The CO2-concentrating mechanism (CCM) is key to allowing robust growth of eukaryotic algae. CCM activation by exposure of cells to low CO2 accompanies induction of genes involved in inorganic carbon uptake and concomitant suppression of many metabolism-related genes. Transcriptome and promoter motif analyses provide insights into CCM-associated regulatory networks.
Abstract
A CO2-concentrating mechanism (CCM) is essential for the growth of most eukaryotic algae under ambient (392 ppm) and very low (<100 ppm) CO2 concentrations. In this study, we used replicated deep mRNA sequencing and regulatory network reconstruction to capture a remarkable scope of changes in gene expression that occurs when Chlamydomonas reinhardtii cells are shifted from high to very low levels of CO2 (≤100 ppm). CCM induction 30 to 180 min post-CO2 deprivation coincides with statistically significant changes in the expression of an astonishing 38% (5884) of the 15,501 nonoverlapping C. reinhardtii genes. Of these genes, 1088 genes were induced and 3828 genes were downregulated by a log2 factor of 2. The latter indicate a global reduction in photosynthesis, protein synthesis, and energy-related biochemical pathways. The magnitude of transcriptional rearrangement and its major patterns are robust as analyzed by three different statistical methods. De novo DNA motif discovery revealed new putative binding sites for Myeloid oncogene family transcription factors potentially involved in activating low CO2–induced genes. The (CA)n repeat (9 ≤ n ≤ 25) is present in 29% of upregulated genes but almost absent from promoters of downregulated genes. These discoveries open many avenues for new research.
INTRODUCTION
In nature, Chlamydomonas reinhardtii and other eukaryotic algae depend on a CO2-concentrating mechanism (CCM) to supply sufficient inorganic carbon (Ci; CO2 or bicarbonate) for photosynthesis-fueled cell growth and proliferation. Mutant cells lacking key components of the CCM molecular machinery or its regulatory system (Moroney and Ynalvez, 2007; Duanmu et al., 2009; Yamano and Fukuzawa, 2009) do not grow or grow poorly unless supplied with high concentrations of CO2 (e.g., >10,000 ppm) that are well above the ambient level of ∼392 ppm. Because the diffusion rate of CO2 in aqueous environments is ∼10,000 times slower than in air, most natural populations of microalgae exist in CO2-limited conditions. This is especially true for dense algal populations growing under abundant sunlight. Under such conditions, CO2 concentrations can become very low (<100 ppm) and cells induce the CCM to maximal levels. CO2 starvation induces the transcription of numerous genes encoding proteins closely associated with the CCM and its activities (Moroney and Ynalvez, 2007; Duanmu et al., 2009; Yamano and Fukuzawa, 2009). Indeed, C. reinhardtii and most other eukaryotic algae have developed a finely tuned regulatory system that suppresses expression of CCM-related genes under conditions of replete CO2 (i.e., >0.1% CO2) and activates expression of these genes when CO2 becomes limiting (Moroney and Ynalvez, 2007; Duanmu et al., 2009; Yamano and Fukuzawa, 2009; also reviewed in the companion article Fang et al., 2012). Previous studies using RNA gel blot analyses and microarray analyses have revealed a number of CCM-associated genes and other CO2-responsive genes whose transcription is tied to the physiological changes that accompany cell acclimation to CO2 stress conditions (Im and Grossman, 2002; Im et al., 2003; Miura et al., 2004; Wang et al., 2005; Yamano et al., 2008; Yamano and Fukuzawa, 2009).
Here, we report an extensive global analysis of the massive transcriptional changes evoked by the deprivation of Ci in C. reinhardtii. We measured these transcriptional events using replicated deep RNA sequencing (RNA-Seq) on the Illumina platform. The highly reproducible RNA-Seq experiments not only confirm earlier observations based on array analyses quoted above but also extend the list of differentially expressed genes from a few hundred to over 4000.
We report the discovery of an extensive system of head-to-head (HTH; also called divergent) gene pairs, many of them sharing bidirectional or connected promoters. HTH conformation and bidirectional or shared promoters frequently perform the highly accurate coregulation of gene pairs encoding subunits of the same protein complex or two proteins of similar or related functions. Here, we focus on those HTH, coregulated, gene pairs that are most relevant to the CCM. Advanced computational techniques also have allowed an extensive evaluation of potential regulatory elements in promoter regions in CO2-responsive genes and the discovery of new elements shared by several of the most highly stimulated CO2-responsive genes. We also report a previously unrecognized pattern of expression for many genes that suggests a significant, but transient, decrease in gene transcription immediately after a shift to very low CO2 conditions (ASVLCO2). Finally, we employ a vastly expanded pool of transcriptomic data to strengthen earlier observations of metabolic and physiological changes that occur when CO2 becomes limiting in the environment, including significant decreases in transcripts encoding proteins involved in photosynthesis, cytoplasmic, chloroplastic, and mitochondrial protein synthesis, energy use, protein transport, and other Gene Ontology (GO) categories (Ashburner et al., 2000).
RESULTS AND DISCUSSION
Overview
Ci deprivation is a major stress that evokes a dramatic transcriptional response in algae. Using EST-based macroarrays, the Fukuzawa laboratory (Miura et al., 2004; Kohinata et al., 2008; Yamano et al., 2008) and Grossman and Weeks laboratories (Wang et al., 2005) pioneered the transcriptional profiling of Ci deprivation. In initiating our studies, our hypothesis was that revolutionary progress in sequencing technology and statistical methodology would allow us to discover a large number of activated or repressed biological processes and individual genes that may have escaped detection using EST arrays. To test this hypothesis, we performed deep RNA-Seq using the Illumina Genome Analyzer II platform at the Joint Genome Institute (JGI) of the Department of Energy. In total, the eight samples collected at four time points (0, 30, 60, and 180 min after Ci deprivation) produced 98.3 million uniquely mapped sequencing reads (12.3 million 71-base-long reads per sample; see Methods). When no more than two mismatches were allowed in the anchor regions, ∼38% of the reads did not map uniquely or contained more than two base errors due to sequencing errors, genomic variability, alternative splicing (Labadorf et al., 2010), a number of recently duplicated genes (Villand et al., 1997), and repetitive DNA elements (Merchant et al., 2007). Even with this conservative approach, RNA-Seq represents a major advance from micro- and macroarrays: It provides an unprecedentedly high coverage of transcripts, eliminates cross-hybridization effects, does not rely on the commercial availability of arrays, and is more robust against errors in predicted exon structures (Margulies et al., 2005). The significantly increased performance of RNA-Seq has been shown specifically for C. reinhardtii (González-Ballester et al., 2010).
The high technological reproducibility of the RNA-Seq measurements performed at the Department of Energy’s JGI is shown by the strong correlations of transcript levels between biological replicates (0.958, 0.965, 0.939, and 0.973 for 0, 30, 60, and 180 min time points after carbon deprivation, respectively). These high Pearson correlation coefficients indicate reproducible and multiplicative (linear) biases and that the nonlinear bias is miniscule. Note that linear, multiplicative, and reproducible bias does not alter fold change values by multiplying the transcript levels both in the numerator and the denominator. Such biases include sequencing reads that match imperfectly to the genome or the transcriptome (Li et al., 2010), amplification, and sequencing biases (Dohm et al., 2008). Additive effects, such as unreal exons, may reduce the extent of differential expression. These effects are due to imperfect gene models, such as those predicted by the Augustus method (Stanke and Waack, 2003) and alternative splicing (Labadorf et al., 2010). Such additive effects remain our primary concern. Recently duplicated genes pose further challenges in mapping the 71-base-long sequencing reads to the transcriptome because these reads contain erroneous base calls, particularly at their 3′ ends. Such gene pairs include major effectors of CO2 concentration, such as four carbonic anhydrases (CAHs), CAH1-CAH2 and CAH4-CAH5, that are recent duplicates. The pair CAH4-CAH5, for example, contains exons that are over 90% identical (Villand et al., 1997).
To avoid mappings to the duplicated genes, rigorous procedures (with one or two mismatches in the anchor regions that connect two exons) are necessary. However, this rigor also drastically reduces the coverage of all genes due to both sequencing errors and polymorphisms. Reduced coverage reduces the number of significantly differentially expressed genes. Therefore, we performed the mapping with both one and two allowed mismatches in the anchor region, as implemented in the tophat program (Trapnell et al., 2009). With one allowed mismatch, fewer but more accurate transcript levels were obtained than with two mismatches. For example, our data, as expected from earlier studies, demonstrated that CAH1 is strongly upregulated at 3 h ASVLCO2. However, in our initial analyses allowing two mismatches, CAH2, which had earlier been reported not to respond to CO2, was falsely classified as upregulated. When reanalyzed using only one mismatch per read, the vast majority of reads in the 60- and 180-min time points were mapped to the CAH1 gene, with few being attributable to CAH2. Quantitative RT-PCR (qRT-PCR) confirmed the results of the more rigorous alignments (see Supplemental Figure 1 online).
The lists of differentially expressed genes may be influenced by the choice of statistical methodology. Therefore, we analyzed our data using three different computational tools, edgeR, DESeq, and baySeq. Because differential expression of a large number (∼16k) of genes is estimated using very few replicates (samples), statistical tools derived from large-sample asymptotic theory do not work. In particular, small sample size affects the correction for overdispersion (greater variability than expected based on Poissonian or other simple models), modeling the empirical distributions, and calculating statistical significance. To solve these issues, edgeR (Robinson et al., 2010) shrinks genewise dispersion estimates toward a constant value using an empirical Bayesian model and performs Fisher’s exact test. DESeq (Anders and Huber, 2010) uses nonparametric regression models to fit the negative binomial variance as a function of the mean, assuming a locally linear relationship between overdispersion and mean expression levels. baySeq (Hardcastle and Kelly, 2010) is free of this assumption and uses a fully empirical Bayesian approach to estimate the posterior probabilities. We compared the numbers of overlapping differentially expressed genes reported by the edgeR, DESeq, and baySeq packages at 180 versus 0 min ASVLCO2 (see Supplemental Figure 2 online). Because the exact test implemented in edgeR calculates lower false discovery rate (FDR) q-values than the other two methods (Lopez et al., 2011), at FDR ≤ 0.01, edgeR, DESeq, and baySeq reported 4222, 2364, and 3248 differentially expressed genes, respectively. The lists of differentially expressed genes are more consistent when the FDR threshold is elevated to 0.05, a still conservative level. All three methods reported differential expression for as many as 3141 genes (see Supplemental Figure 2 online). An additional 702 genes were jointly reported by both edgeR and baySeq, and a further 95 genes were called jointly by edgeR and DESeq. Because of the high overlaps with other methods, and its wider acceptance, below we limit our discussions to the results obtained by the edgeR tool.
