Abstract
In recent years, glycopeptide purification by hydrazide chemistry has become popular in structural studies of glycoconjugates; however, applications of this method have been almost completely restricted to analysis of the N-glycoproteome. Here we report a novel method for analyzing O-glycosylations unique to collagen, which are attached to hydroxylysine and include galactosyl-hydroxylysine and glucosyl-galactosyl-hydroxylysine. We established a hydrazide chemistry-based glycopeptide purification method using (1) galactose oxidase to introduce an aldehyde into glycopeptides and (2) formic acid with heating to elute the bound glycopeptides by cleaving the hydrazone bond. This method allows not only identification of O-glycosylation sites in collagen but also concurrent discrimination of two types of carbohydrate substitutions. In bovine type I and type II collagens, galactosyl-hydroxylysine /glucosyl-galactosyl-hydroxylysine -containing peptides were specifically detected on subsequent comprehensive liquid chromatography (LC)/MS analysis, and many O-glycosylation sites, including unreported ones, were identified. The position of glycosylated hydroxylysine, which is determined by our unambiguous and simple method, could provide insight into the physiological role of the modifications.
Galactosyl-hydroxylysine (GHL)1 and glucosyl-galactosyl-hydroxylysine (GGHL) are O-glycosylations unique to collagen (1). They are also found in several other proteins having a collagen-like sequence, such as the C1q complement protein (2). A monosaccharide or disaccharide is attached to the hydroxylysine residue lying in the Y position of repeating collagenous Gly-X-Y triplets within a triple helix. Specific enzymes add the carbohydrates to hydroxylysine before triple helix formation in the endoplasmic reticulum (3, 4). The carbohydrate content varies with collagen type, tissue, and physiological conditions. There are few glycosylated hydroxylysines in fibrillar-forming collagens. For example, type I collagen alpha 1 chain has approximately one residue per 1000 amino acid residues, the alpha 2 chain has approximately two residues, and type II collagen alpha 1 chain has ∼10 residues (5). In contrast, most lysine residues are glycosylated in network-forming collagens such as type IV collagen. Although disorder-related alterations, such as overglycosylation in osteogenesis imperfecta (6) and spondyloepiphyseal dysplasia (7), have been reported, the biological significance of the carbohydrates remains unclear. The position of glycosylated hydroxylysine in a primary amino acid sequence could provide insight into the physiological role of the modifications.
In the 1970s, extensive structural studies revealed nearly all the primary amino acid sequences of major collagens by automated Edman degradation on a protein sequencer, but a few sites have remained uncertain because of some modifications (8). When the collagen peptides, which are digested by cyanogen bromide (CNBr) or proteases such as trypsin, are sequenced from the N-terminal on the protein sequencer, the cycles of hydroxylysine glycosides appear as “blanks.” Most likely because of their hydrophilicity, glycosylated hydroxylysine derivatives are not recovered into nonpolar solvents (n-butyl chloride and ethyl acetate), which results in no peaks on subsequent reverse-phase chromatography (8). Additional analyses were required to confirm their existence and distinguish between the two types of carbohydrate substitutions. For example, amino acid and carbohydrate analyses of the sequenced peptides were needed; therefore, the position of hydroxylysine glycosides has only been predicted by combining data from several experiments. Later, in the 1990s, Gooley et al. improved the automated Edman degradation methodology used for identification of the sites of N- and O-glycosylation by using polar solvents, such as trifluoroacetic acid and methanol, for transferring the released amino acids (9, 10). However, it is still difficult to comprehensively determine the O-glycosylation sites of collagen with the protein sequencer, and time-consuming separation procedures of each peptide are required before the analysis.
The recent development and high sensitivity of mass spectrometers are invaluable in various glycan studies; however, ionization suppression by co-existing nonglycosylated peptides, and the low ionization efficiency of glycopeptides hamper exhaustive MS glycopeptide analysis without purification/enrichment procedures. To this end, Zhang et al. have recently developed a new method for purification of N-linked glycopeptides using hydrazide chemistry to identify glycosylation sites (11, 12). In their method, glycopeptides are oxidized by sodium periodate to generate aldehydes and then captured on hydrazide resin by forming hydrazone bonds. After removing the nonbinding peptide, the peptide released by PNGase F is subjected to liquid chromatography (LC)/MS analysis. This strategy is becoming popular and is used for analysis of various N-glycoproteome samples (13–16). More recently, applications have been extended to include O-glycosylation analysis. For example, the structure of the carbohydrate and its protein attachment site of N- and O-linked sialylated glycopeptides have been identified after cleavage of the glycosidic bond at the terminal sialic acid by heating with formic acid (17). Another study has analyzed sialylated glycopeptides with an intact glycan by cleaving the hydrazone bond using hydrochloric acid under cooling conditions (18). O-GlcNAc-modified glycopeptides were released from the resin using hydroxylamine thus converting the sugar to an oxime derivative (19). Although various approaches have been adopted, most applications of hydrazide chemistry have been restricted to analysis of the N-glycoproteome.
