Summary
Upon antigen recognition, naive T cells undergo rapid expansion and activation. The energy requirements for this expansion are formidable, and T-cell activation is accompanied by dramatic changes in cellular metabolism. Furthermore, the outcome of antigen engagement is guided by multiple cues derived from the immune microenvironment. Mammalian target of rapamycin (mTOR) is emerging as a central integrator of these signals playing a critical role in driving T-cell differentiation and function. Indeed, multiple metabolic programs are controlled by mTOR signaling. In this review, we discuss the role of mTOR in regulating metabolism and how these pathways intersect with the ability of mTOR to integrate cues that guide the outcome of T-cell receptor engagement.
Keywords: T cells, T-helper cells, cell activation, cell differentiation, anergy
The two signal model, mTOR, and metabolism
As a critical component of the adaptive immune response T cells are characterized by their large diversity of antigen specific receptors and the ability to rapidly respond in a recall response. While T-cell receptor (TCR) engagement heralds recognition, the ultimate outcome of antigen recognition is guided by the integration of multiple signals derived from the immune microenvironment. In this regard, the two signal model provides a framework to understand how a particular antigenic encounter can lead to the appropriate activating or tolerogenic response (1, 2). In this paradigm, antigen recognition in the absence of costimulatory signals can lead to tolerance while antigen recognition in the context of costimulation leads to robust immune responses. TCR engagement in the absence of costimulation leads to activation-induced cell death and anergy (3). The addition of costimulation, for example in the form of CD28 activation, leads to interleukin-2 (IL-2) production and proliferation (4, 5). However, in the past decade, the increased complexity of the response has been better appreciated. For CD4+ T cells, signals from the immune microenvironment dictate the nature of the effector response. Indeed, depending on the cytokine milieu, full activation (Signal 1+2) can lead to a T-helper 1 (Th1), Th2, Th9, or Th17 response (6-8). Alternatively, suboptimal Signal 1 or full stimulation in the presence of transforming growth factor-β (TGF-β) can lead to the induction of regulatory T cells (9).
Our group has proposed that the mammalian target of rapamycin (mTOR) is a central integrator of diverse signals derived from the immune microenvironment that make up ‘Signal 2’ (10, 11). mTOR is an evolutionarily conserved serine/threonine kinase that is a member of the phosphorinositol 3-kinase (PI3K) family. From yeast to mammalian cells, TOR integrates cues from the environment to regulate a diversity of cellular functions including growth, apoptosis, metabolism, actin reorganization, and ribosome biogenesis (12, 13). Early studies demonstrated that by inhibiting mTOR, TCR engagement in the presence of costimulation (Signal 1+2) led to tolerance in the form of T-cell anergy (14). Subsequent studies revealed that in the absence of mTOR signaling, CD4+ T cells fail to differentiate into effector cells under appropriate skewing conditions. Rather, TCR stimulation in the absence of mTOR leads to the generation of forkhead box protein 3 (Foxp3) + regulatory T cells (15). Recent studies have demonstrated that mTOR inhibition promotes the generation of memory CD8+ T cells (16-18). Complementing these observations in T cells are a number of studies demonstrating a critical role for mTOR in integrating signals necessary for antigen-presenting cell (APC) and B-cell differentiation and function (19-23).
The stochastic generation of a diverse array of antigen receptors is an essential component of the ability of T cells to promote immunity. The clonal selection theory provides a model whereby a low frequency of naïve antigen-specific T cells can be expanded to protect the host. Critical to development of T-cell immunity is the extraordinary clonal expansion that accompanies antigen recognition. Such expansion requires energy and the de novo synthesis of proteins, nucleic acids, and lipids rivaling that of cancer cells. As such, it has been appreciated that upon activation, T cells employ metabolic pathways similar to cancer cells (24, 25). Specifically, even in the presence of oxygen, T cells metabolize glucose via glycolytic and mitochondrial pathways, thus not only generating energy but also generating substrates necessary for cellular growth and division (25).
