Summary
Ras proteins play a central role in transducing signals that control cell proliferation, differentiation, motility and survival. The location-specific signaling activity of Ras has been previously shown to be regulated by ubiquitination [1]. However, the molecular machinery that controls Ras ubiquitination has not been defined. Here we demonstrate through biochemical and functional analyses that Rabex-5 (also known as RabGEF1) [2, 3] functions as an E3 ligase for Ras. Rabex-5-mediated Ras ubiquitination promotes Ras endosomal localization and leads to the suppression of ERK activation. Moreover, the Ras effector RIN1 [4, 5] is required for Rabex-5-dependent Ras ubiquitination suggesting a feedback mechanism by which Ras activation can be coupled to ubiquitination. These findings define new elements in the regulatory circuitry that link Ras compartmentalization to signaling output.
Results and Discussion
Rabex-5 Catalyzes Ras Ubiquitination
Ras proteins (HRas, NRas and KRas) are small guanine-nucleotide binding proteins that mediate the transduction of signals from diverse extracellular stimuli to intracellular signaling pathways that control fundamental cellular functions including proliferation, differentiation, and survival. It is now well established that Ras proteins can dynamically partition to different cellular membranes. While all Ras isoforms are found at the plasma membrane, HRas and NRas are also present at the Golgi and endosomes whereas KRas can be found in the endoplasmic reticulum and the outer mitochondrial membrane [6, 7]. Significantly, Ras compartmentalization dictates its signaling capacity and leads to distinct biological outcomes. For example, activation of Ras at the plasma membrane is fast and transient whereas activation of Ras in the Golgi is delayed and sustained [8]. Moreover, in T cells, Ras resides in both the plasma membrane and Golgi, however, it is the activation of the Ras-ERK pathway in the Golgi pool that is essential in specifying the functional T cell population [9]. Lastly, mitochondrial KRas induces apoptosis through its association with Bcl-XL [10].
Several mechanisms have been implicated in the control of Ras intracellular trafficking. KRas which resides predominantly in the plasma membrane is transiently redistributed to intracellular membranes by phosphorylation-dependent alterations of electrostatic interactions with the plasma membrane [10]. HRas and NRas undergo a constitutive depalmitoylation/repalmitoylation cycle, which leads to their continuous shuttling between the plasma membrane and the Golgi [11, 12]. We have shown that in mammalian cells HRas and NRas are subject to mono- and di-ubiquitination and that this modification promotes their endosomal association thereby leading to the attenuation of Ras-ERK signaling [1]. Consistent with these findings, it has been recently reported that in Drosophila, maintaining a threshold of Ras ubiquitination is critical to prevent inappropriate Ras-ERK activation in vivo [13].
To gain insights into the mechanisms that regulate Ras ubiquitination, we sought to determine the identity of the E3 ligase that catalyzes this reaction. We focused on Rabex-5, the mammalian orthologue of yeast Vps9p and a guanine nucleotide exchange factor (GEF) for Rab5 [2, 3]. In addition to its GEF activity, Rabex-5 has been shown to possess an E3 ubiquitin ligase activity mediated by an amino-terminal zinc finger (ZnF) domain which belongs to the A20 ZnF family [14–16]. This catalytic activity along with the observation that Rabex-5 binds to Ras [17] prompted us to investigate the role of Rabex-5 in the regulation of Ras ubiquitination.