Purification methods for collagen O-glycosylations, such as affinity purification using lectin or specific antibodies, have not been reported. In this study, we present a simple and definitive method for purification of peptides containing GHL and GGHL by hydrazide chemistry and subsequent LC/MS analysis. Galactose oxidase was used to introduce an aldehyde instead of sodium periodate oxidation, which is known to destroy O-linked collagen carbohydrates and has been used to produce deglycosylated hydroxylysine (20, 21). Although, in general, the enzyme preferentially oxidizes nonreducing terminal galactose such as GHL, GGHL can also be oxidized despite steric hindrance (1, 22). Galactose oxidase was immobilized on Sepharose 4B to reuse the enzyme and reduce its contaminated proteolytic activity for stabilization of the enzyme, leading to an increase in the total enzyme activity (22). In addition, in the oxidation reaction, we exploited the coordinated addition of catalase and horseradish peroxidase (HRP) to enhance galactose oxidase activity, as has been reported recently (23, 24). Hydrazide resin was heated with 0.1% formic acid to elute binding glycopeptides by cleaving the hydrazone bond; consequently, the eluted peptide possessed the carbohydrate chain, and we could discriminate the two types of carbohydrate substitutions concurrently with determining the modification site. We first optimized this glycopeptide purification method, which is referred to as the “hydrazide method” in this article, based on hydrazide chemistry using purified GHL/GGHL, and practical analysis was performed in bovine type I and type II collagens.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Galactose oxidase, catalase, HRP, tosyl phenylalanyl chloromethyl ketone-treated trypsin, and trypsin-chymotrypsin inhibitor were purchased from Sigma-Aldrich Co. (St. Louis, MO). Sodium borodeuteride and 13C6 15N2-l-lysine were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA), and CNBr-activated Sepharose 4B was purchased from GE Healthcare (Pittsburgh, PA). Affi-gel Hz and Mini Bio-Spin chromatography columns were purchased from Bio-Rad Laboratories, Inc. (Hercules, CA). All other chemicals were purchased from Sigma-Aldrich. Pepsin-solubilized type I collagen was prepared from bovine skin, and pepsin-solubilized type II collagen was prepared from bovine articular cartilage, as reported previously (25, 26). GHL and GGHL were purified from natural sponge (8), and galactose oxidase was immobilized on CNBr-activated Sepharose 4B (22). In brief, 150 U of galactose oxidase was added to 0.4 g of Sepharose 4B and rotated overnight at 4 °C. Unreacted sites were blocked using 1 m glycine at room temperature for 2 h.
Oxidation and Purification of GHL/GGHL Standards by the Hydrazide Method
Both GHL (100 nmol) and GGHL (100 nmol) were dissolved in reaction buffer (100 mm sodium phosphate and 150 mm NaCl, pH 7.2). The samples were incubated with immobilized galactose oxidase (30 U), catalase (115 U), and HRP (1.5 U) in Bio-Spin chromatography columns with end-over-end rotation at 37 °C for 24 h. The samples were collected by centrifugation, and the pH was adjusted to 4–5 with hydrochloric acid. The oxidized samples were coupled to hydrazide resin (200 μl), which was washed with coupling buffer (100 mm sodium acetate and 150 mm NaCl, pH 4.8) before use, in Bio-Spin chromatography columns with end-over-end rotation at 37 °C for 6 h. After the capture reaction, the hydrazide resin was washed with the coupling buffer, 1.5 m NaCl, 100% methanol, and distilled water. The coupling compounds were eluted with 0.1% formic acid by heating at 80 °C for 30 min, and the resin was then washed once with hot 0.1% formic acid to collect the remaining glycopeptides. The samples [preoxidation, postoxidation (oxidant), those unbound to hydrazide resin (flow-through), and postelution (eluant)] were then reduced with 1 mm sodium borodeuteride at room temperature for 1 h in alkaline conditions adjusted by triethylamine. The reduced samples were acidified with formic acid, and 13C6 15N2-lysine was added as an internal standard. The samples were subjected to multiple reaction monitoring (MRM) analysis to calculate the oxidation efficiency and recovery rate of the GHL/GGHL standards.
MRM Analysis of GHL/GGHL and Their Oxidation Products
Analysis was performed on a hybrid triple quadrupole/linear ion trap 3200 QTRAP mass spectrometer (AB Sciex, Foster City, CA) equipped with an electrospray ionization (ESI) source. The instrument was coupled to an Agilent 1200 Series HPLC system (Agilent Technologies, Inc., Palo Alto, CA). The samples were loaded onto a ZIC-HILIC column (5 μm particle size, L × I.D. 150 mm × 2.1 mm; Merck SeQuant, Umea, Sweden) at a flow rate of 200 μl/min and separated by a binary gradient as follows: 90% solvent B (100% acetonitrile) for 5 min, linear gradient of 10–60% solvent A (0.1% formic acid in water) for 5 min, linear gradient of 60–90% solvent A for 15 min, and 90% solvent B for 5 min. The settings for MRM analysis were determined by the compound optimization function provided in Analyst 1.5.1 (AB Sciex). Capillary voltage was 4.5 kV, declustering potential was 25 V, heater gas temperature was 700 °C, curtain gas was 15 psi, nebulizer gas was 60 psi, heater gas was 80 psi, and collision energy was 19 V (GHL and its derivatives), 27 V (GGHL and its derivatives), and 21 V (13C6 15N2-lysine). The following MRM transitions were monitored: GHL (m/z 325.1→163.2), deuterium-labeled GHL (m/z 327.1→163.2), oxidized GHL (m/z 323.1→163.2), peroxidized GHL (m/z 339.1→163.2), GGHL (m/z 487.2→163.3), deuterium-labeled GGHL (m/z 489.2→163.3), oxidized GGHL (m/z 485.2→163.3), peroxidized GGHL (m/z 501.2→163.3), and 13C6 15N2-lysine (m/z 155.1→90.1).
Purification of GHL/GGHL Peptides of Bovine Collagen by the Hydrazide Method
Bovine type I or type II collagen (1 mg) in the reaction buffer was denatured by heating at 60 °C for 30 min and digested by trypsin (50 μg) at 37 °C for 16 h. After heating at 100 °C for 5 min, a portion of each sample was taken and diluted to 0.1 mg/ml with 0.1% formic acid for control samples, which were analyzed by LC/MS with three 10 μl injections for peptide identification without the hydrazide method. The trypsin-chymotrypsin inhibitor (100 μg) was added to the remaining samples. Subsequently, the hydrazide method was used in a manner analogous to that for the GHL/GGHL standards. After elution from the hydrazide resin, the glycopeptides were reduced to their original form using 1 mm sodium borohydride at room temperature for 1 h in alkaline conditions. The glycopeptide solutions were acidified with formic acid and subjected to LC-MS/MS analysis.