While the metabolic changes associated with T-cell activation have been known for some time, it is only relatively recently that the precise mechanisms by which T cells support these changes have begun to be elucidated. Studies by Frauwirth et al. (26, 27) demonstrated that costimulation in the form of CD28 stimulation leads to the upregulation of the metabolic machinery necessary to support increased glycolysis and full T-cell activation. These observations make a critical link between signal 2 and the metabolic function necessary to support T-cell activation. Other studies have revealed the ability of T-cell activation to regulate glutamine metabolism (28). Importantly, these observations suggest that the upregulation of the metabolic machinery is not merely a consequence of T-cell activation but rather play a role in promoting T-cell activation (24). In line with this concept is the observation that blocking leucine metabolism or glucose metabolism can promote T-cell anergy in antigen-stimulated Th1 CD4+ T cells (29). Furthermore, recent studies have demonstrated critical differences between the metabolic requirements and pathways employed by CD8+ effector and memory T cells (30, 31).
Understanding T-cell differentiation and function necessitates understanding the signals that regulate T-cell metabolism. Along these lines, since mTOR plays such a central role in regulating the expression of proteins involved in glycolysis, glucose uptake, lipid and amino acid metabolism, it stands to reason that the ability of mTOR to guide T-cell differentiation and function will be in part through the regulation of metabolism. In this review, we highlight our current knowledge regarding the ability of mTOR to regulate T-cell differentiation and function and the ability of mTOR to regulate metabolism (Fig. 1). In this context, some of the connections between mTOR and metabolism in T cells are well established. Alternatively, we also present data demonstrating the ability of mTOR to regulate metabolic pathways in other systems and in doing so speculate that such pathways may also exist in T cells and thus regulate T-cell function.
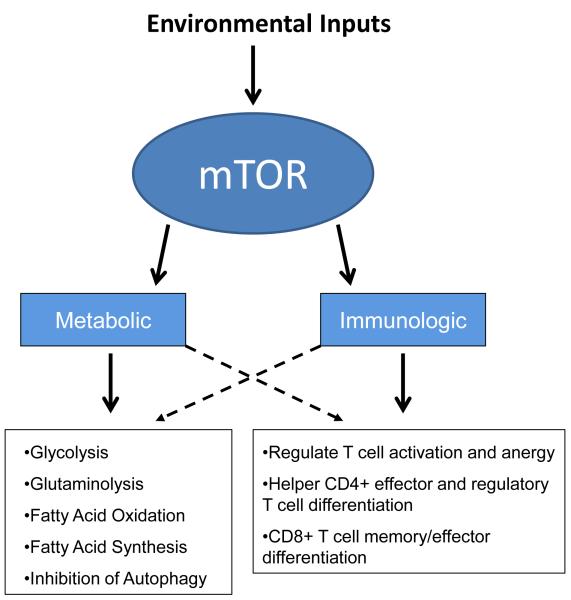
Fig. 1. mTOR integrates environmental signals to coordinate the metabolic and immunologic programs.
mTOR is emerging as a critical integrator of environmental cues guiding T-cell differentiation and function. mTOR also plays a critical role in regulating cellular metabolic programs. In as much as the upregulation of specific metabolic programs is critical for the activation of T cells, mTOR signaling serves to coordinate T-cell differentiation and metabolism.
A primer on mTOR signaling
mTOR forms two functionally distinct signaling complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (12). mTORC1 is comprised of the regulatory-associated protein of mTOR (Raptor), mammalian lethal with Sec13 protein 8 (mLST8), the proline-rich Akt substrate of 40 kDa (PRAS40), and DEP-domain-containing mTOR-interacting protein (DEPTOR). While the mTORC2 complex also contains mLST8 and Deptor, it additionally binds with the rapamycin-insensitive companion of TOR (RICTOR), mSIN1 proteins, and the Protein Observed with RICTOR (PROTOR)(12)
The kinase activity of the mTORC1 complex is positively regulated by the small GTPase Ras homolog enriched in brain (Rheb), whose activity is in turn inhibited by tuberous sclerosis complex 1 (TSC-1) and TSC-2 (32, 33). Phosphorylation of TSC-1/2 by the kinase AKT, whose activity is dependent on PI3K-mediated PDK-1 activation, inhibits the complexes’ GAP activity, thereby facilitating mTORC1 activation by GTP-bound Rheb (34). The kinase AKT can also target PRAS40, which under conditions of low AKT activity inhibits mTOR activity by directly binding with Raptor (35, 36). The activity of the mTORC1 complex is canonically measured by the phosphorylation of its substrates p70 S6-kinase and 4E-BP1 (37). The phosphorylation of 4E-BP1 results in its release of the translation-initiation factor eIF-4e leading to increased protein synthesis. Interestingly, while it has long been thought that mTOR broadly regulates cellular protein translation, detailed proteomic analysis has revealed that a fairly limited number of gene products are directly sensitive to mTOR inhibition. Notably, this includes mRNAs possessing a ‘5′-terminal oligopyrimidine tract’ (5′TOP) sequence (38). Interestingly, the pattern of protein translation inhibition seen in cells treated with allosteric inhibitors of mTOR activity (such as rapamycin) and those treated with selective mTOR kinase inhibitors (such as PP242, Torin1, and INK128) differ significantly (39).
