Abstract
Defective catabolite export from lysosomes results in lysosomal storage diseases in humans. Mutations in the cystine transporter gene CTNS cause cystinosis, but other lysosomal amino acid transporters are poorly characterized at the molecular level. Here we identified the C. elegans lysosomal lysine/arginine transporter, LAAT-1. Loss of laat-1 caused accumulation of lysine and arginine in enlarged, degradation-defective lysosomes. In mutants of ctns-1 (C. elegans homolog of CTNS), LAAT-1 was required to reduce lysosomal cystine levels and suppress lysosome enlargement by cysteamine, a drug that alleviates cystinosis by converting cystine to a lysine analog. LAAT-1 also maintained availability of cytosolic lysine/arginine during embryogenesis. We showed that LAAT-1 is the lysosomal lysine/arginine transporter and suggested a molecular explanation for how cysteamine alleviates a lysosomal storage disease.
Defects in exporting hydrolytic degradation products from lysosomes cause lysosomal storage diseases such as cystinosis, which is characterized by intralysosomal accumulation of free cystine because of mutations in the lysosomal cystine transporter gene CTNS (cystinosin) (1-4). The most effective therapeutic agent for cystinosis, cysteamine (an aminothiol), converts lysosomal free cystine to cysteine and the mixed disulfide of cysteine-cysteamine, which is thought to be exported from lysosomes as a lysine analog through a lysine/cationic amino acid transporter (5-7). The molecular identity of the transporter remains unknown. Although biochemically detected, most mammalian lysosomal amino acid transporters have not been molecularly characterized (1).
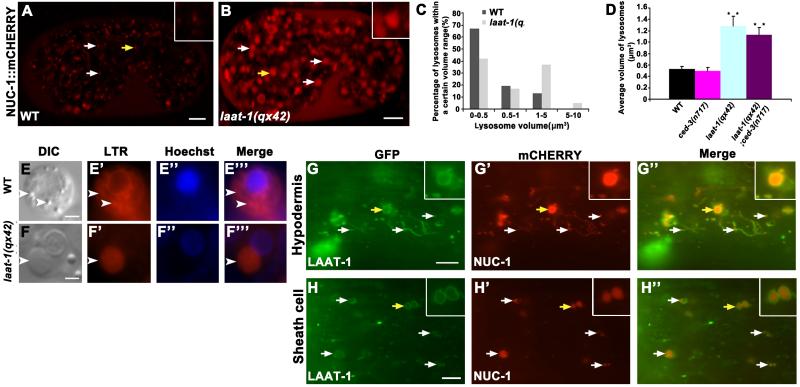
From a forward genetic screen for C. elegans mutants with increased embryonic cell corpses, we isolated a recessive mutant qx42 that accumulated many refractile corpse-like objects and lysotracker-positive puncta, suggestive of abnormal lysosomes (fig. S1, A to G). Using NUC-1::mCHERRY, which labels lysosomes (8, 9), or lysotracker staining, we found that qx42 lysosomes were on average twice the volume of wild type (1.3 versus 0.5 μm3; Fig. 1, A to F”’ and fig. S1, H to K).
Figure 1. laat-1 mutants accumulate enlarged lysosomes.
(A to F”’) Enlarged lysosomes indicated by NUC-1::mCHERRY [(A) and (B), arrows] or lysotracker red (LTR) [(E) to (F”’), arrowheads] were observed in a laat-1(qx42) embryo (B) or cell [(F) to (F”’)] but not wild type [(A), (E)-(E”’)]. Lysosome volumes are quantified in (C) and (D). Average lysosomal volumes (±SEM, n=100) in different strains are shown in (D). **P<0.0001. (G and H) Fluorescent images of hypodermal (G) or sheath cells (H) in wild-type animals expressing LAAT-1::GFP and NUC-1::mCHERRY. In (A), (B), and (G) to (H”), insets show x4 magnification of lysosomes indicated by yellow arrows. Scale bars: 2 μm in (E), (F); 5 μm in other panels.