LC-Tandem MS (MS/MS) Analysis
Samples prepared by the glycopeptide purification procedure were analyzed by LC-electrospray ionization (ESI)-MS/MS. The analysis was performed on a 3200 QTRAP mass spectrometer coupled to an Agilent 1200 Series HPLC system. The sample solutions were loaded onto an Ascentis Express C18 HPLC Column (2.7 μm particle size, L × I.D. 150 mm × 2.1 mm; Supelco, Bellefonte, PA) at a flow rate of 200 μl/min and separated by a binary gradient as follows: 98% solvent A (0.1% formic acid in water) for 5 min, linear gradient of 2–50% solvent B (100% acetonitrile) for 15 min, 90% solvent B for 5 min, and 98% solvent A for 5 min. The eluting peptides were analyzed by the information-dependent acquisition (IDA) method that was operated by selecting the two most intense precursor ions of the prior survey MS scan and then subjecting the precursor ions to MS/MS fragmentation. The collision energy was automatically determined based on the mass and charge state of the precursor ions using rolling collision energy. Capillary voltage was 5.5 kV, declustering potential was 30–50 V, heater gas temperature was 600 °C, curtain gas was 40 psi, nebulizer gas was 50 psi, and heater gas was 80 psi. MS scan and MS/MS acquisition were operated over the m/z range of 400–1300 and 100–1700, respectively.
Database Search of MS/MS Spectra
ProteinPilot software 4.0 (AB Sciex) with the Paragon™ algorithm was used for peptide identification (27). Search parameters included digestion by trypsin, biological modifications ID focus, and 95% protein confidence threshold. Default parameters including number of missed cleavages permitted and mass tolerance for precursor ions and fragment ions were adopted by the software. Two residues of galactosyl hydroxylation and glucosyl galactosyl hydroxylation of lysine (+178 and +340, respectively) were added to the search criteria of post-translational modifications. The probabilities of hydroxylation of proline and lysine were set higher than those of the defaults for collagen analysis. The acquired MS/MS spectra were searched against the UniProtKB/Swiss-Prot database (release 2011_08, on July 2011) for Bos taurus species (5857 protein entries). We defined the confidence threshold of the identified peptides to be 90%.
Sequence Confirmation of GHL/GGHL Peptides
The glycopeptide-containing fraction was collected, and the molecular weight distribution of the fraction was surveyed by MALDI-TOF/MS analysis performed on a Voyager Linear DE apparatus (AB Sciex). The remainder of the fraction was subjected to N-terminal amino acid sequence analysis on a Procise 492 protein sequencer (Applied Biosystems, Invitrogen Co., Carlsbad, CA) in pulsed-liquid mode.
RESULTS
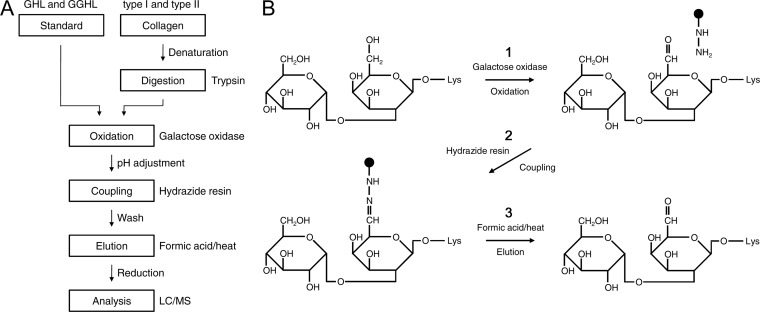
The workflow of the hydrazide method for collagen O-glycosylations is shown in Fig. 1A, and the details of the chemical reactions of GGHL are described in Fig. 1B. GHL/GGHL standards or trypsin-digested glycopeptides of collagen were oxidized by galactose oxidase to generate an aldehyde group in galactose (Fig. 1B-1). After pH adjustment of the solutions to a weak acid for purposes of the coupling reaction, the oxidized molecules were coupled to hydrazide resin by forming a hydrazone bond (Fig. 1B-2). Unbound and nonspecifically bound compounds were removed by extensively washing the resin, and the captured compounds were then released using formic acid and heat (Fig. 1B-3). Finally, the eluted samples were reduced and subjected to LC/MS analysis.
Fig. 1.
Schematic diagram of the hydrazide method for collagen O-glycosylations. A, Purification strategy for standard of hydroxylysine glycosides and collagen. Purified GHL/GGHL and collagen samples, which were denatured and then digested by trypsin, were oxidized by galactose oxidase. The oxidized samples were coupled to hydrazide resin under adjusted weak acidic conditions, and unbound/nonspecifically bound compounds were removed by washing. The bound compounds were released from hydrazide resin by heating with formic acid and were analyzed by LC/MS after reduction treatment. B, Details of the chemical reactions of GGHL using the hydrazide method. The hydroxyl group at the C6 position of galactose was oxidized to the aldehyde group by galactose oxidase (1). The aldehyde group was coupled to hydrazide resin by forming a hydrazone bond (2). The captured GGHL was eluted by heated 0.1% formic acid (3).
Oxidation Efficiency and Recovery Rate of GHL/GGHL Standards
Initially, we optimized the oxidation and elution in the hydrazide method for collagen using purified GHL/GGHL. To enhance the reactivity of galactose oxidase, we immobilized galactose oxidase on agarose beads (22), which increased the generation of oxidized GHL and GGHL ∼threefold and eightfold, respectively (supplemental Fig. S1), as well as added catalase and HRP to further enhance the reactivity (23, 24). The use of a large amount of galactose oxidase enhanced the oxidation efficiency of GGHL, but it resulted in a decrease in oxidized GHL because of the increased peroxidized side product, which was not detected in the GGHL oxidation (supplemental Fig. S2). Hence, the amount of galactose oxidase was determined to be 30 U, which was considered adequate for the concomitant oxidation of both GHL and GGHL. In addition, we determined the conditions for elution to be 0.1% formic acid (pH 2.8) at 80 °C for 30 min, and the elution time seemed to almost reach a plateau for the release of captured GHL and GGHL (supplemental Fig. S3). GHL/GGHL standards were stable during acid/heat treatment under these conditions (GHL, 99.8 ± 6.8%; GGHL, 98.7 ± 6.5%).