While the upstream signals that regulate mTORC1 activity have been very well defined, identification of the precise signals regulating mTORC2 activity is still an active area of investigation. Recent studies have shown that mTORC2 is strongly and specifically activated following association with ribosomes, while its kinase activity is inhibited by ER stress and the glycogen synthetase kinase-3 β (GSK-3 β) (40, 41). Downstream targets of mTORC2 include Akt, serum and glucocorticoid-inducible kinase 1 (SGK-1), and protein kinase C (PKC) (42, 43). Akt acts as both an upstream regulator of mTORC1 activity (as indicated by the PI3K/PDK-1 dependent phosphorylation at the T308 residue) as well as a downstream target of mTORC2 (as indicated by phosphorylation at S473 residue). Akt-dependent inhibition of TSC2 (upstream of mTORC1) does not require mTORC2 in T cells (43-45).
Regulation of mTOR activity
The activity of the mTOR kinase signaling complex is regulated by a wide array of extrinsic immunological and metabolic cues, as well as cell intrinsic stress and danger signals. Ligation of the TCR can induce mTOR activation via the activation of PI3K; however, this activation is potently enhanced by stimulation by the costimulatory receptors CD28, ICOS, and OX-40 (46-52). The T-cell-restricted inhibitory receptors CTLA-4 and PD-1 can antagonize mTOR activity via AKT and PI3K inhibition (53-55). Many soluble factors such as the interleukins, interferons, chemokines, the lysophospholipid S1P1, and the adipokine leptin all possess the ability to induce mTOR activity in T cells expressing the appropriate receptors (56-68). Under conditions of metabolic stress, a decrease in the normally high intracellular ATP:AMP ratio induces the activation of AMP-kinase, which facilitates the inhibition of mTOR by phosphorylating and activating the mTORC1 inhibitory TSC1/2 complex as well as directly phosphorylating the mTORC1 scaffolding protein Raptor (69, 70). Low oxygen tension can also facilitate the inhibition of mTOR activity via the protein REDD1, which facilitates the phosphorylation and stabilization of the TSC1/2 complex (71, 72). Furthermore, cellular stress can induce the activation of the classic tumor suppressor gene p53 in a AMPK-dependent fashion, resulting in enhanced expression of many mTOR-inhibitory proteins including TSC2, PTEN, REDD1, and AMPK (73-75)
mTOR is an important regulator of cellular metabolism
In the presence of oxygen, most mammalian cells rely on the TCA cycle and oxidative phosphorylation to generate ATP. However, as previously mentioned, newly activated T cells (like cancer cells) primarily rely on the processes of aerobic glycolysis for their energetic needs (24). This phenomenon of eschewing the largely efficient process of oxidative phosphorylation following glycolysis (which can produce a total of 30 to 35 molecules of ATP per molecule of glucose) in favor of the much less efficient processes of aerobic glycolysis (a net gain of 2 ATP per glucose) was initially described by Otto Warburg in cancer cells (76). It has been proposed that although such metabolism appears inefficient in its ability to produce energy in the form of ATP, it provides intermediate metabolites that can lead to the generation of structural components necessary for rapidly dividing cells (77). The process of aerobic glycolysis is directly regulated by mTORC1 signaling by the transcriptional regulation of many key glycolytic enzymes (78). Indeed, mTOR activation regulates (in part through HIF and Myc as described below) the transcription of enzymes involved in nearly every step of glucose metabolism from its transport into the cell to the conversion of pyruvate to lactate (79-81).