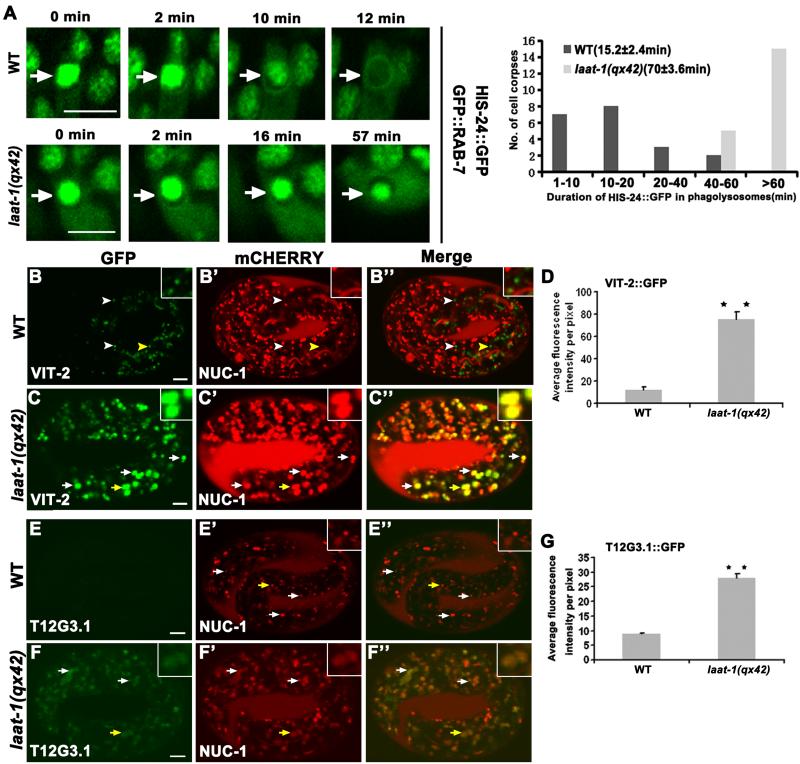
We next examined whether qx42 affected lysosomal cargo degradation. Apoptotic cells are phagocytosed then degraded in lysosomes. Cell death and cell corpse engulfment were normal in qx42 mutants (fig. S2). However, degradation of apoptotic cells in phagolysosomes (indicated by GFP::RAB-7 or NUC-1::mCHERRY) as measured by loss of HIS-24::GFP or H2B::GFP (which label chromatin in all somatic and germ nuclei, including cell corpses, respectively) was severely affected in qx42 mutants, with HIS-24::GFP persisting >4 times as long as in wild type (Fig. 2A and fig. S2, L to O). Yolk lipoprotein is degraded throughout embryogenesis to nourish developing cells (10, 11). In qx42 mutants, intestinal secretion of yolk reporter VIT-2::GFP and uptake by oocytes were normal (fig. S3, A to B’). However, qx42 embryos accumulated significantly more VIT-2::GFP in enlarged puncta, which overlapped with NUC-1::mCHERRY, suggesting defective lysosomal yolk degradation (Fig. 2, B to D and fig. S3, C to H’). Cell surface proteins CAV-1 and RME-2, which are internalized and degraded in wild-type embryos, accumulated in enlarged lysosomes in qx42 embryos (fig. S4) (12). Damaged organelles and protein aggregates are delivered via the autophagy pathway to lysosomes for degradation (13). Autophagy substrates SEPA-1 and T12G3.1 (the C. elegans homolog of mammalian p62) were cleared during embryogenesis in wild type, but persisted in late-stage qx42 mutant embryos and overlapped with NUC-1::mCHERRY, indicating defective autolysosomal degradation (Fig. 2, E to G and fig. S5) (14, 15). Thus, qx42 impairs lysosomal degradation of phagocytic, endocytic and autophagic cargoes.
Figure 2. laat-1 mutants are defective in lysosomal degradation of various cargoes.
(A) Fluorescent images of wild-type and laat-1(qx42) embryos expressing HIS-24::GFP and GFP::RAB-7 at different time points. Arrows indicate phagolysosomes. Quantification is shown in the right panel with average duration (±SEM) shown in parentheses. (B to G) Confocal fluorescent images of wild-type [(B) and (E)] or laat-1(qx42) [(C) and (F)] embryos expressing NUC-1::mCHERRY and VIT-2::GFP [(B) and (C)] or T12G3.1::GFP [(E) and (F)]. Arrows indicate overlapping GFP and mCHERRY; arrowheads indicate non-overlapping GFP. Structures indicated by yellow arrows or arrowheads are magnified x4 in the insets. Quantifications are shown in (D) and (G). At least 10 embryos were scored in each strain. Data are shown as mean±SEM. **P<0.0001. Scale bars: 5 μm.