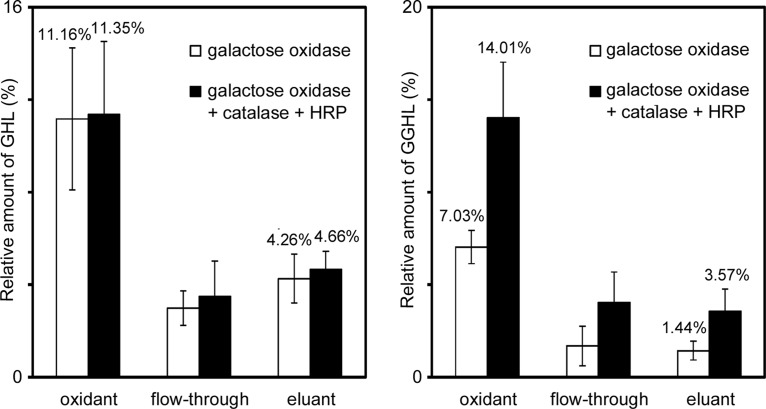
Fig. 2 shows the efficiencies of oxidation and purification of GHL/GGHL in the hydrazide method. The oxidation efficiency of GHL/GGHL was estimated by relative quantification of their oxidants versus preoxidation samples. Similarly, the recovery rate was estimated with relative amounts of eluants. The samples were reduced by sodium borodeuteride before MRM analysis, which permitted a simple comparison of the amounts of original GHL/GGHL with those of their deuterium-labeled oxidation products. The oxidation efficiency of GHL and GGHL was 11.16% and 7.03%, respectively, when oxidized by galactose oxidase only, and the recovery rate of GHL and GGHL was 4.26% and 1.44%, respectively. Substantial amounts of oxidized GHL/GGHL flowed through the resin, but the recovery rate was not improved by increasing the amount of hydrazide resin or the coupling time (data not shown). The particularly low oxidation efficiency and recovery rate of GGHL were presumed to be because of nonreducing terminal glucose that may disturb the interactions of galactose with galactose oxidase and hydrazide resin.
Fig. 2.
Oxidation efficiency and recovery rate of GHL/GGHL standards using the hydrazide method. One sample was oxidized by galactose oxidase only and the other was oxidized by the enzyme with the coordinated addition of catalase and HRP. The oxidized samples were coupled to hydrazide resin and released by heating under acidic conditions following resin washes. The samples (preoxidation, postoxidation (oxidant), those unbound to hydrazide resin (flow-through), and postelution (eluant)) were reduced with sodium borodeuteride. 13C6 15N2-lysine was added as an internal standard, and then the samples were subjected to MRM analysis. The relative amounts of deuterium-labeled oxidation products of GHL/GGHL of each sample were calculated by comparing them to the original GHL/GGHL of preoxidation. The data represent the mean ± standard deviation of five separate experiments.
By adding catalase and HRP, the galactose oxidase activity in the GGHL standard was markedly enhanced by ∼twofold, leading to a striking improvement in the total recovery rate; however, it was less effective for GHL because the peroxidized side product of GHL increased ∼threefold in the oxidant by the coordinated addition of catalase and HRP (data not shown). All the side products shifted to flow-through. The total recovery rate of GHL and GGHL oxidized in the presence of catalase and HRP was 4.66% and 3.57%, respectively. Although the recovery rates appeared to be somewhat low, they were sufficient to identify GHL/GGHL peptides by LC/MS, as described below.
Purification and LC/MS Analysis of GHL/GGHL Peptides in Bovine Collagen
The hydrazide method described in the above section was used on bovine type I and type II collagens. The glycopeptides were purified by the same procedure as for GHL/GGHL standards after trypsin digestion of denatured collagen (Fig. 1A).
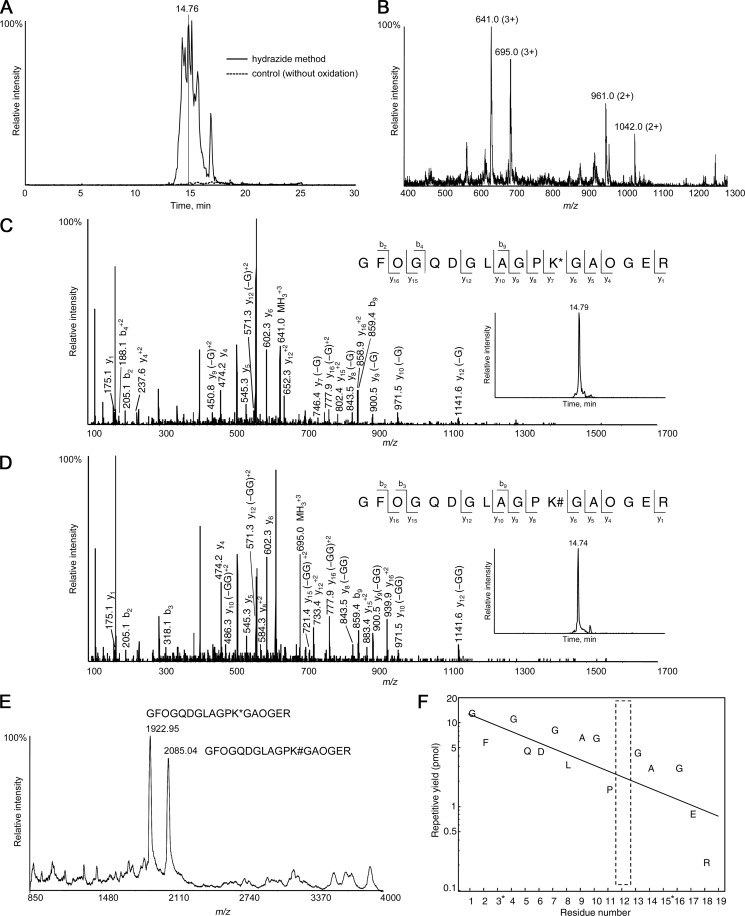
The glycopeptides eluted from the hydrazide resin were analyzed by LC/MS after reducing the galactose to its original form using sodium borohydride. An example of the results of sequence analysis of the GHL/GGHL peptides of type II collagen is shown in Fig. 3. Specific peaks were detected in the total ion current chromatogram (TIC) of LC/MS analysis, whereas no peaks were found in the control nonoxidized sample (Fig. 3A). Purification by the hydrazide method resulted in a decreased number of total peptides, thereby permitting MS/MS acquisitions of most of the glycopeptides (data not shown). Fig. 3B shows the MS spectrum of the survey scan obtained at 14.76 min. The m/z 641.0 ion was subjected to MS/MS analysis (Fig. 3C), which was identified as GFOGQDGLAGPK*GAOGER (charge = 3+, O indicates hydroxyproline and K* indicates GHL). Similarly, the m/z 695.0 ion (Fig. 3D) was identified as the identical peptide with GGHL instead of GHL (GFOGQDGLAGPK#GAOGER; charge = 3+, K# indicates GGHL). The retention time of the GHL peptide on reverse-phase chromatography was somewhat longer than that of the GGHL peptide. A similar tendency was found for all other peptides containing GHL/GGHL. Because collision-induced dissociation preferentially cleaves glycosidic bonds rather than peptide bonds, most of the carbohydrate moieties were lost on MS/MS resulting in the spectra of both the GHL and GGHL peptides closely resembling each other.