Our results reproduced the observed induction of major CCM-associated genes published by Miura et al. (2004), Wang et al. (2005), Yamano et al. (2008), Yamano and Fukuzawa (2009), as well as in the companion article (Fang et al., 2012). In addition, we report a large number of genes that have not been associated with the CCM previously. Some of the notable similarities and differences in gene sets of our studies and those presented in our companion article (Fang et al., 2012) are discussed throughout this section (with special attention to the potential causes of observed differences provided near the end of this section).
Major Transcriptional Changes
Our results greatly extend many aspects of earlier observations of differentially expressed genes following CO2 deprivation. This is indicated by relatively similar lists of induced genes published previously (Miura et al., 2004; Wang et al., 2005; Yamano et al., 2008; Yamano and Fukuzawa, 2009) and by us (see Supplemental Data Set 1 online). In addition, RNA-Seq and modern statistical methodologies allowed us to discover an unexpectedly high 5884 genes that are differentially regulated at either 30, 60, or 180 min ASVLCO2 relative to the 0 min control [FDR ≤ 0.01 and abs(log2(fold change)) ≥ 1] or 3828 genes [FDR ≤ 0.001 and abs(log2(fold change)) ≥ 2].
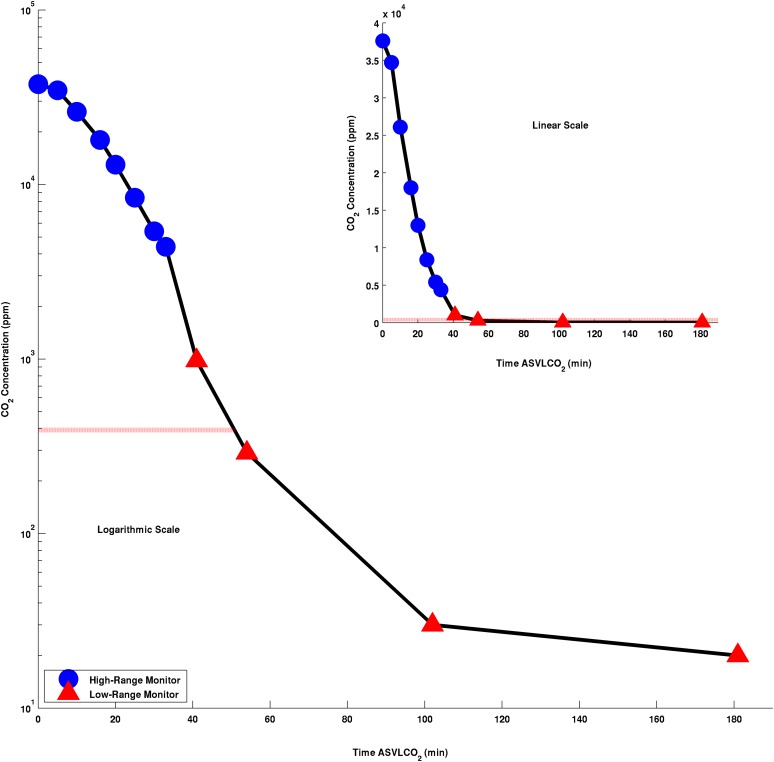
Robust temporal expression patterns emerged under our conditions for imposition of CO2 deprivation. We found that the transcriptional response becomes widespread only after 30 min and increases (or decreases) for many, but not all, genes. The relatively slow onset of significant transcript changes is likely coupled to the relatively slow decline in CO2 concentrations employed in our experiments (Figure 1). At 30 min after deprivation, we found only 37 upregulated and five repressed genes relative to the 0 min control (FDR ≤ 0.001 and abs[log2(fold change) ≥ 2; or in absolute, nonlogarithmic scale, a fourfold increase or decrease) (see Supplemental Data Sets 2 and 3 online). At an hour ASVLCO2, 409 genes are upregulated and 1663 genes are repressed (see Supplemental Data Sets 2 and 3 online). At 3 h ASVLCO2, 981 genes are induced and 1188 genes are repressed. These numbers are approximately doubled at the more typical thresholds (FDR q ≤ 0.01 and abs[log2(fold change) ≥ 1; see Supplemental Data Sets 2 and 3 online). To measure transcript levels by a different method, we performed qRT-PCR analyses on the same RNA samples that were submitted for Illumina sequencing. Three different genes induced by CO2 deprivation (Low CO2 Induced A (LCIA), CAH5, and LCIB) displayed similar expression patterns between RNA-Seq and qRT-PCR, while a fourth gene, CAH2, previously reported as not responding or responding negatively to CO2 depletion (Moroney et al., 2011), showed moderate decreases in transcript levels using both RNA-Seq and RT-PCR measurements (see Supplemental Figure 1 online). Also to be noted (as described above) is the strong correlation of transcript levels in data obtained from biological replicates used for RNA-Seq analyses. Together, these observations confirm the high technological reproducibility of deep RNA-Seq as well as the reproducibility of our biological samples.
Figure 1.
Measurements of CO2 Levels Following a Shift of C. reinhardtii Cells from 5% to 100 ppm CO2.
Two CO2 monitors were used in the fermenter: One was calibrated for high CO2 concentrations (squares), and the other was calibrated to low CO2 concentrations (triangles). CO2 concentrations are plotted on a log10 scale. The horizontal red line represents the 392 ppm concentration of the atmosphere.
Gene Sets Affected by CO2 Deprivation
For brevity, unless otherwise noted, we limit our discussion in this section to transcript differences in cells maintained in high CO2 to transcripts in cells 3 h ASVLCO2. High-level overviews of the massive transcriptional response were obtained using GO categories (Ashburner et al., 2000). PLAZA, an online platform for plant comparative genomics (Proost et al., 2009), assigns GO categories to ∼7700 C. reinhardtii genes. GO categories, like pathways or genes colocalized within a chromosomal band, allow examination of transcriptional changes at the level of gene sets as opposed to individual genes. These gene sets may be enriched in upregulated or downregulated genes. To avoid the subjectivity of interpretations, the statistical significance of enrichment is calculated using gene set enrichment analyses (GSEAs; Subramanian et al., 2005; see Methods). GSEA is capable of handling high-throughput processing of large databases of gene sets, such as GO or the KEGG Database of Biochemical Pathways (Okuda et al., 2008).
The massive downregulation of several fundamental metabolic processes is highly statistically significant as shown by our GSEAs using GO annotations. We found significant repression of translation, ribosomal activities, RNA processing, intracellular protein transport, transport from the endoplasmic reticulum to the Golgi apparatus, nucleic acid binding, ATP synthesis, protein kinase activity, photosynthesis, oxidation reduction processes, protein folding, and unfolded protein binding, etc. (see Supplemental Data Set 4 online). Decreased metabolic activity may reflect the stress/survival mode of metabolism, which is necessary to cope with the stress of CO2 starvation. Upregulated GO categories are less abundant than downregulated ones, biased due to the limited annotations of the CCM- and other plant-specific biological processes in contrast with the basic metabolic processes present in most eukaryotes. Induced GO categories include nucleosome, carbon use, calcium ion transport, and proteolysis-related gene sets (see Supplemental Data Set 4 online). Unexpectedly, five flagella-related categories also are upregulated. GO analyses of several other biochemical pathways provided little additional information, possibly due to limited annotations.
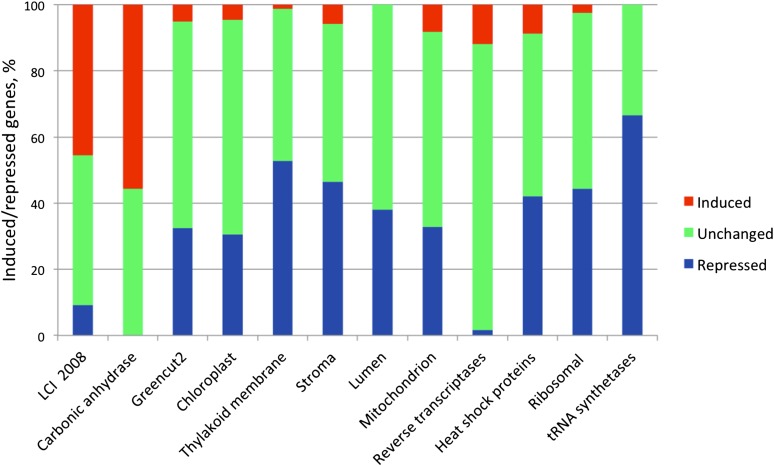
To complement GO categories, we also studied other major functional and subcellular categories (Figure 2). One of these categories is a set of 595 plant-specific (greencut2) genes, which are conserved through diverse representatives of the plant kingdom but have no known relatives in animals, fungi, or prokaryotes (Karpowicz et al., 2011). As many as 193 greencut2 genes were repressed, while only 30 were activated (FDR q-value < 10−256 as calculated by the permutation test implemented in GSEA; Figure 2). To examine whether carbon deprivation reduces photosynthetic activity, we analyzed the 393 genes that code for proteins localized in the chloroplast. Of these nuclear or chloroplast genes, 120 were downregulated and only 18 were induced (FDR q < 10−256).
Figure 2.
Differential Gene Expression in Major Functional and Subcellular Categories in C. reinhardtii Cells before or after a Shift from High to Low CO2.
The percentage of induced genes [red bars; log2(fold change) ≥ 2], repressed genes [blue bars; log2(fold change) ≤ −2] statistically significant at the FDR q ≤ 0.01 level, or unchanged [green bars; log2(fold change) between +2 and −2 and/or not significant]. LCI refers to low CO2–induced genes according to Yamano et al. (2008); greencut2 refers to conserved plant and diatom genes that have no close relatives in other kingdoms and in prokaryotes other than cyanobacteria (Karpowicz et al., 2011).
Using DNA arrays containing oligonucleotides derived from ESTs, the Fukuzawa laboratory (Miura et al., 2004; Yamano et al., 2008; Yamano and Fukuzawa, 2009) and the Grossman and Weeks laboratories (Wang et al., 2005) published several lists of genes differentially regulated by Ci deprivation. In our much more extensive analyses, we reproduced the induction of 40 previously reported genes (38%; see Supplemental Data Set 1 online). LCI1 (Formate/nitrite transporter1.2 [NAR1.2]), a putative plasma membrane anion transporter (Ohnishi et al., 2010), is among the most dramatically induced genes (fold change: 212 ≈ 4000). Fourteen previously designated low CO2-induced genes decreased their expression, including chloroplast geranylgeranyl pyrophosphate synthase (geranylgeranyl diphosphate synthase (GGPS) or LCI14), LCI21, LCI25, and Light Harvesting Complex1 (LHCSR1) (at FDR ≤ 0.01 and log2(fold change) ≤ −1).