The gene affected in qx42, Y43H11AL.2, encodes a conserved protein containing seven predicted transmembrane domains and two internal PQ (Proline-Glutamine) loop repeats, characteristic of lysosomal cystine transporters (LCTs) (16) (fig. S6F). Cystinosin, the archetypal LCT family member, is a lysosomal cystine transporter, abnormal function of which causes cystinosis (4). We named the Y43H11AL.2 gene laat-1 (lysosomal amino acid transporter 1) based on similarity with LCT family proteins and cellular functions (see below). qx42 has an A>T mutation in laat-1 that creates a premature stop codon after Asn127. Other independently isolated laat-1 mutant alleles also caused enlarged lysosome and persistent cell corpse phenotypes (fig. S1, L to R and S2K). laat-1 was expressed in various cell types in embryos, larvae and adults (fig. S7). GFP or mCHERRY fusion of LAAT-1, which fully rescued qx42 defects (fig. S6, A to E), labeled membranes of NUC-1- or lysotracker-positive structures and overlapped with lysosomal membrane protein CTNS-1, the C. elegans homolog of human cystinosin (17), indicating that LAAT-1 localizes to lysosomal membranes (Fig. 1, G to H” and fig. S7, A to C”). LAAT-1(Δ299-304)::GFP, which lacks the C-terminal dileucine-based lysosomal sorting motif (18), stained plasma membranes instead of lysosomes and failed to rescue laat-1(qx42) mutant phenotypes, indicating that LAAT-1 function depends on its lysosomal localization (fig. S6, A to F and S7, D to E”).
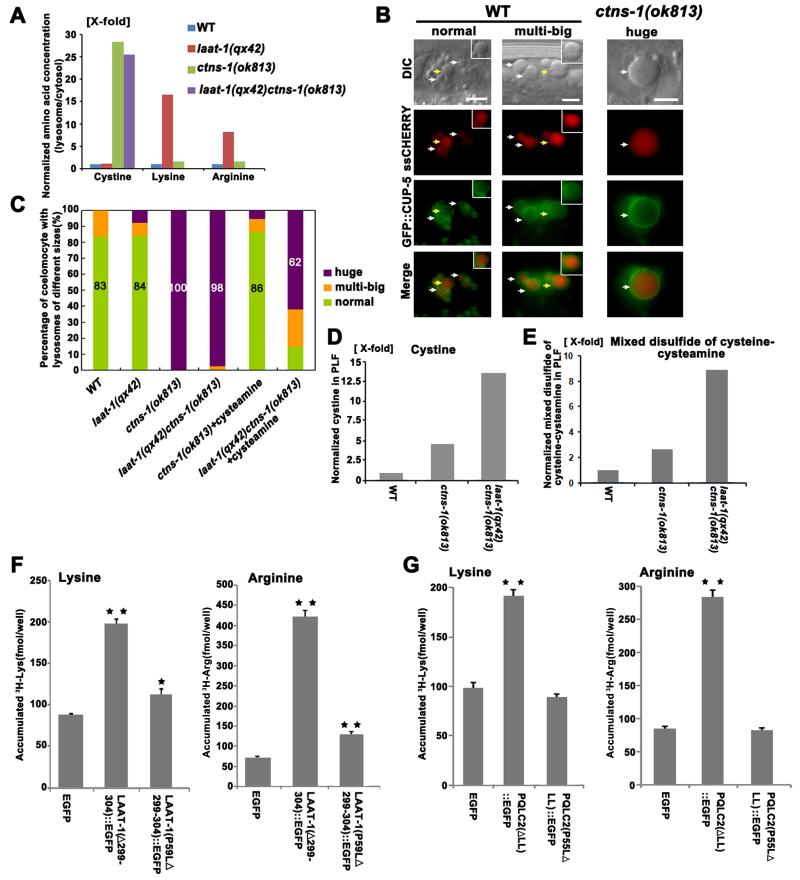
We examined lysosomes purified from C. elegans embryos (fig. S8) and found that loss of CTNS-1 caused cystine accumulation, suggesting that CTNS-1 mediates cystine efflux from lysosomes like human cystinosin (Fig. 3A). In laat-1 mutant lysosomes, cystine levels were normal but lysine and arginine levels were 16 and 8 times as high as wild type, respectively, suggesting that LAAT-1 exports lysine and arginine from lysosomes (Fig. 3A and fig. S9A). Macrophage-like coelomocytes from ctns-1 mutants contained huge granules (>6.5 μm in diameter), which accumulated endocytosed cargo CHERRY and were labeled by lysosomal membrane protein CUP-5 but not endosomal protein RME-8, indicating that they are enlarged lysosomes (19, 20) (Fig. 3, B and C and fig. S9B). Most wild-type and laat-1(qx42) coelomocytes contained small lysosomes (<4.5 μm) or 2-3 bigger ones (4.5 to 6.5 μm) (Fig. 3C). Cysteamine treatment of ctns-1 mutants greatly reduced lysosomal cystine accumulation and almost completely suppressed the enlarged lysosome phenotype (Fig. 3, C and D). In laat-1(qx42)ctns-1(ok813) double mutants, however, cysteamine failed to deplete lysosomal cystine and suppress enlarged lysosomes, which accumulated high levels of cystine and the lysine analog mixed disulfide of cysteine-cysteamine (Fig. 3, C to E). These data strongly suggest that LAAT-1 transports lysine out of lysosomes.