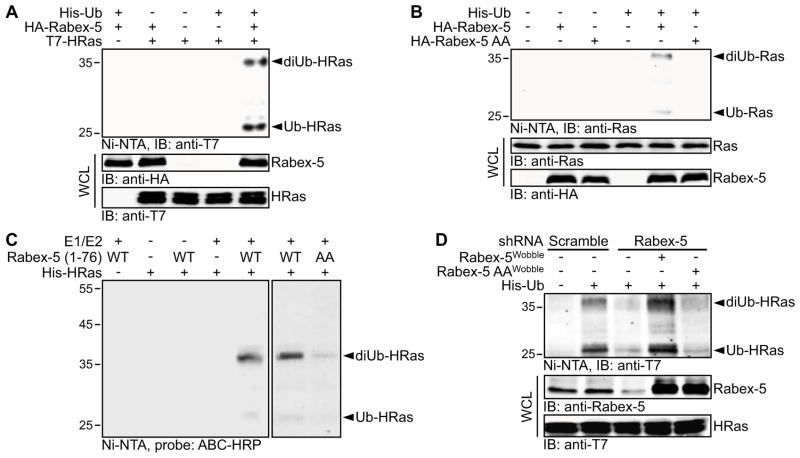
We first examined whether Rabex-5 can promote Ras ubiquitination. As shown, the overexpression of Rabex-5 in HEK 293T cells induced the accumulation of mono- and di-ubiquitinated species of ectopically expressed HRas (Figure 1A and Figure S1A) in agreement with the pattern of Ras ubiquitination reported previously [1]. Similar results were obtained using different cell types [such as COS-1 (Figure S1B) and HeLa (data not shown)] as well as an alternative method to isolate and detect ubiquitinated Ras species (Fig. S1C). Consistent with the ectopic expression data, the mono- and di-ubiquitination of endogenous Ras was also stimulated by the overexpression of Rabex-5 (Figure 1B). To test whether this effect was dependent on the ligase activity of Rabex-5, we used a Rabex-5 mutant (Rabex-5 AA) that is impaired in ligase activity due to Tyr to Ala substitutions in residues 25 and 26 which compromise ubiquitin binding [15]. Rabex-5 AA retained a Ras binding activity (Figure S1D) but was ineffective in stimulating Ras ubiquitination (Figure 1B). Thus, the most plausible interpretation of our findings is that Rabex-5 promotes Ras ubiquitination by functioning as its ubiquitin ligase.
Figure 1. Rabex-5 Ubiquitinates HRas.
(A) HEK 293T cells were co-transfected with expression vectors for His6-tagged ubiquitin (His-Ub), HA-tagged Rabex-5 wild type (WT) and T7-tagged HRas WT as indicated. Ub conjugates were isolated from the transfected cells by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and HRas polypeptides were detected by immunoblotting (IB).
(B) HEK 293T cells were co-transfected with expression vectors for His-Ub and HA-tagged Rabex-5 WT or a ligase-defective Rabex-5 mutant Y25A/Y26A (AA). The ubiquitination of endogenous Ras was analyzed as in (A).
(C) In vitro ubiquitination assay was performed by incubating recombinant GST-tagged Rabex-5 WT or AA (residues 1-76), and recombinant His-tagged HRas in the presence of E1, E2 (UbcH5c) and N-terminal biotinylated Ub. His-tagged HRas was isolated by Ni-NTA affinity chromatography, and biotinylated Ub conjugates were detected by avidin-biotin complex-horseradish peroxidase (ABC-HRP).
(D) COS-1 cells were transfected with constructs encoding a scramble shRNA sequence or shRNA sequences targeting Rabex-5. Following blasticidin S selection, the cells were co-transfected with His-Ub, T7-HRas G12V and the indicated shRNA-resistant Rabex-5 constructs (Wobble). The ubiquitination of HRas G12V was analyzed as in (A).
See also Figure S1. In this and all other figures, levels of endogenous or ectopically expressed proteins were determined by immunoblotting of whole cell lysates (WCL); numbers shown to the left of the blots are molecular masses in kDa; and all results shown are representative of three independent experiments.
To further test this idea, we investigated whether Rabex-5 can directly catalyze the ubiquitination of Ras. To this end, we performed an in vitro ubiquitination assay using recombinant His-HRas and GST-Rabex-5 proteins. It has been reported that when purified as a recombinant protein, full-length Rabex-5 is conformationally unstable [18–20]. Therefore, we employed a recombinant fragment of Rabex-5 (residues 1-76) that encompasses the ubiquitin ligase domain and has been successfully utilized before in in vitro ubiquitination assays [14, 15]. The suitability of this fragment for use in our experiments is further supported by the observations that it contains Ras binding determinants [17] and can interact with Ras directly in vitro (Figure S1E). As shown in Figure 1C, Rabex-5 stimulated the ubiquitination of HRas in vitro whereas Rabex-5 AA was compromised in promoting HRas ubiquitination. Similar results were obtained using a complementary approach (Figure S1F). Notably, Rabex-5 failed to ubiquitinate Rab5, both in vivo (Figure S1G) and in vitro (Figure S1F). Together these findings indicate that HRas is a direct and specific substrate for Rabex-5.