Yamano and Fukuzawa (2009) hypothesized that thylakoid membrane proteins may be involved in the regulation of CCM. We successfully reproduced the earlier reported expression patterns of genes coding for low CO2–inducible chloroplast envelope proteins, including oxygen-evolving proteins, plastid division protein, a number of carbonic anhydrases, photosystem II stability/assembly factor photosystem II stability/assembly factor (HCF136), and peptidyl-prolyl cis-trans isomerase (see Supplemental Data Sets 2 and 3 online).
A Shift of C. reinhardtii Cells from High to Very Low CO2 Triggers Transcription of Several Known Genes Encoding CCM-Associated Proteins and Several New CO2-Responsive Genes
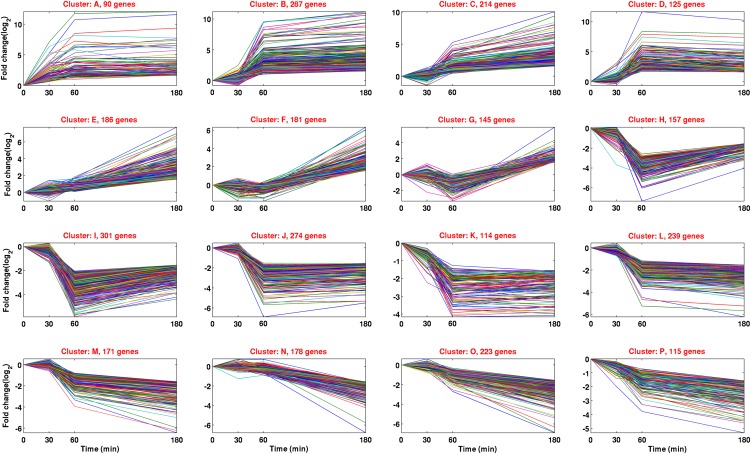
Examination of transcript abundance in this RNA-Seq study for the 106 genes identified in earlier analyses of induced gene transcription triggered by shifts from high to low CO2 conditions (see Supplemental Data Set 1 online) confirms that most are highly induced within an hour ASVLCO2. Among the most highly induced genes at 3 h are several well-recognized CCM-associated or CO2-responsive genes, including LCIA, encoding a putative bicarbonate transporter (4000-fold); LCI1, a gene regulated by the CO2-responsive LCR1 transcription factor (see below) and encoding a transmembrane protein possibly involved in Ci transport (3000-fold); CCP1, a Ci Accumulating5 (CIA5)/CCM1-regulated gene encoding a chloroplast envelope protein (2000-fold); CAH1, encoding a periplasmic carbonic anhydrase (660-fold); LCIE (188-fold); CCP2, encoding a protein closely related to CCP1 (119-fold); LCID, encoding a protein highly similar to LCIB (see below) (89-fold); LCR1, encoding a Myeloid oncogene (Myb)-like transcription factor that regulates expression of CAH1 and LCI1 (52-fold); High Light Acclimation3 (HLA3), encoding a plasma membrane–localized bicarbonate transporter (40-fold); and LCIB, encoding a putative chloroplast CO2-scavenging protein (26-fold) (see Supplemental Data Sets 1 and 2 online). Using cluster analysis of gene expression, we found that expression patterns of these genes during the 3-h time course employed in this study fall primarily into the patterns displayed in clusters A to D (Figure 3). In these clusters, mRNA levels markedly increase by an hour ASVLCO2 and remain high at 3 h.
Figure 3.
Cluster Analysis of Gene Expression Patterns in C. reinhardtii Cells Subjected to a Change from High to Low CO2 Concentrations.
Sixteen clusters were identified by the k-means algorithm. Shown are only the genes that are differentially expressed by a factor of four or more between at least two time points [FDR q ≤ 0.01 and abs(log2[fold change]) ≥ 2]. Time comparisons on the x axis: 0 represents 0-min time point expression levels versus 0 time point expression levels, 30 represents 30-min time point versus 0 time point, 60 represents 60-min time point versus 0 min time point, and 180 represents 180-min time point versus 0 time point. Fold changes in gene expression levels are represented on the y axis in log2 terms.
Strikingly, our deep RNA-Seq analyses increased the number of low CO2-induced (LCI) genes to 1875 (see Supplemental Data Set 2 online), well beyond the 106 previously recognized LCI genes listed in Supplemental Data Set 1 online. The annotation of C. reinhardtii genes has recently been improved (Castruita et al., 2011; Lopez et al., 2011). Still, the function of a number of proteins encoded by differentially regulated genes remains unknown, including 56 out of the 100 most highly induced genes (see Supplemental Data Set 2 online).
A General, but Transient, Decrease in Transcription of Most Genes after a Shift to Very Low CO2 Conditions
Past studies of genes whose transcription changes in response to decreased CO2 levels have focused primarily on those genes whose products are components of the CCM and whose transcription is steadily increased in the first few hours ASVLCO2. Expression patterns typical of such genes are shown in clusters A to D of Figure 3. However, 2399 genes are first downregulated but then induced by 3 h ASVLCO2, forming a distinctive check mark (√) signature of transcript levels. Accordingly, the groups of genes in boxes F to I all display a significant decrease in transcript numbers at 30 and/or 60 min with a subsequent increase at 180 min. Indeed, if one examines the data of Supplemental Data Set 3 online that displays ∼2400 genes downregulated at 180 min ASVLCO2, nearly all of the last 1740 genes display a striking decrease in transcript levels at 60 min, but a marked recovery of transcript levels at 180 min (i.e., the check mark expression pattern; see Supplemental Figure 3 online). This observation suggests that, in general, there is a rapid decrease in gene transcription immediately ASVLCO2. Thus, our studies have revealed that C. reinhardtii cells not only rapidly sense a decrease in CO2 concentrations and respond, as expected, by upregulating CCM-associated genes, but also transition into stress/survival mode by downregulating thousands of genes not directly related to the CCM. This observation opens a new area of study related to transcriptional regulatory networks that orchestrate the rapid and appropriate responses of C. reinhardtii cells to changes in CO2 abundance.
Transient Gene Induction following CO2 Reduction
Antithetic to the check mark pattern of expression discussed above are patterns for 139 genes that are rapidly induced (by a factor of 2 or higher; FDR q ≤ 0.01) during the first hour ASVLCO2 but markedly decrease in transcript levels 2 h later. This caret (∧) expression pattern (see Supplemental Figure 4 online) is observed for the low CO2–induced bestrophin-like protein (516308) regulated by the transcriptional activator CIA5 and, interestingly, for 20 flagellar genes as well. Further experiments will determine if among these genes displaying this transient induction pattern are those encoding transient regulators that control acclimations to long-term restricted CO2 availability. To assess the functions of such transiently expressed genes, mutations or artificial alterations of gene expression (e.g., using RNA interference techniques or gene replacement techniques) will be needed.
Coregulation of Functionally Related Gene Pairs in a HTH Conformation
We discovered an extensive system of HTH (also called divergent) gene pairs, many of which are apparently regulated by bidirectional promoters. In general, when the distance between two start codons is small (in C. reinhardtii, <300 to 500 bp), coregulation is most likely due to a single bidirectional promoter that regulates transcription in both directions. Bidirectional promoters often perform usually tight coregulation of the upstream Crick strand and the downstream Watson strand genes (Trinklein et al., 2004). Bidirectional promoters facilitate the stoichiometric production of subunits of a particular protein complex or members of a particular biochemical or signaling pathway. Such mechanisms have been described in the human pilot-ENCODE project (Birney et al., 2007), yeast (Li et al., 2011b), mouse (Li et al., 2011a), flies, and other organisms. In C. reinhardtii, however, publications are scant regarding the 8852 genes that evolved into the HTH conformation. The few reported coregulated HTH gene pairs include the Argonaut1 and Dicer-like1 genes (Casas-Mollano et al., 2008) and two mitochondrial, β-type carbonic anhydrases CAH4 and CAH5 (then termed ca1 and ca2) reported by Villand et al. (1997). CAH4 and CAH5, like CAH1 and CAH2, recently formed inverted repeats, share very high sequence identity even in introns and promoter regions, and are upregulated under CO2 starvation.
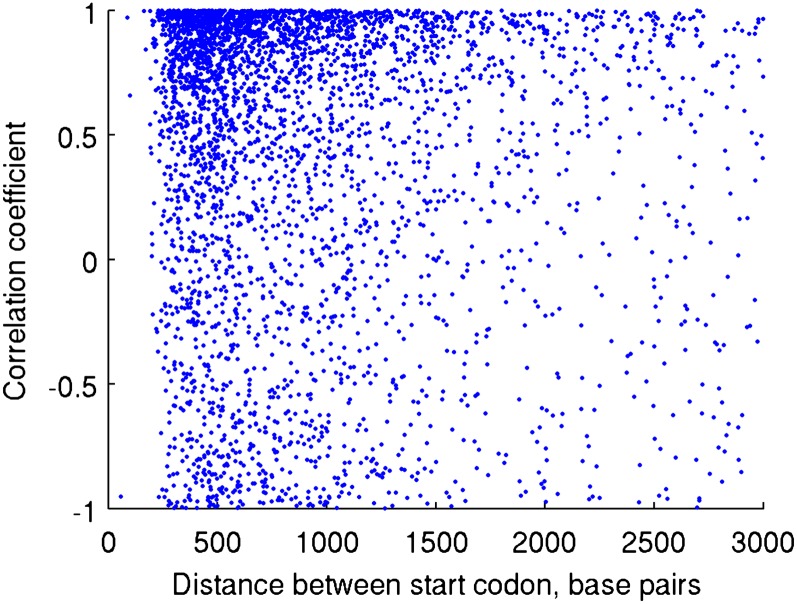
Strong transcriptional correlations within HTH gene pairs may indicate fundamental regulatory mechanisms in C. reinhardtii. Here, we focus on CCM-related HTH gene pairs. Coregulation, frequently by a postulated bidirectional promoter, in many of the 4276 HTH gene pairs in C. reinhardtii is indicated by two observations. First, the median correlation coefficient between transcript levels of the HTH gene pairs is as high as 0.674 compared with 0.522 in all other gene pairs (P < 10−256, Wilcoxon-Mann-Whitney test; Figure 4; see Supplemental Figure 5 online), and for 25% of HTH gene pairs, this correlation exceeds 0.917. For other conformations, the correlation in the top 25% of non-HTH gene pairs is 0.8650 (for the whole distributions, P < 10−256, Wilcoxon-Mann-Whitney test; see Supplemental Figure 5 and Supplemental Data Set 5 online). The high correlation of the top 25% even in the non-HTH genes indicates the existence of other mechanisms of coregulation, including locus control regions, microRNAs, and the binding of similar transcription factors (reviewed in Ladunga, 2010). The second indication for the bidirectional regulation is the short distance between the start codons of HTH gene pairs (median length: 831 ± 3228 bp sd). By contrast, intergenic regions of tail-to-tail, head-to-tail, and tail-to-head gene pairs tend to be more than twice as long (median: 1712 ± 5595 bp, P < 10−256, Wilcoxon-Mann-Whitney test; see Supplemental Figure 5 online). Short intergenic regions of tail-to-tail, head-to-tail, and tail-to-head gene pairs do not necessarily indicate coregulation.