Fig. 3.
Identification of GHL/GGHL peptides of type II collagen. A, TIC of type II collagen. B, Survey MS spectrum obtained at 14.76 min in the TIC. The m/z 641.0 (z = 3+) and 961.0 (z = 2+) ions were both derived from the same GHL peptide, and the 695.0 (z = 3+) and 1042.0 (z = 2+) ions were derived from the identical peptide with GGHL instead of GHL. The labeling numbers represent monoisotopic masses. C, MS/MS spectrum of the precursor ions at m/z 641.0 (z = 3+) and D, m/z 695.0 (z = 3+) selected from the survey MS scan at 14.76 min. The m/z 641.0 ion was identified as GFOGQDGLAGPK*GAOGER (O indicates hydroxyproline and K* indicates GHL), and the m/z 695.0 ion was identified as GFOGQDGLAGPK#GAOGER (K# indicates GGHL). -G represents deglycosylated fragment ions (-162) of GHL peptide, and -GG represents deglycosyated fragment ions (-324) of GGHL peptide. The extracted ion chromatograms of the GHL peptide (m/z 640.96–641.29) and GGHL peptide (m/z 694.98–695.31) were also shown. E, MALDI-TOF mass spectrum of the fraction that contained the peaks shown in Fig. 3B. The peaks at m/z 1922.95 and m/z 2085.04 correspond to peptides GFOGQDGLAGPK*GAOGER and GFOGQDGLAGPK#GAOGER, respectively. F, The result of the N-terminal amino acid sequence analysis of the fraction collected from LC/MS analysis at ∼14.76 min. It was determined as GFOGQDGLAGP(X)GAOGER (X indicates blank). *The repetitive yield of hydroxyproline was not calculated because it divided into two peaks and phenylthiohydantoin-hydroxyproline was not available commercially.
Verification of the Accuracy of the Hydrazide Method
We performed sequence analysis by a protein sequencer to verify the accuracy of the identification of GHL/GGHL peptides using the hydrazide method. The fraction that contained the peaks shown in Fig. 3B was collected, and MALDI-TOF/MS analysis revealed that there were two major peaks in agreement with the molecular weight of the GHL/GGHL peptides identified by LC/MS analysis (Fig. 3E). The remainder of the fraction was analyzed by protein sequencing and determined to be GFOGQDGLAGP(X)GAOGER, where X indicates “blank” and is considered to be GHL or GGHL, as expected (Fig. 3F). These results support the reliability of purification and identification of GHL/GGHL peptides by the hydrazide method.
Database Search of Acquired MS/MS Spectra for Peptide Identification
Whole MS/MS spectra obtained by LC/MS were subjected to peptide identification using a database search against the UniProtKB/Swiss-Prot database for Bos taurus species. Because the structures of GHL and GGHL were already identified, the glycosylations of hydroxylysine were added to the search criteria of post-translational modifications. To exclude possible random database hits, we defined the following three criteria for identification of GHL/GGHL peptides: (1) location of GHL/GGHL as being at the Y position of Gly-X-Y triplets, (2) missed cleavage at GHL/GGHL by trypsin most likely because of steric hindrance (28, 29), and (3) high confidence level of more than 90%.
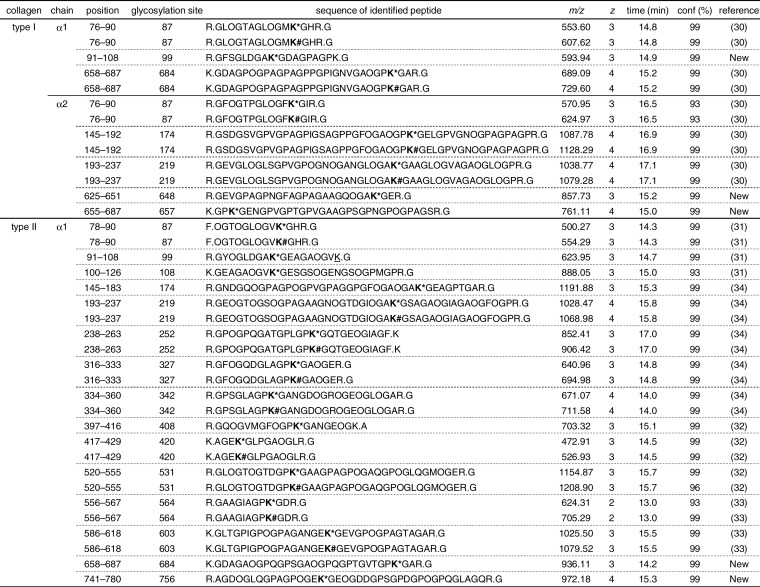
The lists of identified GHL/GGHL peptides and their modification sites are summarized in Table I. The charge state of the glycopeptides was relatively high (ranging from +2 to +4); therefore, high molecular weight peptides resulting from trypsin miscleavage at GHL/GGHL were also identified. We found five GHL/GGHL peptides in type I collagen alpha 1 chain, eight in the alpha 2 chain, and 24 in type II collagen alpha 1 chain. Nearly all the previously reported GHL/GGHL sites, which have been determined by many independent studies (30–34), were identified simultaneously using the hydrazide method. In addition, we found three glycosylation sites in type I collagen and two sites in type II collagen, which had not been reported in previous studies (supplemental Fig. S4). Diverse glycopeptides were identified as both GHL- and GGHL-containing peptides, whereas peptides substituted only with GHL also existed. In contrast, without the hydrazide method, only a few GHL/GGHL peptides were identified despite the high sequence coverage (supplemental Table S1). Thus, identification of GHL/GGHL peptides and exhaustive glycosylation site determination using LC/MS analysis were dramatically enhanced by the hydrazide method.