The pentose phosphate pathway (PPP) is an anabolic program employed in many cells (including activated T cells) that utilizes glucose-6-phosphate generated during glycolysis but yields a different array of metabolic intermediates (82). The primary substrate for the PPP is glucose-6-phosphate produced during the first stage of glycolysis by the enzyme hexokinase 1, but unlike glycolysis, the PPP does not directly generate ATP. During the oxidative phase of the reaction, 2 NADP+ are reduced to NADPH by the enzymes glucose 6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase. NADPH provides reducing equivalents necessary for lipid and nucleotide synthesis and prevents oxidative stress. This anabolic program finally results in the generation of ribose-5 phosphate and erythrose-4-phosphate, which can be utilized by the cell for the generation of nucleic acids or amino acids, respectively. Alternately, these products can feed back into glycolysis, providing more metabolic substrates for the cell. mTORC1 activity directly regulates the expression of enzymes in the PPP, while at the same time facilitates the uptake of glucose via the Glut1 membrane transporter (79). Conversely, inhibition of mTOR activity by rapamycin treatment greatly decreases the expression of these genes (79).
The process of glutaminolysis, the degradation of glutamine to generate substrates for the TCA cycle, is another pathway found in activated T cells. Inhibition of glutaminolysis significantly decreases T-cell proliferation, a phenomenon that can be partially reversed by the addition of supplementary nucleotides and polyamines, two products of glutamine degradation (80). Interestingly, the cellular importation of glutamine via the SLC1A1 Na+ dependent symporter can not only feed into the catabolic glutaminolysis pathway but can also facilitate the induction of mTOR activity. Amino acid availability, especially the branched-chain amino acid leucine, is essential for the induction of mTORC1 activity. Indeed, the lack of plasma-membrane leucine transporters can completely abolish mTORC1 activity, which can be overcome by cytoplasmic injection of this amino acid (83). In part, this phenomenon is thought to be due to the ability of leucine to activate heterodimers of Rag GTPases, which can deliver the mTOR kinase complex to GTP-bound Rheb (84, 85). The import of extracellular leucine occurs via a glutamine antiporter, SLC7A5-SLC3A2. Intracellular glutamine does not itself possess any ability to directly induce mTORC1 activity, but without intracellular glutamine, lecuine influx is mitigated and, therefore, so is mTORC1 activity (86, 87).
Fatty acid oxidation, specifically the process of β-oxidation of free fatty acids (FFA) in the mitochondria, provides an abundant source of ATP for cellular respiration in a process that is highly dependent on the presence of oxygen. The inhibition of mTOR activity by rapamycin treatment decreases the rate of glycolysis, but causes a corresponding increase in the rate of fatty acid oxidation (88). Furthermore, rapamycin-mediated mTOR inhibition blocks the process of fatty acid and complex lipid synthesis by downregulating the expression of acetyl-coenzyme A carboxylase I and mitochondrial glycerol phosphate acyltransferase (89). Fatty acid oxidation appears to play an important role in the metabolic program of a naive resting T cells, while it is significantly down regulated following T-cell activation (80, 90).
The reduced coenzyme NADH/NADPH generated by aerobic glycolysis, the PPP, and the activity of the TCA cycle are utilized by the mitochondrial electron transport chain under aerobic conditions to generate ATP. mTORC1 activity facilitates this process by inducing mitochondrial biogenesis. Inhibition of mTOR activity by rapamycin dramatically reduces mitochondrial oxygen utilization in a process that appears to be independent of S6kinase, while mTORC1 appears to directly control the expression of many mitochondrial genes (91). Recently, it has been shown that mTORC1 regulates the activity of the Yin Yang 1 (YY1) zinc-finger GLI-kruppell class transcription factor, as well as its transcriptional co-activator peroxisome-proliferator-activated receptor coactivator (PGC)-1a, both of which are known to regulate the expression of many mitochondrial genes (92). Furthermore, the inhibition of mTOR activity, either by starvation or pharmacological intervention, can result in a dramatic reduction in cellular mitochondrial load as a result of the induction of autophagy. Therefore, high levels of mTOR activity promote both the generation, as well as the stability, of mitochondria.