Figure 4.
Several HTH Genes Are Coregulated by Bidirectional or Interacting Promoters.
Each dot represents a HTH gene pair where the distance between the start codons of the two genes is represented by the horizontal coordinate and the coexpression of the two genes is represented by the vertical coordinate of the dot. Coexpression is measured by the Pearson correlation coefficients between the two HTH genes for the transcript levels at 0, 30, 60, and 180 min after carbon deprivation.
The correlation coefficient for the transcript levels exceeds 0.9 in 1188 pairs and 0.8 in 1711 pairs (Figure 4; see Supplemental Figure 5 and Supplemental Data Set 5 online). Such correlation exists for both induced and repressed gene pairs.
Several CCM and Related Pathway Genes Exist in HTH Conformation
Among the 1845 significantly induced nonoverlapping genes [FDR q ≤ 0.01, log2(fold) ≥ 1], 212 genes evolved into HTH conformations, in which both genes are upregulated, and an additional 1116 HTH pairs, in which one gene is upregulated. The former include 52 genes previously recognized as regulated by CIA5/CCM1 and associated with CCM (see Supplemental Data Set 1 online). In three pairs, CCP1/ LCIE, CCP2/LCID, and CAH4/CAH5, each gene has been reported as CCM related. The sequence similarity, tight chromosomal linkage, and highly coordinated expression of LCID/CCP2 and LCIE/CCP1 (see Supplemental Figure 6 online) suggest recent gene pair duplications. Another nine known CCM-related genes form pairs with genes that were not reported as CCM related before: LCI1/Flagellar Associated Protein292 (FAP292), LCI6/513965, CAH1/CLR18, LHCSR1/525344, LCI7/525344, LCI7/523113, Early Light Inducible4 (ELI4)/519915, LCI31/517052, and GGPS/512500. Transcript levels in seven of these nine pairs are correlated with r > 0.6.
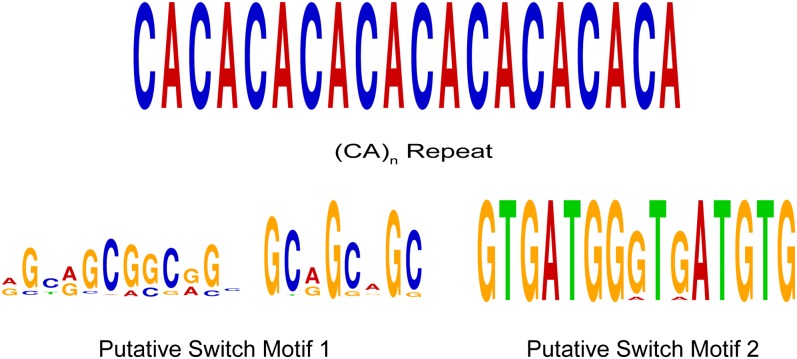
By contrast, gene expression is negatively correlated in 1048 HTH gene pairs, which presents an enigmatic mechanism of transcription regulation (see Supplemental Data Set 6 online). Of these, the transcript levels of 287 pairs had a correlation of −0.8 or lower (see Supplemental Figure 5 and Supplemental Data Set 6 online). Apparently, the extent of negative correlation does not depend on the distance between the two start codons, in sharp contrast with the extent of positive correlations in transcript levels. The most likely explanation for negatively correlated HTH genes is the steric occlusion between overlapping promoter regions, where the direct and indirect binding of transcriptional regulators and the RNA polymerase II to one promoter region excludes binding to the other promoter region. This phenomenon is termed as promoter occlusion or promoter interference (Nakajima et al., 1993). Alternatively, in a minority of negatively correlated HTH gene pairs, switch motifs may account for negatively correlated transcription. We postulated that switch motif(s) might also account for the induction of one gene and the repression of the other. It is also possible that insulator motifs separate the two adjacent but unidirectional and oppositely oriented promoter regions. Using Multiple Expectation Maximization for Motif Elicitation (MEME) and MAST (Bailey et al., 2009), we found two novel putative switch motifs (Figure 5). Putative switch motif 1 is a degenerate but 21-bp-long motif, while putative switch motif 2 is a well-conserved, 15-bp-long motif (Figure 5). Putative switch motif 1 is present in 44 and switch motif 2 is found in nine intergenic regions between negatively correlated HTH genes. Neither switch motif was found in promoter regions of positively correlated HTH genes.
Figure 5.
Putative Transcription Factor Binding Sites and Switch Motifs.
Putative switch motifs were found in intergenic regions between HTH gene pairs with negatively correlated transcription. Motifs are shown as sequence logos where the horizontal axis indicates sequence position and the vertical axis shows information content, related to conservation.
Regulation of Specific Sets of CCM Genes
Carbonic Anhydrases
There are three known C. reinhardtii α-carbonic anhydrase genes, six known β-carbonic anhydrases, and three putative γ-carbonic anhydrases (Moroney et al., 2011) (see Supplemental Data Set 7 online). Four of the nine α- and β-CA genes (CAH1, CAH4/CAH5, and CAH7) were activated (twofold or more) (Figure 2; see Supplemental Data Sets 2 and 7 online) and none were repressed significantly (FDR ≤ 0.01). Transcripts encoding CAH1, an α-type, periplasmic CA and CAH4/CAH5, recently duplicated, mitochondrial, β-type CAs were induced by over 29 (512-fold), similar to RT-PCR observations by Ynalvez et al. (2008). These results are confirmed by the companion article (Fang et al., 2012), including the activation of CAH1 and CAH4/CAH5. As a notable difference to the observations of Ynalvez et al. (2008) and Fang et al. (2012), we found CAH3 slightly upregulated. In the cia5 mutant, Fang et al. found that CAH5, the most highly upregulated CA gene in wild-type cells, is actually repressed. Moreover, in cia5, CAH1, CAH4, CAH6, and CAH8 were weakly induced (Ynalvez et al., 2008), suggesting the possible existence of a minor CCM regulatory mechanism independent of CIA5.
Levels of CAH2, encoding another periplasmic α-type carbonic anhydrase (Rawat and Moroney, 1991) either remained stable or slightly declined in previous RNA gel blot and macroarray transcriptome analyses (Miura et al., 2004; Wang et al., 2005; Yamano et al., 2008; Ynalvez et al., 2008; Yamano and Fukuzawa, 2009). These observations were corroborated in our transcriptome analysis and further confirmed by qRT-PCR analyses (see Supplemental Figure 1 online). These observations strengthen the conclusion that although CAH2 is possibly a recent duplication of the CAH1 gene (or vice versa), it apparently does not have a role in CCM under limiting CO2 conditions (Fujiwara et al., 1990).
Because of its possible localization either inside or immediately outside of the plasma membrane, a recently discovered CA, CAH8, has been considered to be a potential contributor to Ci uptake in C. reinhardtii (Ynalvez et al., 2008; Moroney et al., 2011). The lack of significant induction of the CAH8 gene ASVLCO2 by both the Moroney group (Ynalvez et al., 2008) and this study (see Supplemental Data Set 7 online) would suggest that if this is so, CAH8 either contributes little to enhanced Ci uptake during induction of the CCM or that the CAH8 enzyme is present constitutively in nonlimiting quantities. Quantitative measurements of CAH8 concentrations and activities and/or the availability of CAH8 mutants or knockdown lines are needed to differentiate between these alternatives.
Three putative γ-CA genes are not induced by CO2 starvation (see Supplemental Data Set 7 online). These putative CAs appear to be strongly coupled with the Complex 1 mitochondrial electron transport chain (Klodmann et al., 2010) and therefore may contribute little to the changes in cellular metabolism triggered by CO2 deprivation.
LCIB and LCIC
Although no longer considered as directly involved in Ci transport (because of the lack of transmembrane domains), the soluble chloroplast stromal protein, LCIB (Wang and Spalding, 2006), and the complex of LCIB with its close family member, LCIC (Yamano et al., 2010), have been implicated as involved in CO2 retention in the chloroplast or in assisting the Ci transport system within the chloroplast. The novel CO2-requiring phenotypes of mutants containing defective LCIB genes (Van et al., 2001), the unexpected close functional relationship between LCIB and CAH3 (Duanmu et al., 2009), and the close physical association of LCIB (and supposedly the LCIB/LCIC complex) with the ribulose-1,5-bis-phosphate carboxylase/oxygenase–rich pyrenoid body when cells are exposed to CO2-limiting conditions (Yamano et al., 2010; Wang et al., 2011) have strongly implicated these proteins as key players in the C. reinhardtii CCM. Consistent with this involvement is an observed 20- to 30-fold increase in LCIB and LCIC transcript levels in cells moved from a CO2-replete to a CO2-depleted condition (see Supplemental Data Set 1 online).
HLA3 and Other Putative Ci Transporters
Transcripts encoding HLA3, an ATP-energized HCO3− transporter (Duanmu et al., 2009), increase at 60 min by 22-fold and at 180 min by 40-fold (see Supplemental Data Set 2 online). Increases of LCIA (NAR1.2) transcription are a remarkable 3300-fold at 60 min and 4000-fold at 180 min ASVLCO2. The list of potential Ci transporters also includes the CCP1 gene (induced 2000-fold) and the CCP2 gene (induced 120-fold), two long-recognized CO2-responsive genes whose products have been implicated in potential Ci transport across the chloroplast envelopes. Because these four are among the most highly induced genes (see Supplemental Data Set 2 online), CCM assembly or augmentation in response to CO2 deprivation likely requires the de novo assembly of a number of Ci transporters at several critical subcellular locations. Transporters in the plasma membrane include HLA3 (Duanmu et al., 2009) and LCI1 (Ohnishi et al., 2010); in the chloroplast envelope, they include LCIA (Duanmu et al., 2009), CCP1, and CCP2 (Pollock et al., 2004). Because evidence exists for a critical role of CAH3 in converting HCO3− to CO2 in the lumen of the thylakoid membranes of the chloroplast (Moroney et al., 2011) and the diffusion of the resulting CO2 to ribulose-1,5-bis-phosphate carboxylase/oxygenase in the pyrenoid body, an argument has been made for the existence of a transporter or channel to shuttle bicarbonate from the alkaline chloroplast stroma to the acidic lumen of the thylakoid membrane. A future quest for this hypothetical HCO3− transporter/channel among the strongly induced genes encoding transmembrane proteins may prove rewarding.
