Abstract
We report a genome-wide association study in 10,286 cases and 9,135 controls of European ancestry, in the Cancer Genetic Markers of Susceptibility (CGEMS) initiative, identifying a new association with prostate cancer risk on chromosome 8q24 (rs620861, p=1.3×10-10, heterozygote OR = 1.17, 95% CI 1.10 – 1.24; homozygote OR = 1.33; 95% CI 1.21 – 1.45). This defines a new prostate locus on 8q24, Region 4, previously associated with breast cancer.
Genome-wide association studies (GWAS) have identified single-nucleotide polymorphism (SNP) markers associated with risk for prostate1-5, breast6, colon7-9, and bladder10 cancers for at least five distinct loci within a ∼600kb region of 8q24.21. Though this region contains no known protein-coding genes, an adjacent candidate, the c-myc oncogene (MYC), is attractive because of its established role in carcinogenesis. There is no evidence that common genetic variants in MYC or its proximal promoters are in linkage disequilibrium (LD) with markers identified by GWAS, though current laboratory investigation is focused on the role of genetic variants on chromosome 8q24 and MYC expression and function11.
Three distinct regions within 8q24.21 have been associated with prostate cancer risk (referred to hereafter as “regions 1, 2, and 3”)12. Region 3 (chr8:128473069-128537116) is also associated with risk for colorectal and ovarian cancers7,13, colorectal adenoma14, as well as kidney, larynx, and thyroid cancers15. Centromeric to regions 1 and 3, rs13281615 (chr8: 128424800) was identified in a breast cancer GWAS6. Another locus, telomeric to regions 1 and 3, has been associated with susceptibility to bladder cancer (rs9642880)10. The markers lie within distinct blocks of LD; from centromere to telomere: region 2, breast cancer region, region 3, region 1, bladder cancer region.
In stages 1-3 of the National Cancer Institute's (NCI) Cancer Genetics Markers of Susceptibility (CGEMS) initiative, we genotyped 137 SNPs flanking regions 1 and 3 in 10,286 prostate cancer cases and 9,135 controls of European ancestry in order to determine whether there are additional markers at 8q24.21 associated with prostate cancer risk. SNPs were selected because they either tagged regions identified in cancer GWAS3,4,6 or were previously genotyped in the second stage of CGEMS5. Specifically, we genotyped 82 SNPs across a segment (chr8:128,154,828 – 128,472,696) centromeric to regions 1 and 3 that includes region 2 and the breast cancer region. Telomeric to regions 1 and 3, we genotyped 55 SNPs spanning a region identified in a bladder cancer GWAS (chr8:128,617,860-128,816,653). Cases and controls were drawn from 10 studies from Europe and the United States (Supplementary Methods). Single-SNP association tests were conducted using unconditional logistic regression (2 degrees of freedom (df) genotype tests), adjusted for study, study center (when applicable), and four continuous covariates to account for potential population stratification (see Supplementary Methods).
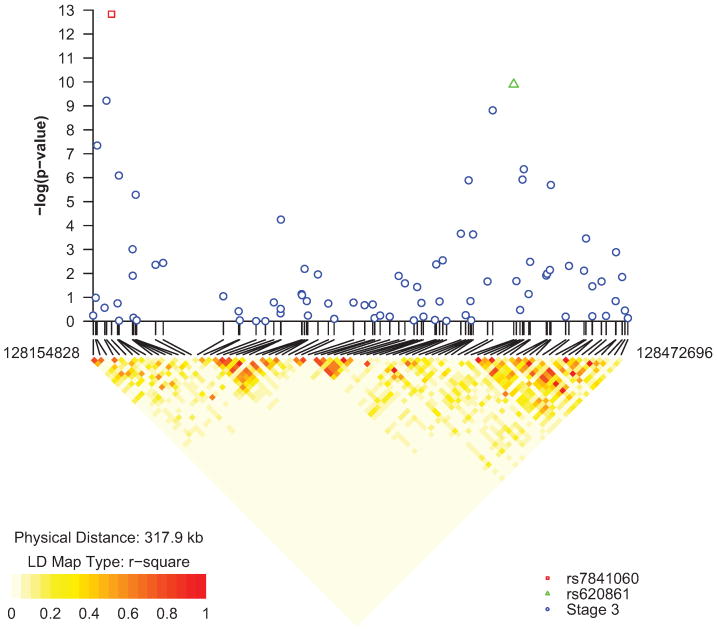
We identified a novel marker, the C allele of rs620861, to be strongly associated with prostate cancer risk (genotype score test, 2 df: p = 1.30 × 10-10, heterozygote OR = 1.17, homozygote OR = 1.33; Table 1, Figure 1 and Supplementary Table 1). rs620861 is centromeric to rs13281615, the marker associated with breast cancer susceptibility6; this region has not been previously implicated in prostate cancer. Modest LD was observed between rs620861 and rs132816156 (r2 = 0.38). In our study, rs13281615 showed a less significant association (p = 0.010, Table 1) and after adjustment for rs620861, rs13281615 was unremarkable (p = 0.142), indicating it does not confer risk independently of rs620861. An additional SNP (rs445114) within this region also was significantly associated with prostate cancer risk (p = 1.54 × 10-9; Table 1), but most likely points towards the same locus because of its strong correlation with rs620861 among controls (pairwise r2 = 0.94).