To determine whether Rabex-5 is essential for Ras ubiquitination in vivo, we investigated whether Rabex-5 silencing suppresses Ras ubiquitination. The capacity to detect a significant level of Ras ubiquitination under conditions of endogenous Rabex-5 expression is a prerequisite for this assay. Because the ubiquitination signal of wild-type HRas is barely detectable under basal conditions (Figure 1A and Figure S1B), we resorted to the use of activated HRas (HRas G12V) which, for reasons delineated below (see Figure 4F), displays a robust ubiquitination even in the absence of Rabex-5 overexpression. Transfection of shRNA against Rabex-5 was accompanied by suppression of HRas ubiquitination and this effect could be rescued by the overexpression of shRNA-resistant Rabex-5 WT but not Rabex-5 AA constructs (Figure 1D). We conclude that Ras ubiquitination in vivo is predominantly controlled by Rabex-5.
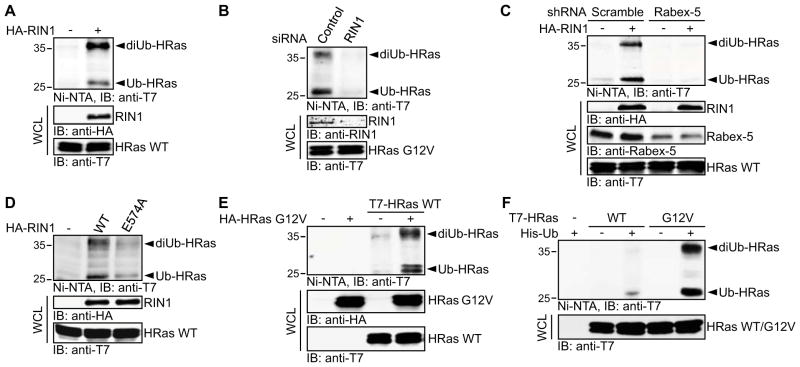
Figure 4. RIN1 Is Required for Rabex-5-mediated HRas Ubiquitination.
(A-F) COS-1 cells were transfected as indicated and HRas ubiquitination was analyzed as in Figure 1A.
(A) Cells were co-transfected with His-Ub and T7-HRas WT in the absence or presence of HA-tagged RIN1.
(B) Cells were first transfected with non-targeting (Control) or RIN1 siRNA duplexes and then with His-Ub and T7-HRas G12V.
(C) Cells were transfected with scramble or Rabex-5 shRNAs. Following blasticidin S selection, the cells were transfected with His-Ub and T7-HRas WT in the absence or presence of HA-RIN1 construct.
(D) Cells were co-transfected with His-Ub, T7-HRas WT and RIN1 WT or the GEF-defective RIN1 mutant E574A.
(E) Cells were transfected with T7-tagged HRas WT and HA-tagged HRas G12V as indicated, along with His-Ub.
(F) Cells were transfected with His-Ub and T7-tagged HRas WT or HRas G12V constructs as indicated.
See also Figure S4.
Rabex-5-mediated Ubiquitination Controls the Endosomal Partitioning of Ras
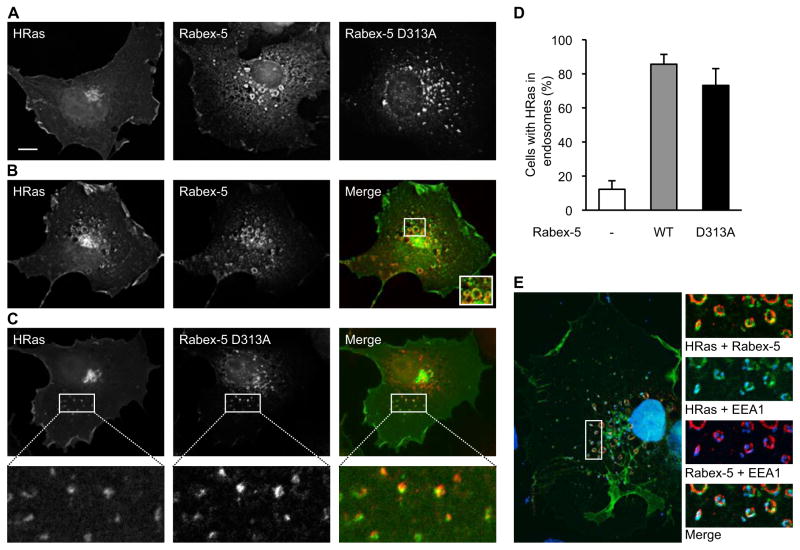
Mono- and di-ubiquitination of plasma membrane proteins serves as a signal for endosomal sorting [21, 22]. Accordingly, we have shown that the ubiquitination of HRas promotes its endosomal association [1]. Since Rabex-5 regulates the ubiquitination of HRas, we predicted that the partitioning of Ras to the endocytic compartment would be augmented in the presence of Rabex-5. To test this prediction, we analyzed the effect of Rabex-5 overexpression on HRas localization by fluorescence microscopy. Under basal conditions, GFP-HRas displayed a characteristic distribution pattern with prominent presence on the plasma membrane and Golgi (Figure 2A). Upon Rabex-5 overexpression, a significant enhancement of endosome-associated GFP-HRas was detected (Figures 2B and 2D). Conversely, depletion of Rabex-5 by shRNA resulted in reduced levels of endosomal GFP-HRas (Figure S2A). The endosomal identity of the GFP-HRas-labeled vesicular structures was ascertained using the early endosomal marker EEA1 (Figure 2E). The majority of the GFP-HRas-labeled endosomes were also Rabex-5 positive (Figures 2B and 2E) consistent with the documented physical interactions between the two proteins [17] and the notion that Ras is a substrate of Rabex-5. Moreover, since Rabex-5 resides mostly in the early endosomes (Figure 2E) [23], it is possible that the ubiquitination of Ras takes place at the endosomal membrane, and, if so, would function as an endosomal retention rather than endosomal targeting signal.
Figure 2. Rabex-5 Increases the Endosomal Pool of HRas.
(A) COS-1 cells were transfected with expression vectors for GFP-HRas, HA-tagged Rabex-5 WT or a GEF-defective Rabex-5 mutant (Rabex-5 D313A) and immunostained with mouse anti-HA.
(B) COS-1 cells were co-transfected with GFP-HRas and HA-Rabex-5 (red) and immunostained as in (A). The inset in the merge panel represents an enlargement of the boxed area.
(C) COS-1 cells were co-transfected with GFP-HRas and HA-Rabex-5 D313A (red) and immunostained as in (A). The lower panels represent enlargements of the boxed areas in the upper panels.
(D) Quantification of Rabex-5-dependent endosomal recruitment of HRas. The percentage of cells with HRas localized to the endocytic compartment is shown. Values represent means ± S.D. from three independent experiments.
(E) COS-1 cells transfected with GFP-HRas and HA-Rabex-5 WT were stained with rabbit anti-HA (red), mouse anti-EEA1 and DAPI (blue).
Images for all panels represent a single 0.2 μm deconvolved Z-section. Scale bar, 10μm. See also Figure S2.
As a GEF for Rab5, Rabex-5 promotes endosomal fusion [2, 18] as reflected by the accumulation of enlarged endosomes in Rabex-5 expressing cells (Figures 2A, 2B, 2E and Figure S2B). To rule out the possibility that the Rabex-5-dependent accumulation of HRas in the endosomes is due to a general stimulatory effect of Rabex-5 on endosomal trafficking, we examined the localization of GFP-HRas in the presence of a Rabex-5 mutant that was rendered defective in GEF activity by mutation in the Vps9 domain [3, 23]. As previously demonstrated [23], this mutant (Rabex-5 D313A) was localized to endosomes albeit of smaller size relative to Rabex-5 WT-containing endosomes (Figure 2A). Significantly, this mutant, which possesses Ras ubiquitination activity (Figure S2C), retained the ability to enhance Ras endosomal association (Figures 2C and 2D). Hence, by acting as a ubiquitin ligase for Ras, Rabex-5 tightly controls the pool size of endosomal Ras.