Figure 3. LAAT-1 is a lysosomal lysine and arginine transporter.
(A) The ratio of amino acid concentration in lysosomal versus cytosolic fractions prepared from embryonic lysates was determined and normalized as 1 fold in wild type (y-axis). (B) DIC and fluorescent images of wild-type and ctns-1(ok813) coelomoctyes expressing secreted CHERRY (ssCHERRY) and the lysosomal marker GFP::CUP-5. Lysosomes are labeled by CHERRY and CUP-5 (arrows). Insets show lysosomes indicated by yellow arrows. Scale bars: 5 μm. Quantification is shown in (C). (D and E) Cystine (D) and mixed disulfide of cysteine-cysteamine (E) was determined in purified lysosomal fractions (PLF) after cysteamine treatment and normalized as 1 (fold) in wild type. (F and G) Lysine and arginine uptake was determined in (F) LAAT-1- or (G) PQLC-2-expressing COS-7 cells. Data are shown as mean±SEM. **P<0.0001,*P<0.05; all other points had P>0.05. Data in (A), (D), (E), (F), and (G) are representative of at least 3 independent experiments.
We tested whether LAAT-1 or its human counterpart PQLC2 transported lysine and arginine using a whole cell-based transporter assay (4). Wild-type PQLC2::GFP localized to lysosomes in COS-7 cells, while PQLC2 (ΔLL)::GFP, which lacks the lysosomal sorting motif, associated with plasma membranes, indicating that PQLC2 is a lysosomal membrane protein like LAAT-1 (fig. S6F and fig. S9, C to H”). Expression of plasma membrane-targeted LAAT-1 [LAAT-1(Δ299-304)::GFP] or PQLC2 [PQLC2(ΔLL)::GFP] caused increased uptake of lysine and arginine, which was almost completely abolished when the invariant Pro in the first PQ loop was mutated to Leu (Fig. 3, F and G and fig. S6F). Uptake of histidine but not alanine, glutamic acid, cystine or cysteine was increased in LAAT-1- or PQLC2-expressing cells, suggesting specific transport of cationic amino acids (fig. S10). laat-1 lysosomes did not significantly accumulate histidine, indicating that LAAT-1 is probably not the major histidine transporter in vivo (fig. S9A).