In our RNA-Seq study, GGPS (LCI14) (part of the ∼15-element chlorophyll a biosynthesis pathway in higher plants) (Robinson et al., 2010), LCI25 (encoding a protein homologous to stress induced one-helix protein in Arabidopsis thaliana), and LHCSR1 (encoding a stress-related chlorophyll a/b binding protein) are all downregulated ASVLCO2. Because both GGPS and LHCSR1 are induced by the transcriptional regulator CIA5/CCM1 (Wang et al., 2005), their downregulation may be due to other transcription factors or noncoding RNAs.
Mitochondrial Functions
Our transcriptome analyses suggest that carbon deprivation suppressed most genes involved in classical mitochondrial functions. Transcripts from 24 of the 73 annotated mitochondrial proteins were suppressed and only six were increased (see Supplemental Data Sets 2 and 3 online; Figure 2). On the whole, the decreased transcription of genes producing mitochondrial components is reflective of the decreased metabolic activity of cells exposed to CO2-limiting conditions.
Regulation of the Response to Very Low CO2
Despite considerable efforts, most regulators of the CCM remain unknown or incompletely characterized. Indeed, very few C. reinhardtii transcription factor binding sites have been determined experimentally (Kucho et al., 2003; Yoshioka et al., 2004; Sommer et al., 2010; Kropat et al., 2011). Inferences from other photosynthetic organisms are also limited due to the scarcity of known transcription factor binding sites (Yilmaz et al., 2011). Previous studies and our companion article (Fang et al., 2012) have focused on CIA5/CCM1, the master regulator of the transcriptional response to low CO2, and the putative transcription factor LCR1, itself regulated by CIA5 (Kucho, Yoshioka et al. 2003). The CIA5/CCM1 orthologs in C. reinhardtii and Volvox carteri share 51% identity at the amino acid level (Yamano et al., 2011). This conservation extends to two zinc-finger domains (a mutation that gives rise to the CCM defective phenotype of cia5), a multifunctional protein–protein interaction domain characteristic of transcriptional regulators (Plevin et al., 2005), putative nuclear export and localization signals, and a sumoylation site. We reported earlier (Wang et al., 2005) that regardless of the ambient CO2 levels, the CIA5/CCM1 gene is constitutively expressed and the protein is also constitutively localized in the nucleus. Given that there is no evidence for CIA5/CCM1 binding to DNA (Kohinata et al., 2008), our hypothesis is that the atypical zinc-finger domains (Kohinata et al., 2008) of CIA5/CCM1 do not bind directly to DNA but regulate other transcription factors as a transcriptional activator via direct or indirect interactions. We postulate that, once activated, these latter factors may bind to cis-regulatory elements in the promoter regions of CO2-responsive genes to enhance transcription.
To examine this hypothesis, we searched for potential transcription factor binding sites in the putative promoter regions of two gene sets. The first gene set is the union of low CO2–induced genes identified from previous publications and/or from our study. The second set is the potential CIA5/CCM1-regulated genes identified by the comparative transcriptomic studies of the cia5 mutant and wild-type C. reinhardtii cells in our companion article (Fang et al., 2012). For these and other sets of promoter regions, we performed DNA sequence motif discovery (see Methods). We found several relatively conserved DNA sequence motifs that, as discussed below, are candidates for transcription factor binding sites. Because none of the binding sites is present in more than a quarter of the low CO2–induced genes, the CCM is apparently regulated by multiple transcription factors, cofactors, and possibly other regulators. From its high level in the regulatory hierarchy, CIA5/CCM1 appears to regulate many or most subordinate regulators, but not all of them.
LCR1, whose binding (enhancer) sites are essential for the induction of the CAH1 gene, is a prime example of a CIA5/CCM1-regulated transcription factor. Using gel mobility shift assays, the Fukuzawa laboratory identified two enhancer elements (EE’s) in the promoter region of CAH1 (Kucho et al., 2003; Yoshioka et al., 2004). These elements, EE-1 (5′-AGATTTTCACCGGTTGGAAGGAGGT-3′) and EE-2 (5′-CGACTTACGAA-3′), share the DNA sequence GANTTNC (referred to here as Motif 1A). Later, this motif was shown to be the binding site for LCR1 (Miura et al., 2004). LCR1 is an outlier of the 1R-Myb subfamily; its Myb domain is only ≈20% identical to its relatives. We confirmed the extremely strong induction of the gene coding for the transcription factor LCR1 following CO2 deprivation (see Supplemental Data Set 2 online). We found for the GANTTNC motif (and its reverse complement) in the upstream 500-bp regions upstream of the start codon of the 893 genes that were upregulated in our experiments using very stringent upregulation criteria: log2FC ≥ 1 and FDR q ≤ 0.0001 3 h after CO2 deprivation. A total of 476 promoter regions (53%) matched this low information content motif or its reverse complement (see Supplemental Table 1 online). Surprisingly, however, 697 of the 1227 downregulated genes (78%) and 7141 of the 13,380 genes (53%) that were not significantly differentially regulated by the above criteria also matched Motif 1A. To increase the specificity of the low information content Motif 1A, we searched for its supersets EE-1 and EE-2 in the promoter regions of upregulated genes but found no exact matches other than the previously identified single copy in the CAH1 promoter region. Therefore, we performed de novo motif discovery using a strategy described in detail earlier (Ladunga, 2010). Although no chromatin immunoprecipitation, protein binding array, or protein–protein interaction data are available for C. reinhardtii, diverse algorithms for promoter sequence analysis, each with different advantages, knockout mutants, and expression data allowed us to obtain insight into the regulatory network of CCM-associated genes (see Methods).
Putative Novel cis-Regulatory elements: (CA)n Repeats
From the promoter regions of all upregulated genes in our experiments, the MEME tool (Bailey et al., 2009) reported new motifs. The most remarkable is a (CA)n repeat (Motif 2), where n ranges from 9 to over 20. (CA)n is present in 29% of the promoters of upregulated genes ASVLCO2, but it is almost absent from the promoters of downregulated genes. Indications of potential transcription activating mechanisms associated with (CA)n repeats can be found in mammalian and yeast systems. In the human genome, 19.4 CA repeats occur per mega base pair, representing the most common simple-sequence repeat motif (Waterston et al., 2002). Among all dinucleotides, the structure of CA dinucleotides are the most stable regardless of their environment, and the stability of the DNA structure enhances the stability of the chromatin as well (Fujii et al., 2007). This stable chromatin structure allows the binding of numerous transcriptional activators and splicing regulators. Heterogeneous nuclear RiboNuclearProtein L (hnRNP L) in mammals binds to an intronic polymorphic CA repeat region in the human endothelial nitric oxide synthase gene (Hui et al., 2005). hnRNP L has a role in determining alternative splicing and splicing efficiency. A homologous nuclear ribonucleoprotein exists in C. reinhardtii as well (Johnston et al., 1999). In humans, the length of the (CA)n repeats positively correlates with the expression of the interferon γ gene (Pravica et al., 2000) and the integrin α2 gene. Similarly, alleles with longer (CA)n repeats in the upstream regulatory region of the integrin α2 gene enhance the specific binding of poly(ADP-ribose)polymerase-1, a molecular nick sensor, and KU80 polypeptide (Ku80)/70, two components of transcription coactivator complexes (Cheli et al., 2010). C. reinhardtii genes encode for proteins remotely similar to Poly(ADP-ribose) polymerase-1 (PARP-1), including the chloroplast tscA maturation factor (Protein ID: 525840, XP_001694431). The Ku80-like domain is also present in two proteins (509698, XP_001702579; 515111, XP_001699098). These observations suggest the possibility that the C. reinhardtii (CA)n motif may recruit transcription activator complexes similar to the mammalian PARP-1 Ku80/70–containing complexes.
Transcription Factors That May Bind to the (CA)n Motif
Searching against all known motifs in the TRANSFAC Professional Database (Matys et al., 2006), we found that (CA)n and similar sequences are bound both in vivo and in vitro by yeast Ras-related protein1 (Rap1), another Myb family repressor-activator protein. In yeast, the general transcription factors Rap1 and autonomously replicating sequence-binding factor1 (Abf1) can create local nucleosome-free regions by evicting nucleosomal histones, thereby facilitating the binding of more specific transcription factors (Castruita et al., 2011). Rap1 was observed to bind to 5′-CACACCCACACACC-3′ motifs as well, and even low affinity binding sites for Abf1 and Rap1 play a role in determining nucleosome occupancy (Ganapathi et al., 2007). Rap1 is also implicated in silencing at telomeres and silent mating type loci in yeast (Iglesias et al., 2011). A BLAST search could not detect any homologs of Rap1 in the C. reinhardtii proteome. However, more sensitive hidden Markov model searches as implemented in the hmmsearch tool (Johnson et al., 2010) against the PFAM Database of protein domains (Punta et al., 2012) indicated that, at the very least, 55 C. reinhardtii proteins carry some combinations of the Myb-, Sant-, BRCT domain of the BRAC1 oncogene, or homeodomain-related domains that are characteristic of Rap1. Of these, 35 genes are slightly upregulated at 180 min after CO2 deprivation. Based on domain similarity with Rap1 and the upregulation of their genes, our best candidate for (CA)n binding is the predicted protein 515479 (XP_001692722), conserved in V. carteri, Chlorella vulgaris NC64A, Ostreococcus tauri, Physcomitrella patens, and Micromonas pusilla. Other candidates include two Myb2-like transcription factors (Protein IDs 511319 [XP_001690083] and 519163 [XP_001699726]), each of them harboring as many as 11 to 12 Sant DNA binding domains conserved across many eukaryotes, even in mammals. Myb2 transcription factors, associated with calmodulins, coregulate salt and dehydration response in Arabidopsis (Yoo et al., 2005) and numerous other biological processes. LCR1, despite its robust induction, is a less likely candidate because it shares only the Myb domain with RAP1. In summary, the presence of (CA)n repeat elements in 29% of the promoters of upregulated genes and their complete absence from the promoters of downregulated genes (see Supplemental Data Set 3 online) combined with knowledge of the activator role these elements in the human integrin α2 and interferon γ genes discussed above, suggest that the (CA)n motif in C. reinhardtii may also serve as binding sites for transcriptional activator proteins.