Rabex-5-mediated Ubiquitination Attenuates Ras-induced ERK Activation
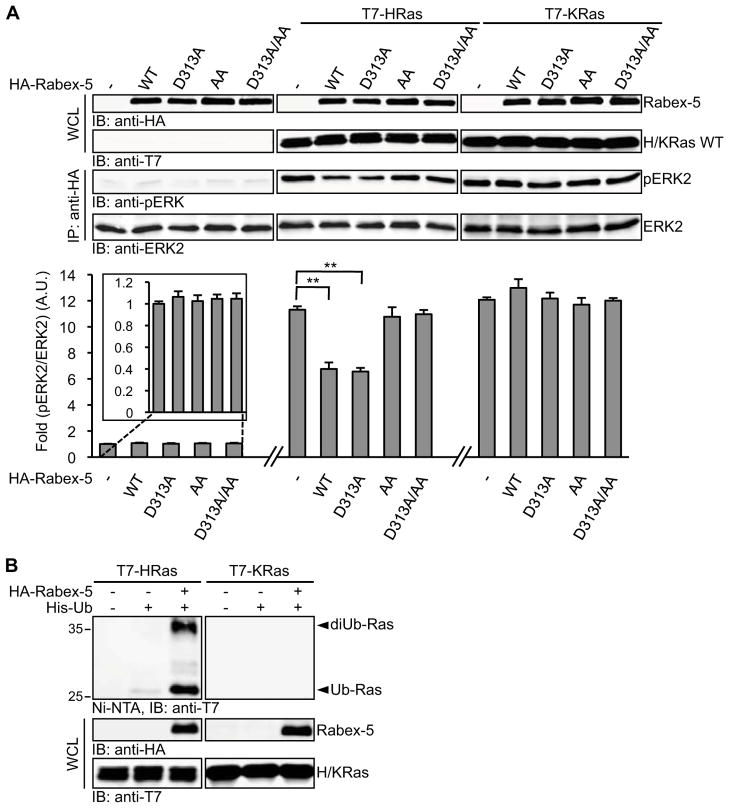
We have previously demonstrated that HRas ubiquitination and the resulting endosomal retention negatively regulates Ras-dependent recruitment of Raf to the plasma membrane and ERK activation [1]. Given the role of Rabex-5 in promoting HRas ubiquitination and endosomal association, we surmised that ERK activation by Ras would be attenuated by overexpressing Rabex-5. As illustrated in Figures 3A and S3A, the overexpression of Rabex-5 WT or Rabex-5 D313A led to the suppression of HRas-induced ERK activation indicating that this effect is independent of the GEF activity of Rabex-5. In contrast, the ligase activity of Rabex-5 is essential for its attenuating effect on HRas-induced ERK activation as indicated by the observation that this effect was not exhibited by Rabex-5 AA or Rabex-5 D313A/AA mutants (Figure 3A and Figure S3A).
Figure 3. Rabex-5 Attenuates Ras-induced ERK Activation.
(A) COS-1 cells were transfected with HA-tagged ERK2 and the indicated Rabex-5 constructs in the absence or presence of T7-tagged HRas or KRas. Transfected ERK2 was isolated by immunoprecipitation (IP), and activation was determined by immunoblotting (IB) with rabbit anti-phospho-ERK antibodies (pERK2) and mouse anti-ERK2 antibodies (ERK2) on the same membranes. Quantification was carried out by two-color Odyssey infrared imaging system (lower panel, and see also Figure S3A). Values shown represent means ± S.D. from three independent experiments. A.U., arbitrary unit. A paired Student’s t-test was performed using the two-tailed distribution. ** p<0.01.
(B) COS-1 cells were transfected with His-Ub, HA-Rabex-5 and T7-tagged HRas or KRas as indicated, and Ras ubiquitination was analyzed as in Figure 1A. See also Figure S3B.
To substantiate the link between Rabex-5-mediated Ras ubiquitination and downstream ERK signaling, we took advantage of a previous finding that Ras ubiquitination displays isoform specificity. In particular, we have shown that KRas is not modified by ubiquitination [1]. Consistent with this finding, the overexpression of Rabex-5 did not induce KRas ubiquitination (Figure 3B). Moreover, KRas-mediated ERK activation was not affected by Rabex-5 overexpression (Figure 3A) confirming that the negative effect exerted by Rabex-5 on HRas signaling is mediated by its HRas ubiquitination activity. It should be noted that NRas, which has been previously shown to be ubiquitinated [1], is also a target for Rabex-5-mediated ubiquitination (Figure S3B) and thus, in principle, could be subjected to a similar mode of regulation.