Table 1.
Prostate cancer risk association results for the novel locus (Region 4) and Region 2, previously known as the breast cancer locus.
| 8q24 Prostate Cancer region12 | Marker (risk allele) | Location | MAF CONTROLS | MAF CASES | Score χ2 | p | Het OR (95% CI) | Hom OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| 4 (novel) | rs620861 (C) | 128,404,855 | 0.372 | 0.338 | 45.56 | 1.30 × 10-10 | 1.17 (1.10 – 1.24) | 1.33 (1.21 – 1.45) |
| rs445114 (T) | 128,392,363 | 0.370 | 0.337 | 40.58 | 1.54 × 10-9 | 1.15 (1.08 – 1.23) | 1.32 (1.20 – 1.44) | |
| rs13281615 (A) | 128,424,800 | 0.412 | 0.396 | 9.16 | 0.010 | 1.03 (0.97 – 1.11) | 1.14 (1.05 – 1.24) | |
| 2 | rs7841060 (G) | 128,165,659 | 0.211 | 0.246 | 59.09 | 1.48 × 10-13 | 1.19 (1.12 – 1.26) | 1.52 (1.33 – 1.74) |
| rs4871008 (C) | 128,162,723 | 0.431 | 0.399 | 42.42 | 6.16 × 10-10 | 1.17 (1.10 – 1.25) | 1.30 (1.19 – 1.41) | |
| rs6470494 (T) | 128,157,086 | 0.281 | 0.309 | 33.84 | 4.49 × 10-8 | 1.09 (1.03 – 1.16) | 1.36 (1.22 – 1.51) |
The effect of rs620861 is comparable in aggressive (Gleason 7+ or Stage 2/3) and non-aggressive cases based on a combined joint analysis by polytomous logistic regression (Supplementary Table 1). The genotype association test with 2 degrees of freedom adjusted for study, center (when applicable, for PLCO, CeRePP, and EPIC) and four eigenvectors determined by significant principal component analysis per study (using 1400 SNPs with r2 < 0.004 in CGEMS control data set of 9,135). For regions 1 and 3, rs4242382 and rs6983267 remained the most highly significant markers (p = 6.66 × 10-21 and p = 5.76 × 10-23, respectively)
Figure 1. Linkage disequilibrium (LD) and Association Results for 82 SNPs Genotyped Over ∼314kb of Chromosome 8q24.
SNPs included were tagged with r2 > 0.8 in HapMap CEU (build 26) or included in Stage 2 of CGEMS5 across the region defined for chr8:128,154,828 – 128,472,696. The LD shown was determined on the basis of the 9,135 control individuals.
The two markers for regions 1 and 3, rs4242382 and rs6983267, were most significant, similar to our previous report5. rs620861 was not correlated with either SNP. Based on data from 9,135 control individuals (and per geographic region and by study, see Supplementary Table 2), LD between rs620861 and rs4242382 (region 1)5 and rs6983267 (region 3)5 is negligible (r2 = 7.8 × 10-4 and, 9.6 × 10-3, respectively). Similarly, rs620861 is not correlated with markers tested in region 2 (all pairwise r2 ≤ 9 × 10-3). rs620861 remained strongly associated (p = 1.13 × 10-8) with prostate cancer risk after adjustment for the most significant SNPs marking regions 1, 2 and 3 (rs4242382, rs7841060, and rs6983267, Supplementary Table 3).
We genotyped 18 SNPs across region 2, and observed three markers significantly associated with prostate cancer risk (rs7841060 G, p = 1.48 × 10-13; rs4871008 C, p = 6.16 × 10-10, and rs6470494 T, p = 4.49 × 10-8, Table 1, and Supplementary Table 4). We compared our results with those of a previously-reported rare 14-SNP haplotype (MAF = 2%, known as HapC)3 that contains the A allele of a low frequency (3.8%) SNP, rs16901979, exhibiting a significant association with prostate cancer risk. Four SNPs (rs6470494, rs1016342, rs1378897, and rs1456305) of the 14-SNP HapC were tested in the present study3; based on HapMap CEU data (build 26), an additional five markers were correlated at an r2 > 0.8 with HapC SNPs, including rs7841060 (r2 = 1 with HapC:rs1016343) and rs4871008 (r2 = 0.96 with HapC:rs1551510); thus, rs7841060 is a surrogate for a component of HapC3.