A fourth member of the Myb family of transcription factors is the also induced Myb11 (Protein ID 511940, XP_002945989). Its Arabidopsis ortholog regulates the biosynthesis of flavonols that absorb potentially damaging UV-B radiation (Stracke et al., 2010a, 2010b). In Arabidopsis, intense light conditions induce Myb11, even under normal CO2 conditions (Stracke et al., 2010b). In C. reinhardtii, because relatively intense light appears to be necessary for the activation of carbon-concentrating mechanisms (Yamano et al., 2008), Myb11 and other light-induced Myb transcription factors may be necessary for low CO2 response.
The discovery of these new, putative, transcription factors should facilitate the discovery of the transcription factor network(s) that regulate several CCM-associated genes. The development of synthetic gene promoters containing these and other putative transcription factor binding sites (motifs) and the isolation of the transcription factors that bind these sites are but two approaches by which the C. reinhardtii community can begin to understand the detailed mechanisms by which algal cells sense and respond to external changes, such as altered CO2 abundance.
Biological and Experimental Reproducibility
The companion study by Fang et al. (2012) focusing on both the effects of CO2 deprivation and the role of the CIA5 gene in regulating the C. reinhardtii CCM nicely complement this article. Their findings support many of our prime observations and conclusions in regard to areas in which our studies overlap (i.e., in comparison of transcriptome changes associated with CO2 deprivation). Nonetheless, the two studies were conducted completely independently and involved a number of significant differences in conditions (detailed below). Thus, it is not unexpected in the comparison of our two data sets there are differences in the patterns and the magnitude of gene expression levels (i.e., it is to be expected that there will be considerably lower interlaboratory reproducibility than intralaboratory reproducibility). Key differences in the two studies include the strains employed, transcript level measurement technologies, the tempo and mode of reducing CO2 levels, temperature, illumination, and cell densities.
Tempo and Mode of CO2 Deprivation. In our earlier microarray studies, we rapidly shifted cells from high to ambient CO2 levels (Wang et al., 2005). In the Fukuzawa laboratory’s EST-based macroarray studies (Miura et al., 2004; Yamano et al., 2008; Yamano and Fukuzawa, 2009), CO2 levels were shifted gradually. In this study, we initially maintained high CO2 levels by sparging with 5% CO2. At 0 time, we switched the input to a mixture of nitrogen and 100 ppm CO2, which, within the rapidly stirred fermenter, resulted in a slow decline in CO2 concentration: at 50 min, to ambient levels; at 75 min, to 100 ppm; and finally, at 180 min, to ∼20 ppm (Figure 1). By contrast, Fang et al. (2012) measured the effect of CO2 deprivation 1 h later than we did, 4 h ASVLCO2. Given the rapid increases and decreases in transcript levels seen for many genes during the 3-h time course of CO2 starvation (Figure 3), it is expected the extra hour of CO2 deprivation in the studies of Fang et al. (2012) might well lead to at least some of the differences observed.
Light and Temperature. Our C. reinhardtii cultures grew in a rapidly stirred fermenter at 25°C under white light illumination at 200 μmol photons s−1 m−2, 2 times higher than in our companion article (Fang et al., 2012). In C. reinhardtii cultures, a light intensity of 200 μmol photons s−1 m−2 is sufficiently high to potentially cause oxidative damage to pigments, such as chlorophyll (Peers et al., 2009), damage that is much less likely to occur at a light intensity of 100 μmol s−1 m−2. In addition, Fang et al. maintained cultures at room temperature, whereas our cultures were warmed to a constant 25°C.
Strains of C. reinhardtii May Differ in Splicing and Genomic Polymorphisms. A high proportion of sequencing reads from a nonstandard strain cannot be mapped to the reference transcriptome and genome or to the probes of a microarray, and fewer reads may bias the estimates of differential gene expression. For example, when one or more exons of a multiexon gene are not expressed but the others are, this bias decreases the expression difference that is calculated for a gene model incorrect for the strain.
Quantitative Analysis of Reproducibility
In assessing the potential causes of differences between results in our study and those in our companion article by Fang et al., we feel the above-mentioned variations in strains and culture conditions likely account for the bulk of differences in gene expression levels and patterns. Because previous studies reported almost exclusively genes induced by CO2 deprivation (Miura et al., 2004; Wang et al., 2005; Yamano et al., 2008, 2010), our analysis will necessarily need to be limited to induced genes. To ensure conservative estimates of differential gene expression, our analyses based on the edgeR tool (Robinson et al., 2010), an FDR threshold of 10−4 is applied. Our experiments reproduced 166 of the 393 genes (42%) upregulated by CO2 deprivation in our companion study (Fang et al. 2012) as shown by a Venn diagram (see Supplemental Figure 1 online). We also reproduced 40 genes (38%) upregulated in previous experiments (Miura et al., 2004; Wang et al., 2005; Yamano et al., 2008, 2010), and Fang et al. (2012) reproduced 39 genes (37%). Importantly, 27 of the 106 previously published genes (25%) were reproduced by both Fang et al. (2012) and us. Indeed, the expression patterns of genes most critical to CCM are similar across these three gene sets (see Supplemental Data Set 1 online). In the order of fold change 180 versus 0 min, these genes include carbonic anhydrases 1, 4, 5; NAR1 (LCIA), low carbon–induced genes (LCI) 1, 23, 11, D, B, 15, C, 22, 31, and 19, as well as CCP1 and 2, LHCSR2 and 3, HLA3, guanine deaminase 1 (GUD1), Chloroplast DnaJ-like protein3 (CDJ3), ELI4 (LCI16), and Alanine Aminotransferase1 (AAT1).
Perspectives
The RNA-Seq data presented here relating changes in environmental CO2 abundance to changes in gene expression levels greatly expand our knowledge and understanding of the CCM compared with data obtained in more limited RNA gel blot and microarray studies conducted earlier. Some general conclusions made in the earlier investigations have been confirmed, such as the expected decline in photosynthetic capacity as CO2 availability becomes limited. Other observations, such as the marked downregulation of a vast number of genes immediately after CO2 decrease have not been noted earlier and may provide the foundation for a more detailed examination of how algal cells perceive and respond so rapidly to changes in environmental conditions that demand prompt changes in cellular metabolism and physiology. Likewise, our studies revealed an unexpectedly large number of gene pairs involved in similar cellular activities that are oriented in a HTH conformation, where a shared bidirectional promoter coordinately regulates the response to changes in CO2 levels. Investigations of common regulatory elements now become a potentially fruitful area of investigation. Our results from a time-course study of changes in the transcriptomes at critical time points after imposition of CO2 starvation builds on results from the accompanying publication of the Spalding laboratory (Fang et al., 2012) regarding the pivotal regulatory role of CIA5/CCM1 in response to CO2 deprivation.
Our data confirm that synthesis of many of the key components of the CCM are triggered when CO2 levels decrease and reveal an even greater number of candidate genes whose function may be essential or helpful to assembly of a fully functional CCM. These genes become potential targets for chromatin immunoprecipitation and deep sequencing (Wang et al., 2011) or for inactivation through RNA interference techniques (Cerutti et al., 2011), insertional gene inactivation (Ermilova et al., 2000), or targeted gene knockout using zinc-finger nucleases or newly developed techniques, such as TAL effector nucleases (Li et al., 2011a). The vast array of genes induced or repressed as a result of CO2 changes point to a sophisticated set of regulatory networks that must effectively govern multiple cellular responses, an area of study that appears fruitful for future exploration.
METHODS
Ci Deprivation
Chlamydomonas reinhardtii wild-type strain CC124 was used for analysis. Briefly, cells were grown in 2 liters of Tris Phosphate medium at 25°C and 3% CO2 to a density of 1 × 106 cells/mL (Harris, 1989) before being transferred to a 3-liter autoclavable glass bioreactor (Applikon Biotechnology) that was connected with EZ control for analysis of temperature, pH, and dissolved oxygen. The bioreactor was illuminated with a light intensity of 200 µmol photons m−2 s−1, and an input gas containing 5% CO2 was introduced. Algal cells were allowed to equilibrate with the new environment for 1 h. Following a sampling of the culture, the input gas for the bioreactor was shifted to 100 ppm CO2, which was monitored in the culture using two CO2 transmitters (Vaisala; models GMT221 and GMT222). Samples were taken at 15, 30, 60, and 180 min following the shift to 100 ppm CO2. During the experiment, pH was maintained at 7.2 using 3 M KOH.
RNA Preparation
Cellular samples were taken from the bioreactors in 100-mL volumes and transferred directly to a sterilized 2-liter Erlenmeyer flask submerged in an ice water bath. The flask was agitated within the ice water for 2 min to rapidly decrease the temperature of the algal culture to reduce the degree of transcriptional changes preceding removal from the bioreactor. The cellular samples were then centrifuged at 2000g for 5 min at 4°C. The supernatant was discarded and the cellular pellet was frozen by immersing the centrifuge tube in liquid nitrogen. Samples were stored at −80°C until RNA extraction.
RNA extraction was performed using TRIzol LS reagent (Life Technologies). Briefly, cellular pellets composed of ∼5 × 107 cells were resuspended and incubated for 5 min at room temperature with Trizol LS reagent. Chloroform was added to the samples, agitated for 15 s by tube inversion, and allowed to incubate at room temperature for 15 min. Samples were centrifuged at 12,000g for 15 min at 4°C. The aqueous phase was recovered and transferred to new tubes. Isopropanol was added to the aqueous phase, mixed, and centrifuged at 12,000g for 10 min at 4°C to pellet RNA. Isopropanol was removed and the RNA pellet was washed with 75% ethanol. Following further centrifugation (7500g for 5 min at 4°C), ethanol was removed and the RNA pellets were allowed to air dry for 5 min. RNA pellets were solubilized in 500 μL nuclease-free water. RNA was further purified by precipitation with lithium chloride. An equal volume (500 µL) of 4 M LiCl was added with mixing, and samples were incubated at −20°C for 60 min. Following this incubation, samples were centrifuged at 16,000g for 20 min at 4°C. The aqueous phase was removed and discarded and the RNA pellet was washed twice with 75% ethanol. Following air drying, the pellet was resuspended in 50 μL of nuclease-free water. RNA samples were then analyzed using a NanoDrop 2000 spectrophotometer (NanoDrop Products/Thermo Fisher Scientific) to verify RNA quantity and purity.