RIN1 Is Required for Rabex-5-mediated HRas Ubiquitination
Rabex-5 has been shown to bind to the Rab5 effector Rabaptin-5 [2]. This interaction serves to recruit Rabex-5 to early endosomes in a Rab5 dependent manner via the interaction of Rabaptin-5 with Rab5-GTP [18]. In mammalian cells, the levels of Rab5-GTP are positively regulated not only by Rabex-5, but also by the Rab5 GEF proteins of the RIN family, the best characterized of which is RIN1 [24]. Therefore, we sought to determine whether RIN1 participates in the regulation of Ras ubiquitination. RIN1 overexpression stimulated the ubiquitination of HRas (Figure 4A), and conversely, HRas ubiquitination was reduced to basal levels upon siRNA-mediated suppression of RIN1 expression (Figure 4B). Moreover, RIN1-mediated HRas ubiquitination is Rabex-5 dependent as indicated by the failure of RIN1 to enhance HRas ubiquitination upon suppression of Rabex-5 (Figure 4C). Thus, RIN1 appears to be an essential component of the regulatory cascade that controls Ras ubiquitination. This newly identified function for RIN1 might account, at least in part, for its reported activity as a negative regulator of Ras signaling [4].
To further investigate the mechanism by which RIN1 participates in the regulation of HRas ubiquitination we tested the effect of a RIN1 mutant that is defective in Rab5 GEF activity, RIN1 E574A [25], on HRas ubiquitination. As illustrated in Figure 4D, the Rab5 GEF activity of RIN1 was required for its promoting effect on HRas ubiquitination. Hence, it appears that RIN1-mediated activation of Rab5 serves as a prerequisite for Rabex-5-dependent HRas ubiquitination. This idea is supported by the finding that the dependence of HRas ubiquitination on RIN1 could be obviated by the expression of a constitutively active form of Rab5, Rab5 Q79L (Figure S4A). Moreover, Rab5 Q79L stimulated HRas ubiquitination in a Rabex-5-dependent manner (Figure S4B). Together this data is consistent with a model in which RIN1, through its function as a Rab5 GEF, promotes the Rab5-GTP-dependent recruitment of Rabaptin-5/Rabex-5 complex to endosomal sites where HRas ubiquitination takes place.
In addition to its function as a GEF for Rab5, RIN1 is a Ras effector [4, 5]. It directly interacts with GTP-bound Ras through a canonical Ras association domain, and this interaction potentiates RIN1 GEF activity [26]. The role of RIN1 in promoting Ras ubiquitination along with its Ras effector function raises the intriguing possibility that Ras ubiquitination might be subject to feedback regulation through the activation of the RIN1/Rab5/Rabex-5 axis. To test this idea, HRas WT and HRas G12V were differentially tagged with T7 and HA epitopes, respectively, and co-expressed in COS-1 cells. As illustrated in Figure 4E, the level of ubiquitination of HRas WT was significantly enhanced in the presence of HRas G12V indicating that HRas activation can promote HRas ubiquitination. The link between HRas activation and the status of HRas ubiquitination also predicts that HRas G12V would be preferentially ubiquitinated relative to HRas WT. Indeed, at basal conditions the level of HRas G12V ubiquitination is appreciably higher in comparison to HRas WT (Figure 4F). A similar difference has been observed in HEK 293T and HeLa cells (data not shown). HRas WT and HRas G12V displayed similar in vivo interactions with Rabex-5 (Figure S4C) ruling out differential Rabex-5 binding as the mechanism underlying the preferential ubiquitination of HRas G12V. Of note, this result is at variance with our previous demonstration of no significant differences between the level of ubiquitination of HRas WT and HRas G12V in CHO cells [1]. As we have proposed previously, it is conceivable that in these cells the differences might be obscured because of altered relative stoichiometries of one or more of the regulatory components that control Ras ubiquitination.
In summary, our study identifies Ras as a target of the E3 ligase Rabex-5. By promoting Ras ubiquitination and endosomal association, Rabex-5 can function to dynamically shift the partitioning of Ras between different membrane compartments thereby contributing to the spatial control of Ras-dependent cellular responses. The Rab5 GEF activity of Rabex-5 has been also implicated in the control of receptor-mediated Ras signaling through the modulation of receptor endocytosis [23]. Hence, the dual function of Rabex-5 as an E3 ligase for Ras and as a GEF for Rab5 provides a molecular platform for the integration of endocytic trafficking and signaling.