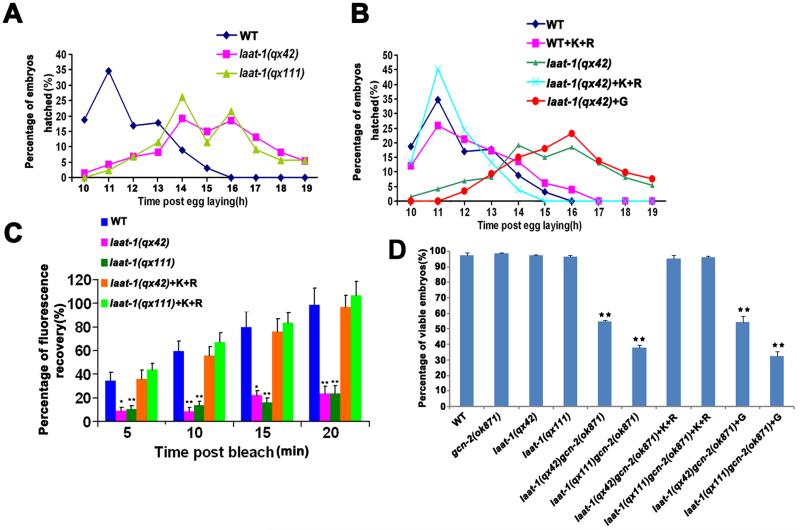
laat-1 mutants were viable but developed slowly (Fig. 4A). External supplements of both lysine and arginine completely rescued retarded embryonic development (Fig. 4B and fig. S11, A and B) but did not reverse the enlarged lysosome or defective yolk degradation phenotypes in laat-1 mutants (fig. S11C). Thus, loss of laat-1 affects lysosomal export of lysine/arginine, which limits their cytoplasmic availability and thereby retards embryonic development. When deprived of amino acids, eukaryotic cells trigger the amino acid response (AAR) pathway through activation of GCN2 protein kinase, leading to repression of global protein synthesis (21). Consistent with this, laat-1 embryos showed reduced protein synthesis, which was efficiently rescued by supplementing lysine and arginine (Fig. 4C and fig. S11D) (22). The AAR pathway is essential for survival during amino acid deprivation (23, 24). gcn-2(ok871) embryos developed normally but died when laat-1 was defective. The synthetic lethality was completely rescued by supplying both lysine and arginine but not glycine (Fig. 4D). Thus, loss of laat-1 limits cytosolic lysine and arginine, causing embryonic lethality when the GCN-2-mediated AAR pathway is impaired (fig. S11E).
Figure 4. LAAT-1 maintains lysine and arginine availability for normal embryonic development.
(A and B) Retarded embryonic development in laat-1 mutants is rescued by external lysine and arginine supplements. At least 88 embryos were examined. (C) Protein synthesis rates determined by fluorescence recovery after photobleaching in wild-type, laat-1(qx42) and laat-1(qx111) embryos expressing Plaat-1mCHERRY with or without externally supplied lysine and arginine. At least 20 embryos were quantified in each strain/treatment. (D) Loss of laat-1 and gcn-2 causes synthetic embryonic lethality. The y-axis indicates the percentage of viable embryos in each strain/treatment. 3 independent experiments were performed with at least 95 embryos examined in each. In panels (C) and (D), data are shown as mean±SEM. **P<0.0001. In panels (B) to (D), lysine (K) and arginine (R) were supplied at 100 mM each and glycine (G) was supplied at 200 mM.
We have identified LAAT-1 and its human homolog PQLC2 as the lysosomal lysine/arginine transporter. Cysteamine treatment significantly reduced lysosomal free cystine and efficiently suppressed the enlarged lysosome phenotype in ctns-1(lf) single mutants but not laat-1(lf) ctns-1(lf) double mutants, which accumulated the lysine analog mixed disulfide of cysteine-cysteamine in lysosomes, suggesting that LAAT-1 (and probably PQLC2) may mediate cystine depletion by cysteamine. It is thus important to examine whether loss of PQLC2 affects mammalian lysosome function and causes lysosome-related diseases. Our finding that defective lysosomal export of lysine/arginine leads to retarded embryonic development reveals the role of lysosomal amino acid transporters in maintaining cytosolic amino acid availability during embryonic development, providing insights into the pathogenesis of lysosomal transport disorders.
Supplementary Material
Acknowledgements
We thank Drs B. Zhu and X.Wang for discussion and critical reading of the manuscript, Dr. M. Dong for antibodies, Drs. B. Grant, H. Fares, D. Xue, H. Zhang and the C. elegans Genetic Center (CGC) for strains, the Moerman lab (University of British Columbia) for performing the comparative genomic hybridization array and I. Hanson for editing services. Data described in the paper are presented in the main text and the supporting online material. This work was supported by a Ministry of Science and Technology grant (2010CB835201) to X.W. and a CR-UK Career Development Award (C11852/A4500), a CR-UK Project Grant (C11852/A5991) and a Wellcome Trust Senior Research Fellowship (0909444/Z/09/Z) awarded to A.G..