Preliminary Analysis of RNA Samples
To confirm induction of the carbon-concentrating mechanism, preliminary analysis of RNA samples was performed using qRT-PCR. RNA samples were prepared for analysis using the Plexor Two-Step qRT-PCR system (Promega). qRT-PCR analysis was performed using a 7500 Real-Time PCR System (Life Technologies). The genes LCIA (AB168092), LCIB (XM_001698292), and mitochondrial carbonic anhydrase (CAH4, XM_001695951) were chosen for analysis as they have been observed to increase in expression during carbon deprivation (Miura et al., 2004; Yamano et al., 2008). CAH2 (X54488) was also selected as a control gene reported as displaying a moderate decrease in expression in response to carbon deprivation. CIA5/CCM1 (AF317732) was used as a positive control as it shows constitutive expression during carbon deprivation. Fluorescently labeled primer pairs were designed for each of the aforementioned genes. Quantitative PCR analysis was performed using a 7500 Real-Time PCR System by measuring the threshold cycle (Ct) of each gene. Using the Ct values of CIA5/CCM1 for each RNA sample as a baseline control, the change in Ct for each gene could be used to calculate the fold change response of each gene throughout the time course (see Supplemental Figure 1 online).
RNA Sequencing, Mapping, and the Analyses of Gene Expression
From the qRT-PCR data, it was determined that four time points should be analyzed by RNA-Seq. A 15-min ASVLCO2 sample was omitted from RNA-Seq as qRT-PCR analysis of this sample showed limited induction of the aforementioned genes. C. reinhardtii equilibrated at ∼5% CO2 was used as the 0 time control, and three time points ASVLCO2 (30, 60, and 180 min) were also analyzed. To provide for biological replicates, RNA samples from two individual bioreactor runs were analyzed. In total, eight RNA samples were submitted for RNA-Seq. Prior to submission, RNA samples were treated with DNase and resuspended in 8.3 mM Tris-HCl and 4.2 mM EDTA. RNA-Seq was performed at the JGI using an Illumina Genome Analyzer II.
Sequencing reads were mapped to the C. reinhardtii version 4 genome (Department of Energy JGI) as well as to the processed Augustus5 (Stanke and Waack, 2003) exon structure predictions using the tophat and cufflinks software (Trapnell et al., 2009). No more than two mismatches per sequencing read were allowed. Analyses of differential expression including FDR calculations were performed using three independent Bioconductor packages: edgeR (Robinson et al., 2010), DESeq (Anders and Huber, 2010), and baySeq (Hardcastle and Kelly, 2010).
Time series analysis of the transcriptional response was performed by k-means cluster analysis (MacQueen, 1967). This method partitions fold change patterns into k clusters where each fold change time series belongs to the cluster with the nearest mean. The clusters are iteratively refined. Such analyses have been used to identify temporal expression patterns of a large numbers of genes.
Gene Ontology Analyses
Complex functional patterns of the differentially regulated genes emerge at the level of photosynthetic categories, low CO2–regulated genes, and plant and diatom genes that have no close relatives in other kingdoms or in prokaryotes other than cyanobacteria (Karpowicz et al., 2011). We extended these analyses to GO (Ashburner et al., 2000), a system for the hierarchical annotation of homologous gene and protein sequences in multiple organisms using a common, controlled vocabulary. GO allows the practical, high-throughput interpretation of experiments including RNA-Seq. To avoid the subjectivity inherent in the ad hoc interpretations for less than obvious patterns, a rigorous method was employed to assess the statistical significance of expression patterns, called GSEA (Subramanian et al., 2005). Briefly, GSEA ranks genes by fold changes and calculates enrichment scores for each set. Then, primarily upregulated gene sets are assigned high positive enrichment scores and primarily downregulated sets are assigned low negative scores. For the statistical significance of these enrichment scores, FDR (Benjamini and Hochberg, 1995) is calculated.
De Novo Motif Discovery of Putative Transcription Factor Binding Sites
Our complex strategy for the discovery and limited confirmation of the transcriptional regulatory network was described earlier (Ladunga, 2010). Even in the almost complete absence of chromatin immunoprecipitation, protein binding array, or protein–protein interaction observations for algae, an array of motif discovery algorithms for promoter sequence analysis, each complementing the others, knockout mutants of transcription factors, and RNA-Seq data allowed us to better understand the CCM regulatory network. A key tool is the MEME package (Bailey et al., 2009) for the identification of statistically overrepresented variable sequence motifs. We searched all promoter regions for all motifs represented as positional weight matrices in the commercial version of the TRANSFAC Database (Matys et al., 2006) using its advanced search tool. Conversely, all identified motif were queried against the TRANSFAC motifs.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers (sequences are from C. reinhardtii unless otherwise noted): AAT1, XM_001698466; Saccharomyces cerevisiae Abf1, M29067; Argonaut1, XM_001694788; CAH1, D90206; CAH2, X54488; CAH3, U40871; CAH4, XM_001695951; CAH5, XM_001700718; CAH6, AY463239; CAH7, EU045569; CAH8, EU045570; CAH9, XM_001700857; CAH10, XM_001703185; CAH11, AY538680; CAH12, AY463241; CCP1/LCIE, XM_001692145; CCP2/LCID, XM_001692236; CDJ3, XM_001700205; CIA5/CCM1, AF317732; CLR18, XM_001692143; Dicer-like1, EU707797; ELI4/LCI16, XM_001694932; FAP292, XM_001703500; GGPS/LCI14, XM_001694932; GUD1, XM_001695571; HCF136, XM_001696142; HLA3, XM_001699988; human integrin α2, NM_002203; human interferon γ2, NM_005534; Ku80-like domain, XM_001702527; LCI1, XM_001703335; LCI11, XM_001697911; LCI15, XM_001691879; LCI19, XM_001698768; LCI21, XM_001692906; LCI22, XM_001702321; LCI23, XM_001695392; LCI25, XM_001696602; LCI31, XM_001691740; LCI6, XM_001693972, AB168091; LCI7, XM_001696515; LCIA, AB168092; LCIB, XM_001698292; LCIC, AB168094; LCID, DQ657195; LCIE, DQ649007; LCR1, AB168089; LHCSR1, XM_001696086; LHCSR2, XM_001696012; LHCSR3, XM_001696086; Arabidopsis thaliana Myb11, AF062863; Myb2, XM_001690031; human PARP1, NM_001618; Saccharomyces cerevisiae Rap1, M18068; tcsA, XM_001694379. All RNA-Seq data are available at the National Center for Biotechnology Information Sequencing Read Archive at http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP004215.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. qRT-PCR Support for RNA Sequencing Results.
Supplemental Figure 2. Overlaps among the Lists of Differentially Expressed Genes Reported by the edgeR, DESeq, and baySeq Tools.
Supplemental Figure 3. Genes Showing a Transient Decrease in Response to CO2 Deprivation.
Supplemental Figure 4. Genes Showing a Transient Increase in Response to CO2 Deprivation.
Supplemental Figure 5. Spacing and Expression of HTH Conformation Genes.
Supplemental Figure 6. Comparison of Transcript Levels from Two Closely Linked Head-To-Head Gene Pairs.
Supplemental Figure 7. Overlaps Among Sets of Genes Induced by CO2 Deprivation.
Supplemental Table 1. Widespread DNA Sequence Motifs as Putative Transcription Factor Binding Sites in the Upstream Regions of Genes.
Supplemental Data Set 1. Comparison of Differential Regulation Observations between RNA-Seq (This Study) and Arrays.
Supplemental Data Set 2. The Statistically Significantly Upregulated Genes for 180 versus 0, 60 versus 0, and 30 versus 0 Min ASVLCO2.
Supplemental Data Set 3. The Statistically Significantly Downregulated Genes for 180 versus 0, 60 versus 0, and 30 versus 0 Min ASVLCO2.
Supplemental Data Set 4. Gene Ontology Categories (Ashburner et al., 2000) Significantly Enriched in Upregulated and Downregulated Genes, Respectively.
Supplemental Data Set 5. The Most Highly Correlated HTH Gene Pairs Potentially Regulated by Bidirectional Promoters.
Supplemental Data Set 6. Anticorrelated Expression in HTH Gene Pairs with a Pearson Correlation Coefficient r ≤ −0.8.
Supplemental Data Set 7. Expression Profiles of the 12 Carbonic Anhydrase Genes.
Supplementary Material
Acknowledgments
We thank Christa Pennachio, Erika Lindquist, and Feng Chen at JGI for sequencing, Paul Blum and Kemp Horken for help with the biofermenter, Martin H. Spalding, Wei Fang, Sabeeha S. Merchant, and Matteo Pellegrini for the access to their results and for reviewing the article, and Jean-Jack M. Riethoven and Kartik Vedalaveni for assistance. D.P.W. acknowledges support from the Department of Energy JGI (Award JGI 2010 CSP-146), the National Science Foundation (Grants MCB-0952533 and EPSCoR-1004094), and Consortium for Algal Biofuels Commercialization (Department of Energy Award DE-EE0003373). I.L. and D.P.W. received support from an interdisciplinary grant from the University of Nebraska–Lincoln. D.C. is supported by a grant to Sabeeha S. Merchant from the Air Force Office of Science Research (FA 9550-10-1-0095). The views presented in this work are not endorsed by the sponsors.
AUTHOR CONTRIBUTIONS
D.P.W., I.L., and A.J.B. designed the research. I.L., A.J.B., and D.S.G. performed the research. D.P.W., I.L., D.S.G., A.J.B., M.F.C., and D.C. analyzed the data. D.C. analyzed third-party data. I.L., D.P.W., and A.J.B. wrote the article.