Table I. Results of Comprehensive LC-MS/MS Analysis of O-glycosylated Peptides of Collagen.
Purified GHL/GGHL peptides were identified by LC/MS analysis based on the following three criteria: (i) location of GHL/GGHL as being at the Y position of Gly-X-Y triplets, (ii) missed cleavage at GHL/GGHL by trypsin, and (iii) high confidence (conf) level of more than 90%. The numbering of residues begins with the triple-helical portion of the chains. First residue corresponds to residue 178 of P02453 (type I collagen alpha 1 chain), residue 89 of P02465 (type I collagen alpha 2 chain), and residue 201 of P02459 (type II collagen alpha 1 chain). O indicates hydroxyproline, K* indicates GHL, K# indicates GGHL, and K indicates hydroxylysine. The m/z values are monoisotopic.
DISCUSSION
In this study, we established the hydrazide method for O-glycosylation analysis of collagen. To avoid degradation of labile carbohydrate moieties in GHL/GGHL by periodate oxidation, galactose oxidase was used to introduce an aldehyde into GHL/GGHL peptides. Because galactose oxidase activity especially in the GGHL standard was markedly enhanced by the coordinated addition of catalase and HRP (23, 24), we applied this three-enzyme system. We used acid/heat treatment to cleave the hydrazone bond so that the eluted peptides contained entire carbohydrate chains, which has also been previously achieved with alternative methods (18, 19). Using the hydrazide method, we could identify the glycan structures concurrently with determining the position of O-glycosylated hydroxylysine. In addition, the LC/MS equipment enabled comprehensive and high-sensitivity sequence analysis compared with use of a protein sequencer, which has been used for sequence analysis of collagen in past studies (8).
During oxidation of the GHL standard, we observed a peroxide that has been reported as a main side product of galactose oxidase (23), whereas it was not detected in the GGHL reaction. Use of a large amount of galactose oxidase or the coordinated addition of catalase and HRP enhanced the oxidation efficiency, as shown for the GGHL standard. In contrast, excessive enhancement of the enzyme activity seemed to increase the peroxide generation, thereby reducing the oxidation efficiency for the GHL standard. The elution time was determined to be 30 min with acid/heat treatment because it seemed to almost reach a plateau for the release of GHL/GGHL, and a longer heating time under acidic conditions led to the smaller recovery rate of GHL/GGHL peptides, which was probably because of peelings of the carbohydrate or peptide cleavages (supplemental Fig. S3). In addition, two glycopeptides containing the Asp-Pro (Hyp) bond were identified among the three sites in type I and type II collagens, although the Asp-Pro bond was reported to be hydrolyzed by acid/heat treatment (17, 35). Thus, the elution conditions seemed to be adequate for the O-glycopeptide identification of collagen despite the possible specific peptide cleavages.
There were few nonspecific peptides that did not contain the GHL/GGHL modification site from the peptide identification by the hydrazide method. A large number of GHL/GGHL peptides were identified and the modification sites were consistent with those reported previously. For normal peptide identification with trypsin digestion, only 1/50 protein concentration was required compared with the hydrazide method, and approximately half of the total sequences were identified in type I and type II collagens. However, only a few GHL/GGHL peptides were identified with the direct LC/MS analysis because a number of co-eluted peptides competitively hampered the MS/MS acquisition of the glycopeptides in the IDA method. These data indicate that the hydrazide method efficiently purified GHL/GGHL peptides and enhanced the identification of the glycosylation sites. In addition, the hydrazide method provides greatly enhanced signal-to-noise ratio of the glycopeptides (data not shown), which would offer a big advantage for quantification analysis, such as stable isotope labeling by amino acids in cell culture (SILAC). Interestingly, some new glycosylation sites, which had not been reported in previous sequence analyses using protein sequencing, were also identified. Most O-glycosylation sites of collagen are partially glycosylated and exist as mixtures of lysine, hydroxylysine, GHL, and GGHL; therefore, a possible O-glycosylation site, which is partially substituted by lysine or hydroxylysine, could be identified as lysine or hydroxylysine on N-terminal sequence analysis in some cases. It is presumed that several GHL/GGHL modification sites have been missed, which further demonstrates the advantage of the hydrazide method in collagen analysis.
Within the GHL/GGHL peptides identified with a confidence level of more than 90%, most of the glycosylated lysine residues lie in the Y position of Gly-X-Y triplets, and trypsin-missed cleavages were observed at the residues, which is consistent with our prediction. High molecular weight peptides resulting from trypsin-missed cleavage at glycosylated hydroxylysine were identified with a high charge state of the precursor ions. However, the peptide containing consecutive GHL/GGHL is presumably over the MS range, and thus, the modification site could not yet be identified. Previously reported GHL/GGHL sites of type I collagen were fully covered in this study, but a few sites of type II collagen were not found (site 264, 270, 648, and 657), probably because of the above mentioned or other factors.