The metabolic programs described thus far play an important role during the tremendous growth phase that characterizes T-cell activation. Alternatively, autophagy is a tightly controlled catabolic metabolic process during which a nutrient-deprived or stressed cell degrades its own organelles, cytoplasm, lipids, or proteins to derive energy. Not surprisingly, the process of autophagy is a finely balanced calculation, weighing the benefits of degrading critical cellular components to insure continued cell survival during nutrient deprivation against the possibility of inducing cell death. In addition, the process of autophagy provides a cell with a pathway by which to dispose of damaged organelles and aggregation-prone proteins independently of nutrient availability (93). Therefore, autophagy is both a pro- and anti- survival process, depending on the context of initiation. While there are several autophagic pathways and programs, including macroautophagy, microautophagy, and chaperone-mediated autophagy, the process is commonly quantified by the accumulation of intracellular LC3B coated vesicles (94). LC3 is ubiquitously expressed in mammalian cells during all metabolic states, but it rapidly accumulates on autophagic vesicles following nutrient withdrawal (95).
Autophagy and the formation of autophagic vesicles are directly regulated by mTOR signaling in a process primarily involving the mTORC1 complex (96, 97). The addition of rapamycin to a cell culture rapidly induces autophagy, while the constitutive activation of mTORC1 by the deletion of the negative regulator protein TSC-2 renders a cell resistant to autophagy induction (98). The initiation of autophagy in mammalian cells is dependent on the assembly and activity of a complex containing the kinase ULK1, the adapter protein autophagy-related gene-13 (Atg13), and the 200 kDa focal adhesion kinase family-interacting protein (FIP200) (97). Unlike their yeast homologs, the ULK1:Atg13:Fip200 complex in mammalian cells does not appear to disassemble under condition of high mTOR signaling and abundant nutrient availability (99). Rather, the mTOR kinase complex appears to directly phosphorylate inhibitory sites on the kinase ULK1 and the adapter protein Atg13, thereby inhibiting the induction of autophagy. In contrast, under conditions of low mTOR activity, ULK1 appears to autophosphorylate as well as directly phosphorylate Atg13 and FIP200, facilitating the activation of the complex and the induction of autophagy (100, 101). Inhibition of this process by siRNA-mediated knockdown of ULK1 renders cells resistant to rapamycin-induced autophagy (102).
mTOR-mediated transcription intersects with T-cell transcriptional programs
The ability of mTOR to regulate metabolic programs is mediated through the regulation of key transcriptional regulators. These same factors implicated in the regulation of metabolism also play roles in T-cell function. Thus, in this fashion, mTOR plays a role in coordinating the cellular immune response with metabolic demands (Fig. 2).
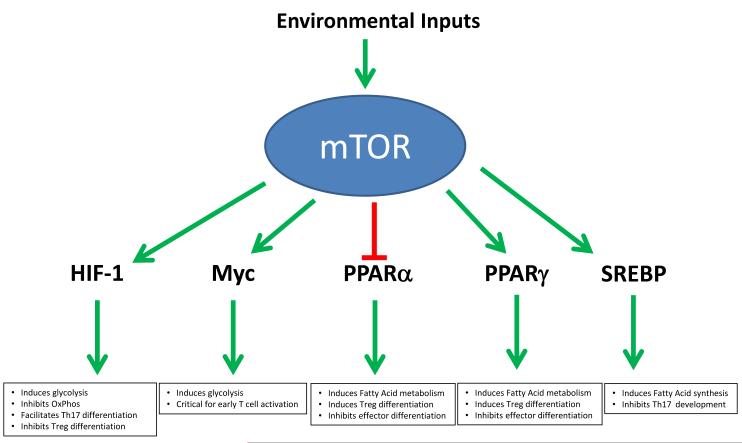
Fig. 2. mTOR regulates expression of metabolic genes via transcriptional programs.
Following activation by intrinsic and extrinsic cues, mTOR regulates transcription factors involved in coordinating glycolysis (HIF-1α, Myc), fatty acid oxidation (PPAR α, PPARγ), as well as fatty acid synthesis (SREBP). These same factors have been implicated in regulating T-cell differentiation and function upon antigen recognition.