Glossary
- CCM
CO2-concentrating mechanism
- Ci
inorganic carbon
- RNA-Seq
RNA sequencing
- ASVLCO2
after a shift to very low CO2 conditions
- JGI
Joint Genome Institute
- FDR
false discovery rate
- qRT-PCR
quantitative RT-PCR
- GO
Gene Ontology
- GSEA
gene set enrichment analyses
- MEME
Multiple Expectation Maximization for Motif Elicitation
- Ct
threshold cycle
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., et al. The Gene Ontology Consortium (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue): W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple hypothesis testing. J. R. Statist. Soc. B 57: 289–300 [Google Scholar]
- Casas-Mollano J.A., Rohr J., Kim E.J., Balassa E., van Dijk K., Cerutti H. (2008). Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics 179: 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M., Casero D., Karpowicz S.J., Kropat J., Vieler A., Hsieh S.I., Yan W., Cokus S., Loo J.A., Benning C., Pellegrini M., Merchant S.S. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Ma X., Msanne J., Repas T. (2011). RNA-mediated silencing in Algae: Biological roles and tools for analysis of gene function. Eukaryot. Cell 10: 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y., Williams S.A., Ballotti R., Nugent D.J., Kunicki T.J. (2010). Enhanced binding of poly(ADP-ribose)polymerase-1 and Ku80/70 to the ITGA2 promoter via an extended cytosine-adenosine repeat. PLoS ONE 5: e8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J.C., Lottaz C., Borodina T., Himmelbauer H. (2008). Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 36: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D., Miller A.R., Horken K.M., Weeks D.P., Spalding M.H. (2009). Knockdown of limiting-CO2-induced gene HLA3 decreases HCO3- transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 106: 5990–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., et al. ENCODE Project Consortium (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermilova E.V., Zalutskaya Z.M., Gromov B.V., Häder D.P., Purton S. (2000). Isolation and characterisation of chemotactic mutants of Chlamydomonas reinhardtii obtained by insertional mutagenesis. Protist 151: 127–137 [DOI] [PubMed] [Google Scholar]
- Fang W., Si Y., Casero D., Merchant S.S., Pellegrini M., Ladunga I., Liu P., Spalding M.H. (2012). Genome-wide changes in Chlamydomonas gene expression regulated by carbon dioxide and the CO2 concentrating mechanism regulator CIA5/CCM1. Plant Cell 24: 1876–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Kono H., Takenaka S., Go N., Sarai A. (2007). Sequence-dependent DNA deformability studied using molecular dynamics simulations. Nucleic Acids Res. 35: 6063–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Fukuzawa H., Tachiki A., Miyachi S. (1990). Structure and differential expression of two genes encoding carbonic anhydrase in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87: 9779–9783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi M., Singh G.P., Sandhu K.S., Brahmachari S.K., Brahmachari V. (2007). A whole genome analysis of 5′ regulatory regions of human genes for putative cis-acting modulators of nucleosome positioning. Gene 391: 242–251 [DOI] [PubMed] [Google Scholar]
- González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S.S., Grossman A.R. (2010). RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle T.J., Kelly K.A. (2010). baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook. (San Diego, CA: Academic Press)
- Hui J., Hung L.H., Heiner M., Schreiner S., Neumüller N., Reither G., Haas S.A., Bindereif A. (2005). Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 24: 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N., Redon S., Pfeiffer V., Dees M., Lingner J., Luke B. (2011). Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 12: 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im C.S., Grossman A.R. (2002). Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. Plant J. 30: 301–313 [DOI] [PubMed] [Google Scholar]
- Im C.S., Zhang Z., Shrager J., Chang C.W., Grossman A.R. (2003). Analysis of light and CO(2) regulation in Chlamydomonas reinhardtii using genome-wide approaches. Photosynth. Res. 75: 111–125 [DOI] [PubMed] [Google Scholar]
- Johnson L.S., Eddy S.R., Portugaly E. (2010). Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.D., Lew J.E., Berman J. (1999). Gbp1p, a protein with RNA recognition motifs, binds single-stranded telomeric DNA and changes its binding specificity upon dimerization. Mol. Cell. Biol. 19: 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz S.J., Prochnik S.E., Grossman A.R., Merchant S.S. (2011). The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286: 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J., Sunderhaus S., Nimtz M., Jänsch L., Braun H.P. (2010). Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohinata T., Nishino H., Fukuzawa H. (2008). Significance of zinc in a regulatory protein, CCM1, which regulates the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 49: 273–283 [DOI] [PubMed] [Google Scholar]
- Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D. (2011). A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66: 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K., Yoshioka S., Taniguchi F., Ohyama K., Fukuzawa H. (2003). Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 133: 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadorf A., Link A., Rogers M.F., Thomas J., Reddy A.S., Ben-Hur A. (2010). Genome-wide analysis of alternative splicing in Chlamydomonas reinhardtii. BMC Genomics 11: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladunga I. (2010). An overview of the computational analyses and discovery of transcription factor binding sites. In Computational Biology of Transcription Factor Binding, I. Ladunga, ed (New York: Humana Press), pp. 1–22
- Li B., Ruotti V., Stewart R.M., Thomson J.A., Dewey C.N. (2010). RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26: 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Jiang W.Z., Wright D., Spalding M.H., Weeks D.P., Yang B. (2011a). TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 39: 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Zhao X., Wright D.A., Carpenter S., Spalding M.H., Weeks D.P., Yang B. (2011b). Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39: 6315–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D., Casero D., Cokus S.J., Merchant S.S., Pellegrini M. (2011). Algal Functional Annotation Tool: A web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics 12: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen J.B. (1967). Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, L.M. Le Cam and J. Neyman, eds (Berkeley, CA: University of California Press), pp. 281–297
- Margulies M., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 Erratum. Nature 441: 120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., et al. (2006). TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34(Database issue): D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Yamano T., Yoshioka S., Kohinata T., Inoue Y., Taniguchi F., Asamizu E., Nakamura Y., Tabata S., Yamato K.T., Ohyama K., Fukuzawa H. (2004). Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 135: 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney J.V., Ma Y., Frey W.D., Fusilier K.A., Pham T.T., Simms T.A., DiMario R.J., Yang J., Mukherjee B. (2011). The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: Intracellular location, expression, and physiological roles. Photosynth. Res. 109: 133–149 [DOI] [PubMed] [Google Scholar]
- Moroney J.V., Ynalvez R.A. (2007). Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot. Cell 6: 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Ikenaka K., Nakahira K., Morita N., Mikoshiba K. (1993). An improved retroviral vector for assaying promoter activity. Analysis of promoter interference in pIP211 vector. FEBS Lett. 315: 129–133 [DOI] [PubMed] [Google Scholar]
- Ohnishi N., Mukherjee B., Tsujikawa T., Yanase M., Nakano H., Moroney J.V., Fukuzawa H. (2010). Expression of a low CO2-inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. Plant Cell 22: 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S., Yamada T., Hamajima M., Itoh M., Katayama T., Bork P., Goto S., Kanehisa M. (2008). KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 36(Web Server issue): W423–W426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G., Truong T.B., Ostendorf E., Busch A., Elrad D., Grossman A.R., Hippler M., Niyogi K.K. (2009). An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Plevin M.J., Mills M.M., Ikura M. (2005). The LxxLL motif: A multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 30: 66–69 [DOI] [PubMed] [Google Scholar]
- Pollock S.V., Prout D.L., Godfrey A.C., Lemaire S.D., Moroney J.V. (2004). The Chlamydomonas reinhardtii proteins Ccp1 and Ccp2 are required for long-term growth, but are not necessary for efficient photosynthesis, in a low-CO2 environment. Plant Mol. Biol. 56: 125–132 [DOI] [PubMed] [Google Scholar]
- Pravica V., Perrey C., Stevens A., Lee J.H., Hutchinson I.V. (2000). A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: Absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum. Immunol. 61: 863–866 [DOI] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. (2009). PLAZA: A comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40(Database issue): D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat M., Moroney J.V. (1991). Partial characterization of a new isoenzyme of carbonic anhydrase isolated from Chlamydomonas reinhardtii. J. Biol. Chem. 266: 9719–9723 [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Kropat J., Malasarn D., Grossoehme N.E., Chen X., Giedroc D.P., Merchant S.S. (2010). The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Waack S. (2003). Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (suppl. 2): ii215–225 [DOI] [PubMed] [Google Scholar]
- Stracke R., Favory J.J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R. (2010b). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33: 88–103 [DOI] [PubMed] [Google Scholar]
- Stracke R., Jahns O., Keck M., Tohge T., Niehaus K., Fernie A.R., Weisshaar B. (2010a). Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 188: 985–1000 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M. (2004). An abundance of bidirectional promoters in the human genome. Genome Res. 14: 62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K., Wang Y., Nakamura Y., Spalding M.H. (2001). Insertional mutants of Chlamydomonas reinhardtii that require elevated CO(2) for survival. Plant Physiol. 127: 607–614 [PMC free article] [PubMed] [Google Scholar]
- Villand P., Eriksson M., Samuelsson G. (1997). Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Biochem. J. 327: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Duanmu D., Spalding M.H. (2011). Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: Inorganic carbon transport and CO2 recapture. Photosynth. Res. 109: 115–122 [DOI] [PubMed] [Google Scholar]
- Wang Y., Spalding M.H. (2006). An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103: 10110–10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun Z., Horken K.M., Im C.S., Xiang Y., Grossman A.R., Weeks D.P. (2005). Analyses of CIA5, the master regulator of the carbon-concentrating mechanism in Chlamydomonas reinhardtii, and its control of gene expression. Can. J. Bot. 83: 765–779 [Google Scholar]
- Waterston R.H., Lander E.S., Sulston J.E. (2002). On the sequencing of the human genome. Proc. Natl. Acad. Sci. USA 99: 3712–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T., Fujita A., Fukuzawa H. (2011). Photosynthetic characteristics of a multicellular green alga Volvox carteri in response to external CO2 levels possibly regulated by CCM1/CIA5 ortholog. Photosynth. Res. 109: 151–159 [DOI] [PubMed] [Google Scholar]
- Yamano T., Fukuzawa H. (2009). Carbon-concentrating mechanism in a green alga, Chlamydomonas reinhardtii, revealed by transcriptome analyses. J. Basic Microbiol. 49: 42–51 [DOI] [PubMed] [Google Scholar]
- Yamano T., Miura K., Fukuzawa H. (2008). Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 147: 340–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T., Tsujikawa T., Hatano K., Ozawa S., Takahashi Y., Fukuzawa H. (2010). Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 51: 1453–1468 [DOI] [PubMed] [Google Scholar]
- Yilmaz A., Mejia-Guerra M.K., Kurz K., Liang X., Welch L., Grotewold E. (2011). AGRIS: The Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 39(Database issue): D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ynalvez R.A., Xiao Y., Ward A.S., Cunnusamy K., Moroney J.V. (2008). Identification and characterization of two closely related beta-carbonic anhydrases from Chlamydomonas reinhardtii. Physiol. Plant. 133: 15–26 [DOI] [PubMed] [Google Scholar]
- Yoo J.H., et al. (2005). Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 280: 3697–3706 [DOI] [PubMed] [Google Scholar]
- Yoshioka S., Taniguchi F., Miura K., Inoue T., Yamano T., Fukuzawa H. (2004). The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell 16: 1466–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.