Although the amino acid sequences were clearly determined by LC/MS analysis, most GHL/GGHL sites were determined by the predicted molecular weight of the modifications. To dispel uncertainty about the site assignment, the accuracy of this collagen O-glycosylation analysis was verified by N-terminal amino acid sequence analysis. Thus, the hydrazide method is considered to be a highly accurate way of identifying O-glycopeptides of collagen. There are a few peptides that are cleaved at sites not accepted universally most likely by other protease activity, which has been reported to be contaminated in commercially available galactose oxidase (22, 36). Despite the fact that the proteolytic activity of galactose oxidase could be suppressed by immobilization to agarose beads and co-addition of the trypsin-chymotrypsin inhibitor, slight proteolytic activity may have existed. Repurification of the enzyme or use of recombinant galactose oxidase may be required for more precise analysis.
Recently, localization of hydroxylysine glycosides in the collagenous domain of the C1q complement protein has been studied by comprehensive proteomic analysis (37). In that study, hydroxylysine glycosides are clearly identified without glycopeptide purification procedures. Use of a highly sensitive nano-LC/linear quadrupole ion trap-orbitrap instrument and the relatively small size of the collagenous domain of the C1q protein are considered to permit direct analyses of the GHL/GGHL-containing peptides. In comparison, our hydrazide method permits comprehensive identification of hydroxylysine glycosides of the collagen macromolecule, which contains about 1000 amino acid residues per individual chain, with conventional LC/MS.
In this study, we developed a simple procedure in which captured glycopeptides were eluted from hydrazide resin by heating with 0.1% formic acid. Because the purpose of this study was to determine GHL/GGHL modification sites, this simple purification method was used. There are several other ways to release glycopeptides from hydrazide resin. One is to convert the glycopeptides to oxime derivatives with aminooxy reagents on hydrazide resin (19). Although our hydrazide method enabled us to identify the O-glycosylation sites of collagen, the low oxidation efficiency and recovery rate of GHL/GGHL remain challenges for precise quantitative analysis. We would further develop the method to establish quantitative analysis concurrently with identification of the glycosylation site by optimizing the conditions and, for example, using isobaric labeling on hydrazide resin or metabolic amino acid labeling by SILAC. GHL/GGHL sites of human type I collagen, which was purified from the culture supernatant of skin fibroblasts, have also been identified by using the hydrazide method (data not published). Development of a quantitative hydrazide method may enable us to identify the association of hydroxylysine glycosides with disorders, such as overglycosylation in osteogenesis imperfecta, and the physiological role of the modifications.
Acknowledgments
We would like to thank Tomomi Kiriyama, Takayuki Ogura, and Naomi Hirayama (Nippi) for providing collagen samples and GHL/GGHL standards.
Footnotes
 This article contains supplemental Figs. S1 to S4 and Table S1.
This article contains supplemental Figs. S1 to S4 and Table S1.
1 The abbreviations used are:
- GHL
- galactosyl-hydroxylysine
- GGHL
- glucosyl-galactosyl-hydroxylysine
- HRP
- horseradish peroxidase
- CNBr
- cyanogen bromide
- MRM
- multiple reaction monitoring
- IDA
- information-dependent acquisition
- TIC
- total ion current chromatogram
- SILAC
- stable isotope labeling by amino acids in cell culture.
REFERENCES
- 1. Spiro R. G. (1967) The structure of the disaccharide unit of the renal glomerular basement membrane. J. Biol. Chem. 242, 4813–4823 [PubMed] [Google Scholar]
- 2. Shinkai H., Yonemasu K. (1979) Hydroxylysine-linked glycosides of human complement subcomponent C1q and various collagens. Biochem. J. 177, 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang C., Luosujärvi H., Heikkinen J., Risteli M., Uitto L., Myllylä R. (2002) The third activity for lysyl hydroxylase 3: galactosylation of hydroxylysyl residues in collagens in vitro. Matrix Biol 21, 559–566 [DOI] [PubMed] [Google Scholar]
- 4. Schegg B., Hülsmeier A. J., Rutschmann C., Maag C., Hennet T. (2009) Core glycosylation of collagen is initiated by two beta(1-O)galactosyltransferases. Mol. Cell Biol. 29, 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kivirikko K. I., Myllylä R. (1979) Collagen glycosyltransferases. Int Rev Connect Tissue Res 8, 23–72 [DOI] [PubMed] [Google Scholar]
- 6. Tenni R., Valli M., Rossi A., Cetta G. (1993) Possible role of overglycosylation in the type I collagen triple helical domain in the molecular pathogenesis of osteogenesis imperfecta. Am. J. Med. Genet. 45, 252–256 [DOI] [PubMed] [Google Scholar]
- 7. Murray L. W., Bautista J., James P. L., Rimoin D. L. (1989) Type II collagen defects in the chondrodysplasias. I. Spondyloepiphyseal dysplasias. Am. J. Hum. Genet. 45, 5–15 [PMC free article] [PubMed] [Google Scholar]
- 8. Butler W. T. (1982) Methods in Enzymology, vol. 82, 339–346, Academic Press, New York [Google Scholar]
- 9. Gooley A. A., Classon B. J., Marschalek R., Williams K. L. (1991) Glycosylation sites identified by detection of glycosylated amino acids released from Edman degradation: the identification of Xaa-Pro-Xaa-Xaa as a motif for Thr-O-glycosylation. Biochem. Biophys. Res. Commun. 178, 1194–1201 [DOI] [PubMed] [Google Scholar]
- 10. Zachara N. E., Gooley A. A. (2000) Identification of glycosylation sites in mucin peptides by edman degradation. Methods Mol. Biol. 125, 121–128 [DOI] [PubMed] [Google Scholar]