References and Notes
- 1.Sagne C, Gasnier B. Molecular physiology and pathophysiology of lysosomal membrane transporters. J Inherit Metab Dis. 2008;31:258. doi: 10.1007/s10545-008-0879-9. [DOI] [PubMed] [Google Scholar]
- 2.Gahl WA, Bashan N, Tietze F, Bernardini I, Schulman JD. Cystine transport is defective in isolated leukocyte lysosomes from patients with cystinosis. Science. 1982;217:1263. doi: 10.1126/science.7112129. [DOI] [PubMed] [Google Scholar]
- 3.Jonas AJ, Greene AA, Smith ML, Schneider JA. Cystine accumulation and loss in normal, heterozygous, and cystinotic fibroblasts. Proc Natl Acad Sci U S A. 1982;79:4442. doi: 10.1073/pnas.79.14.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalatzis V, Cherqui S, Antignac C, Gasnier B. Cystinosin, the protein defective in cystinosis, is a H(+)-driven lysosomal cystine transporter. EMBO J. 2001;20:5940. doi: 10.1093/emboj/20.21.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thoene JG, Oshima RG, Crawhall JC, Olson DL, Schneider JA. Cystinosis. Intracellular cystine depletion by aminothiols in vitro and in vivo. J Clin Invest. 1976;58:180. doi: 10.1172/JCI108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gahl WA, Tietze F, Butler JD, Schulman JD. Cysteamine depletes cystinotic leucocyte granular fractions of cystine by the mechanism of disulphide interchange. Biochem J. 1985;228:545. doi: 10.1042/bj2280545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisoni RL, Thoene JG, Christensen HN. Detection and characterization of carrier-mediated cationic amino acid transport in lysosomes of normal and cystinotic human fibroblasts. J Biol Chem. 1985;260:4791. [PubMed] [Google Scholar]
- 8.Guo P, Hu T, Zhang J, Jiang S, Wang X. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc Natl Acad Sci U S A. 2010;107:18016. doi: 10.1073/pnas.1008946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu YC, Stanfield GM, Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536. [PMC free article] [PubMed] [Google Scholar]
- 10.Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol. 1983;96:189. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- 11.Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 10:1999. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Audhya A, McLeod IX, Yates JR, Oegema K. MVB-12, a fourth subunit of metazoan ESCRT-I, functions in receptor downregulation. PLos One. 2007;2:e956. doi: 10.1371/journal.pone.0000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Tian Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 16.Zhai Y, Heijne WH, Smith DW, Saier MH., Jr. Homologues of archaeal rhodopsins in plants, animals and fungi: structural and functional predications for a putative fungal chaperone protein. Biochim Biophys Acta. 2001;1511:206. doi: 10.1016/s0005-2736(00)00389-8. [DOI] [PubMed] [Google Scholar]
- 17.Mangahas PM, Yu X, Miller KG, Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008;180:357. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009;1793:605. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Treusch S, et al. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci U S A. 2004;101:4483. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Grant B, Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol Biol Cell. 2001;12:2011. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dever TE, et al. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 22.Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 23.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riddle DL, Blumenthal T, Meyer BJ, Priess JR. Introduction to C. elegans. C. elegans II. Cold Spring Harbor Laboratory Press; Plainview, New York: 1997. [PubMed] [Google Scholar]
- 27.Nukazuka A, Fujisawa H, Inada T, Oda Y, Takagi S. Semaphorin controls epidermal morphogenesis by stimulating mRNA translation via eIF2alpha in Caenorhabditis elegans. Genes Dev. 2008;22:1025. doi: 10.1101/gad.1644008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato K, et al. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell. 2006;17:3085. doi: 10.1091/mbc.E06-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 30.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumacher B, et al. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ. 2005;12:153. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- 32.Maydan JS, Okada HM, Flibotte S, Edgley ML, Moerman DG. De Novo identification of single nucleotide mutations in Caenorhabditis elegans using array comparative genomic hybridization. Genetics. 2009;181:1673. doi: 10.1534/genetics.108.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 34.Shen Q, et al. Adenine nucleotide translocator cooperates with core cell death machinery to promote apoptosis in Caenorhabditis elegans. Mol Cell Biol. 2009;29:3881. doi: 10.1128/MCB.01509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, et al. Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLos Genet. 2010;6:e1001235. doi: 10.1371/journal.pgen.1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong M, et al. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 38.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 39.Hersh BM, Hartwieg E, Horvitz HR. The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc Natl Acad Sci U S A. 2002;99:4355. doi: 10.1073/pnas.062065399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Odera S, Chuang CH, Lu N, Zhou Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 42.Chen D, et al. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- 43.Liu QA, Hengartner MO. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell. 1998;93:961. doi: 10.1016/s0092-8674(00)81202-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.