HIF-1α
HIF-1 is a hetrodimeric protein consisting of the constitutively expressed β subunit and an inducible α subunit. The α subunit undergoes rapid proteasomal degradation under normoxic conditions due to prolyl 4-hydroxylase dependent modification of the Pro-402/564 residues, resulting in its being targeted for destruction by E3 ubiquitin ligases (103). However, under conditions of low oxygen tension, this protein degradation pathway is strongly inhibited, thereby allowing the accumulation of HIF-1α and the formation of a transcriptionally active HIF-1 dimer (104, 105). As would be expected in light of its mode of activation, HIF-1 controls the expression of many genes critical for cellular adaptation to a low-oxygen metabolic environment. As glycolysis is the primary metabolic program utilized by oxygen-starved cells, HIF-1 directly upregulates the expression of gene products along the entire glycolytic pathway, including the plasma membrane glucose transporter Glut1 and lactate dehydrogenase A (LDH-A) (81, 106). Furthermore, HIF-1 indirectly inhibits the ability of pyruvate produced as the product of glycolysis to enter into the TCA cycle by inducing the activity of pyruvate dehydrogenase kinase 1 (PDK1) (107). This enzyme antagonizes the activity of pyruvate dehydrogenase, which converts pyruvate to acetyl-CoA and CO2, thereby further enforcing an anaerobic and glycolytic metabolic program.
In addition to the turnover rate of HIF-1α being regulated by the availability of oxygen, HIF-1α transcription is enhanced by mTORC1 activity (108). This effect is readily apparent under low-oxygen conditions, where the addition of rapamycin can abrogate many of the pro-survival effects mediated by HIF-1 expression (109). In T cells, HIF-1α, expression is induced by a number of immunologic cues even in the presence of normoxia. This includes TCR stimulation as well as the presence of IGF-1 and glucose availability (110-112). This control can be quite complex. For example, as mentioned previously the HIF-1α protein is stabilized under hypoxic conditions. However, mTOR activity, and therefore HIF-1α transcription, is inhibited under conditions of low oxygen tension via the protein REDD-1 (71, 72, 113). Thus, hypoxic conditions simultaneously stabilize the HIF protein and inhibit the transcriptional expression of HIF-1α.
Myc
The oncogenic transcription factor Myc regulate many key metabolic pathways critical for cellular growth and proliferation (80, 114, 115). These pathways include glycolysis, glutaminolysis, and fatty acid oxidation (80). T cells rapidly induce Myc expression following TCR stimulation to facilitate the marked metabolic shift required for rapid proliferation (80). Additionally, Myc is among the most commonly overexpressed genes in many tumors due to its pro-survival/growth activity (116, 117). The kinase activity of mTORC1 appears to facilitate Myc translation by directly phosphorylating the translational repressor 4E-BP, which results in the release of eIF-4e. This effect is in part due to the tertiary structure of the 5′-UTR of the Myc mRNA, which has been shown to contain an internal ribosome entry site (118-122). Additionally, there is evidence that AKT/mTOR activity can regulate Myc expression at the transcriptional level via AIP4/ITCH-mediated degradation of the transcriptional repressor JunB (123). Interestingly, Myc is a transcriptional repressor of TSC2 gene expression, meaning that high levels of Myc expression facilitates a feed-forward loop resulting in less TSC2 protein, higher mTOR activity, and even more Myc expression (124). In addition to directly regulating the expression of Myc protein, high levels of mTOR activity synergizes with Myc expression by cooperatively enhancing protein translation and elongation (124). Indeed, many tumor cells that exhibit resistance to rapamycin show a compensatory increase in Myc expression, and Myc null cells are significantly more sensitive to rapamycin-induced inhibition of proliferation (124, 125).