- 11. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 12. Tian Y., Zhou Y., Elliott S., Aebersold R., Zhang H. (2007) Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2, 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramachandran P., Boontheung P., Xie Y., Sondej M., Wong D. T., Loo J. A. (2006) Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome. Res. 5, 1493–1503 [DOI] [PubMed] [Google Scholar]
- 14. Pan S., Wang Y., Quinn J. F., Peskind E. R., Waichunas D., Wimberger J. T., Jin J., Li J. G., Zhu D., Pan C., Zhang J. (2006) Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J. Proteome. Res. 5, 2769–2779 [DOI] [PubMed] [Google Scholar]
- 15. Cao J., Shen C., Wang H., Shen H., Chen Y., Nie A., Yan G., Lu H., Liu Y., Yang P. (2009) Identification of N-glycosylation sites on secreted proteins of human hepatocellular carcinoma cells with a complementary proteomics approach. J. Proteome. Res. 8, 662–672 [DOI] [PubMed] [Google Scholar]
- 16. Zeng X., Hood B. L., Sun M., Conrads T. P., Day R. S., Weissfeld J. L., Siegfried J. M., Bigbee W. L. (2010) Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. J. Proteome. Res. 9, 6440–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nilsson J., Rüetschi U., Halim A., Hesse C., Carlsohn E., Brinkmalm G., Larson G. (2009) Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 6, 809–811 [DOI] [PubMed] [Google Scholar]
- 18. Kurogochi M., Matsushista T., Amano M., Furukawa J., Shinohara Y., Aoshima M., Nishimura S. (2010) Sialic acid-focused quantitative mouse serum glycoproteomics by multiple reaction monitoring assay. Mol. Cell Proteomics 9, 2354–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klement E., Lipinszki Z., Kupihár Z., Udvardy A., Medzihradszky K. F. (2010) Enrichment of O-GlcNAc modified proteins by the periodate oxidation-hydrazide resin capture approach. J. Proteome. Res. 9, 2200–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spiro R. G. (1967) Studies on the renal glomerular basement membrane. Nature of the carbohydrate units and their attachment to the peptide portion. J. Biol. Chem. 242, 1923–1932 [PubMed] [Google Scholar]
- 21. Michaelsson E., Malmström V., Reis S., Engström A., Burkhardt H., Holmdahl R. (1994) T cell recognition of carbohydrates on type II collagen. J. Exp. Med. 180, 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Light N. D. (1986) Use of galactose oxidase in labelling hydroxylysine glycosides of collagen. Connect Tissue Res. 15, 221–233 [DOI] [PubMed] [Google Scholar]
- 23. Parikka K., Tenkanen M. (2009) Oxidation of methyl alpha-D-galactopyranoside by galactose oxidase: products formed and optimization of reaction conditions for production of aldehyde. Carbohydr Res. 344, 14–20 [DOI] [PubMed] [Google Scholar]
- 24. Parikka K., Leppanen A. S., Pitkänen L., Reunanen M., Willför S., Tenkanen M. (2010) Oxidation of polysaccharides by galactose oxidase. J. Agric Food Chem. 58, 262–271 [DOI] [PubMed] [Google Scholar]
- 25. Miller E. J. (1972) Structural studies on cartilage collagen employing limited cleavage and solubilization with pepsin. Biochemistry 11, 4903–4909 [DOI] [PubMed] [Google Scholar]
- 26. Ueno T., Tanaka K., Kaneko K., Taga Y., Sata T., Irie S., Hattori S., Ogawa-Goto K. (2010) Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion. J. Biol. Chem. 285, 29941–29950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 28. Morgan P. H., Jacobs H. G., Segrest J. P., Cunningham L. W. (1970) A comparative study of glycopeptides derived from selected vertebrate collagens. A possible role of the carbohydrate in fibril formation. J. Biol. Chem. 245, 5042–5048 [PubMed] [Google Scholar]
- 29. Wu G. Y., Pereyra B., Seifter S. (1981) Specificity of trypsin and carboxypeptidase B for hydroxylysine residues in denatured collagens. Biochemistry 20, 4321–4324 [DOI] [PubMed] [Google Scholar]
- 30. Fietzek P. P., Kühn K. (1976) The primary structure of collagen. Int. Rev. Connect Tissue Res. 7, 1–60 [DOI] [PubMed] [Google Scholar]
- 31. Butler W. T., Miller E. J., Finch J. E., Jr. (1976) The covalent structure of cartilage collagen. Amino acid sequence of the NH2-terminal helical portion of the alpha 1 (II) chain. Biochemistry 15, 3000–3006 [DOI] [PubMed] [Google Scholar]
- 32. Butler W. T., Finch J. E., Jr., Miller E. J. (1977) Covalent structure of cartilage collagen. Amino acid sequence of residues 363–551 of bovine alpha1(II) chains. Biochemistry 16, 4981–4990 [DOI] [PubMed] [Google Scholar]
- 33. Francis G., Butler W. T., Finch J. E., Jr. (1978) The covalent structure of cartilage collagen. Amino acid sequence of residues 552–661 of bovine alpha1(II) chains. Biochem. J. 175, 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seyer J. M., Hasty K. A., Kang A. H. (1989) Covalent structure of collagen. Amino acid sequence of an arthritogenic cyanogen bromide peptide from type II collagen of bovine cartilage. Eur. J. Biochem. 181, 159–173 [DOI] [PubMed] [Google Scholar]
- 35. Piszkiewicz D., Landon M., Smith E. L. (1970) Anomalous cleavage of aspartyl-proline peptide bonds during amino acid sequence determinations. Biochem. Biophys. Res. Commun. 40, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 36. Hatton M. W., Regoeczi E. (1976) The proteolytic nature of commercial samples of galactose oxidase. Purification of the enzyme by a simple affinity method. Biochim. Biophys. Acta 438, 339–346 [DOI] [PubMed] [Google Scholar]
- 37. Pflieger D., Przybylski C., Gonnet F., Le Caer J. P., Lunardi T., Arlaud G. J., Daniel R. (2010) Analysis of human C1q by combined bottom-up and top-down mass spectrometry: detailed mapping of post-translational modifications and insights into the C1r/C1s binding sites. Mol. Cell Proteomics 9, 593–610 [DOI] [PMC free article] [PubMed] [Google Scholar]