962;4:39. doi: 10.1016/s0022-2836(62)80115-6. [DOI] [PubMed] [Google Scholar]
- Bray GA. Anal Biochem. 1960;1:279. [Google Scholar]
- Bryan WP, Nielsen SO. Biochim Biophys Acta. 1960;42:552. doi: 10.1016/0006-3002(60)90842-8. [DOI] [PubMed] [Google Scholar]
- Eaker DL. PhD thesis. The Rockefeller Institute; New York: 1962. [Google Scholar]
- Elliott A. In: Polyamino Acids, Polypeptides and Proteins. Stahmann MA, editor. Madison: University of Wisconsin Press; 1962. p. 119. [Google Scholar]
- Flodin P. J Chromatog. 1960;5:103. [Google Scholar]
- Flodin P. Dextran Gels and Their Applications in Gel Filtration, obtainable from. A. B. Pharmacia; Uppsala, Sweden: 1962. [Google Scholar]
- Haggis GH. Biochim Biophys Acta. 1957;23:494. doi: 10.1016/0006-3002(57)90368-2. [DOI] [PubMed] [Google Scholar]
- Harrington WF, Schellman JA. Compt Rend Trav Lab Carlsberg, Sér Chim. 1956;30:21. [PubMed] [Google Scholar]
- Harrington WF, Schellman JA. Compt Rend Trav Lab Carlsberg, Sér Chim. 1957;30:167. [PubMed] [Google Scholar]
- Hermans J, Jr, Scheraga HA. Biochim Biophys Acta. 1959;36:534. doi: 10.1016/0006-3002(59)90197-0. [DOI] [PubMed] [Google Scholar]
- Hvidt A. Biochim Biophys Acta. 1955;18:306. doi: 10.1016/0006-3002(55)90083-4. [DOI] [PubMed] [Google Scholar]
- Hvidt A, Linderstrøm-Lang K. Biochim Biophys Acta. 1955;16:168. doi: 10.1016/0006-3002(55)90200-6. [DOI] [PubMed] [Google Scholar]
- Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Philip DC, Shore VC. Nature. 1960;185:422. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- Lang ARG, Mason SG. Can J Chem. 1960;38:373. [Google Scholar]
- Leach SJ, Springell PH. Australian J Chem. 1962;15:350. [Google Scholar]
- Lenormant H, Blout ER. Nature. 1953;172:770. doi: 10.1038/172770a0. [DOI] [PubMed] [Google Scholar]
- Linderstrøm-Lang K. Chem Soc (London), Spec Publ 2, Sympos on Peptide Chem. 1955. [Google Scholar]
- Linderstrøm-Lang K. In: Sym Protein Struct. Neuberger A, editor. London: Methuen and Co; 1958. p. 23. [Google Scholar]
- Lobunez W, Karush F. J Am Chem Soc. 1959;81:795. [Google Scholar]
- Mandelkern L, Roberts DE. J Am Chem Soc. 1961;83:4292. [Google Scholar]
- Nielsen SO. Biochim Biophys Acta. 1960;37:146. doi: 10.1016/0006-3002(60)90090-1. [DOI] [PubMed] [Google Scholar]
- Nielsen SO, Bryan WP, Mikkelsen K. Biochim Biophys Acta. 1960;42:550. doi: 10.1016/0006-3002(60)90842-8. [DOI] [PubMed] [Google Scholar]
- Ottesen M, Stracher A. Compt Rend Trav Lab Carlaberg. 1960;31:457. [Google Scholar]
- Rosenheck K, Doty P. Proc Nut Acad Sci. 1961;47:1775. doi: 10.1073/pnas.47.11.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut CL, Scheraga HA. J Am Chem Soc. 1960;82:58. [Google Scholar]
- Stracher A. Compt Rend Trav Lab Carlsberg. 1960;31:468. [Google Scholar]
- Tomita K, Rich A, de Lozé C, Blout ER. J Mol Biol. 1961;4:83. [Google Scholar]
- Urnes P, Doty P. Advan Protein Chem. 1961;16:509. doi: 10.1016/s0065-3233(08)60033-9. [DOI] [PubMed] [Google Scholar]
- Weinberg I, Zimmerman JR. J Chem Phys. 1955;23:748. [Google Scholar]
- Wishnia A, Saunders M. J Am Chem Soc. 1962;84:4235. [Google Scholar]