Experimental Procedures
Human Rabex-5 and RIN1 cDNAs were cloned from MIA PaCa-2 and HeLa cells respectively. COS-1 and HEK 293T cells were transfected using Fugene 6 Reagent (Roche Applied Science) and Lipofectamine 2000 (Invitrogen), respectively. After 1 day, the transfected cells were harvested and in vivo Ras ubiquitination was analyzed by Ni-NTA affinity chromatography as previously described [1]. In vitro ubiquitination assays were carried out by incubating the following components at 30°C for 2 h: biotin-N-terminal ubiquitin, ubiquitin activating enzyme (UBE1), E2 (UbcH5C), human recombinant GST-Rabex-5 (1-76) and His-HRas. Following purification by Ni-NTA affinity chromatography, the biotinylated ubiquitin conjugates were then probed with ABC-HRP (Vector Laboratories, Inc.) and detected using ECL (Pierce). Rabex-5 was knocked down by transfecting COS-1 cells with DNA-directed shRNA using pcDNA/SUPER vector and selecting with blasticidin S. RIN1 was knocked down by transfecting COS-1 cells with a pool of siRNA duplexes (Dharmacon, Inc.). For immunofluorescence analyses, COS-1 cells were transfected, fixed, and stained as previously described [1]. ERK activation assay was performed by immunoprecipitation of HA-ERK2, followed by immunoblot analysis using phospho-ERK antibody as previously described [1]. A detailed description of constructs, antibodies and experimental methods is reported in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Rabex-5 functions as an E3 ubiquitin ligase for HRas and NRas, but not KRas.
Rabex-5-mediated ubiquitination controls the endosomal partitioning of Ras.
Rabex-5-mediated ubiquitination attenuates Ras-induced ERK activation.
The Ras effector RIN1 is required for Rabex-5-dependent Ras ubiquitination.
Acknowledgments
We thank Dr. Michele Pagano for advice and reagents and members of our laboratory for helpful suggestions and discussions. This work was supported by a research grant from National Institutes of Health to D.B.-S. (CA55360).
Footnotes
Supplemental Data include four figures, Supplemental Experimental Procedures, and Supplemental References and can be found with this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21:679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 3.Delprato A, Merithew E, Lambright DG. Structure, exchange determinants, and family-wide Rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 2004;118:607–617. doi: 10.1016/j.cell.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Han L, Colicelli J. A human protein selected for interference with Ras function interacts directly with Ras and competes with Raf1. Mol Cell Biol. 1995;15:1318–1323. doi: 10.1128/mcb.15.3.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Natl Acad Sci U S A. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 7.Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu Rev Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- 8.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 9.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 10.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan H, Chin ML, Horvath EA, Kane EA, Pfleger CM. Impairment of ubiquitylation by mutation in Drosophila E1 promotes both cell-autonomous and non-cell-autonomous Ras-ERK activation in vivo. J Cell Sci. 2009;122:1461–1470. doi: 10.1242/jcs.042267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattera R, Tsai YC, Weissman AM, Bonifacino JS. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J Biol Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Tam SY, Tsai M, Snouwaert JN, Kalesnikoff J, Scherrer D, Nakae S, Chatterjea D, Bouley DM, Galli SJ. RabGEF1 is a negative regulator of mast cell activation and skin inflammation. Nat Immunol. 2004;5:844–852. doi: 10.1038/ni1093. [DOI] [PubMed] [Google Scholar]
- 18.Lippé R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esters H, Alexandrov K, Iakovenko A, Ivanova T, Thoma N, Rybin V, Zerial M, Scheidig AJ, Goody RS. Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol. 2001;310:141–156. doi: 10.1006/jmbi.2001.4735. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H, Zhu G, Liu J, Liang Z, Zhang XC, Li G. Rabaptin-5-independent membrane targeting and Rab5 activation by Rabex-5 in the cell. Mol Biol Cell. 2007;18:4119–4128. doi: 10.1091/mbc.E07-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 23.Kalesnikoff J, Rios EJ, Chen CC, Alejandro Barbieri M, Tsai M, Tam SY, Galli SJ. Roles of RabGEF1/Rabex-5 domains in regulating FcεRI surface expression and FcεRI-dependent responses in mast cells. Blood. 2007;109:5308–5317. doi: 10.1182/blood-2007-01-067363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Hu H, Milstein M, Bliss JM, Thai M, Malhotra G, Huynh LC, Colicelli J. Integration of transforming growth factor β and RAS signaling silences a RAB5 guanine nucleotide exchange factor and enhances growth factor-directed cell migration. Mol Cell Biol. 2008;28:1573–1583. doi: 10.1128/MCB.01087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.