We explored the effect of rs7841060 after adjusting for rs16901979, a surrogate for HapC. Since rs16901979 was not included in the Stage 3 assay, we genotyped rs16901979 using TaqMan in a subset of subjects from Stages 1 – 2 (4,888 controls, 5,005 cases), and observed a significant association with prostate cancer (p = 6.27 × 10-8) assuming a multiplicative risk model. In evaluating the same subset of cases and controls for association with rs7841060, we obtained a p-value of 1.41 × 10-6 under the same model. In a logistic regression of both SNPs assuming log-additive effects and no interaction, each remained significant when adjusted for the other (rs7841060, p = 5.5 × 10-4; rs16901979, p = 4.1 × 10-5). A test for non-multiplicative interaction was not significant (p = 0.17). Although the risk alleles of the common SNP, rs7841060 (MAF=22.3%) and rare SNP rs16901979 (MAF=3.8%) tend to cosegregate (D′=0.87, r2=0.11), an additional copy of the G allele at rs7841060 appears to confer further risk for prostate cancer. The identification of rs7841060 suggests that the genomic architecture underlying this region could be complex and may also indicate the presence of additional lower risk variants.
No evidence of association with prostate cancer was observed for 57 SNPs telomeric of prostate cancer region 1 (Supplementary Figure 1 and Supplementary Table 4).
In our large data set, we explored the possibility of a non-multiplicative effect between six pairs of the four independent loci (including the novel one reported herein) that have been associated with prostate cancer. We observed departures from a multiplicative odds ratios model between rs4242382 (region 1)5 and rs620861 (p= 2.2 × 10-3 based on a 1 df test for non-multiplicative effect) (Supplementary Figure 2); this observation remained noteworthy after Bonferroni adjustment (p = 0.013). There was no clear pattern of association between rs4242382 and prostate cancer in the absence of the C risk allele for rs620861, but we did observe strong trends in risk by allele count of rs4242382 in the presence of either one or two copies of risk alleles for rs620861. Furthermore, the association trend between rs620861 and prostate cancer risk is stronger in the presence of the two at-risk alleles for rs4242382. There was no evidence for significant non-multiplicative effects for regions 2 and 3.
In follow-up of GWAS findings for 8q24, we identified a novel SNP marker (rs620861) in a locus previously associated with breast cancer risk, which remains highly significant (p = 1.13 × 10-8) after adjustment for other known 8q24 SNPs. Our data also provide evidence that multiple variants may independently contribute to the risk of prostate cancer in region 2; rs7841060, in conjunction with and perhaps also independently of rs16901979, contributes to risk. Our findings underscore the importance of exploring regions of association with dense genotyping in follow-up studies and expand the number of loci on 8q24 independently associated with prostate cancer risk. Further investigation is warranted to determine the molecular basis of each of the prostate susceptibility loci at 8q24 and the possible interaction between loci.
Supplementary Material
Acknowledgments
This project has been funded in part using grants CA129684 to J.X. and CA131338 to S.L.Z. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Author Contributions
M.Y., N.C., and S.J.C drafted the manuscript; N.C., J.C., K.B.J., Z.W., and M.Y. performed statistical analyses; K.B.J, N.C., J.G-B., P.K., S.W., N.O., K.Y. and L.A. made important suggestions to the analytic plan and/or aided interpretation of results; A.H. led the genotyping efforts; R.B.H., S.B., H.S.F., M.J.T., W.R.D., D.A., J.V., S.W., F.R.S., G.C.-T., O.C., A.V., G.L.A., E.D.C., C.A.H., B.H., L.K., L.L., A.S., E.R., T.J.K., R.K., W.I., S.I., K.E.W., H.G., F.W., P.S., J.X., S.L.Z., J.S., L.J.V., K.H., and M.K. provided DNA or genotyping data for study subjects; M.T., D.S.G., R.N.H., J.F.F., D.J.H., and G.T. participated in revising the manuscript and provided important intellectual contributions.
References
- 1.Amundadottir LT, et al. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Freedman ML, et al. Proc Natl Acad Sci U S A. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudmundsson J, et al. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 4.Haiman CA, et al. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas G, et al. Nat Genet. 2008 [Google Scholar]
- 6.Easton DF, et al. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haiman CA, et al. Nat Genet. 2007;39:954–6. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanke BW, et al. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 9.Tomlinson I, et al. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 10.Kiemeney LA, et al. Nat Genet. 2008;40:1307–12. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pomerantz MM, et al. Nat Genet. 2009 doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witte JS. Nat Genet. 2007;39:579–80. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 13.Ghoussaini M, et al. J Natl Cancer Inst. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berndt SI, et al. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wokolorczyk D, et al. Cancer Res. 2008;68:9982–6. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.