PPARα
The peroxisome proliferator-activated receptor α (PPARα) is a nuclear hormone receptor that is thought to regulate fatty acid metabolism as well as glucose homeostasis by acting as a intracellular sensor of endogenous fatty acids (126). The precise endogenous ligand for PPARα is still an active area of investigation, but it has been shown that mono/polyunsaturated fatty acids, long-chain fatty acyl-CoAs, and saturated fatty acids can all bind and activate the receptor (127-130). The binding of these natural ligands to PPARα facilitates the nuclear translocation of the protein, displacement of transcriptional repressors from target genes, and the recruitment of transcriptional activators. PPARα possesses the ability to regulate a wide range of metabolic processes, including mitochondrial fatty acid β-oxidation (131, 132), fatty acid uptake (133), and glucose homeostasis (134). However, PPARα also possess the ability to directly repress the expression of several pro-inflammatory transcription factors, including AP1 and NF-κB (135-137). While the transcriptional activity of PPARα has long been thought to be primarily dictated by nuclear translocation, it has recently been shown that PPARα protein levels and transcriptional activity is inhibited by mTORC1 activity downstream of TSC1/2 (138). This was determined by the observation that the livers of fasted mice show very low levels of mTORC1 activity relative to fed controls but exhibit complementarily high level of PPARα and PPARα-target gene expression. This phenomenon could be reversed by the genetic deletion of TSC-1, which eliminated the fasting-induced upregulation of PPARα expression and transcriptional activity (138).
PPARγ
Like its homolog PPARα, PPARγ is a nuclear hormone receptor that plays a critical role in regulating adipogenesis, as well as lipid metabolism and glucose homeostasis in cells. There are three known mRNA isoforms of PPARγ, with isoform 1 showing widespread tissue expression, while isoforms 2 and 3 appear to be exclusively expressed in adipose tissue (139-142). It has been shown that polyunsaturated acids (such as linoleic acid and aracidonic acid), eicosanoids, and many synthetic ligands (such as TZDs and tyrosine agonists) can bind to PPARγ with high affinity and induce nuclear translocation (128, 143, 144). Interestingly, unlike PPARα, PPARγ transcriptional activity appears to be enhanced by mTOR activity. The addition of rapamycin to cultured 3T3-L1 pre-adipocytes profoundly represses the rate of adipogenisis in a process that appears to be dependent on the disruption of a feed-forward loop between PPARγ and another metabolically important transcription factor C/EBPα (145). Furthermore, the development of mature adipocytes in vitro is dependent on the availability of extracellular amino acids. Stimulation of AKT/mTOR activity by the addition of amino acids to the culture conditions significantly enhances PPARγ activity in an mTOR dependent fashion, thereby increasing the rate of adipogensis. The genetic deletion of the mTORC1 inhibitory protein TSC in mouse embryonic fibroblasts and 3T3-L1 adipocytes additionally results in a significantly enhanced rate of adipogenesis in an mTORC1 and PPARγ dependent fashion (146).
SREBP
The sterol regulatory element binding proteins (SREBPs) family of basic helix-loop-helix transcription factors are master regulators of cellular lipogenesis, facilitating the transcription of anabolic enzymes involved in fatty acid and cholesterol synthesis (147). The activity of SREBP-1, the canonical member of the transcription factor family which is expressed in T cells, is regulated by several mechanism, including cellular localization, post-translational modification, degradation, and transcription in a process that is dependent on mTORC1 activity downstream of AKT (148, 149). The transcription of SREBP is suppressed by rapamycin-mediated mTORC1 inhibition, inducing a feed-forward process due to the fact that SREBP binds to its own promoter and facilitates its own expression (150, 151). Furthermore, mTORC1 signaling facilitates SREBP transcriptional activity by inducing the ER to Golgi translocation of the immature membrane-bound form as well as the cleavage of the immature factor (79, 150). Lastly, mTORC1 can facilitate the DNA binding activity of SREBP by the direct phosphorylation and subsequent nuclear-exclusion of the phosphatidic acid phosphatase Lipin-1, which under conditions of low-mTOR activity antagonizes the nuclear accumulation of SREBP (152).
mTOR, T cells, and metabolism
Following a 24-36 h period of activation, T cells are capable of undergoing a round of replication every 8 h (153). To facilitate this energy-intensive process, activated T cells utilize a wide range of metabolic processes to derive energy as well as metabolic intermediates required for cellular growth. However, it is clear that different types of T cells exhibit different metabolic demands. In this section, we focus on the intersection of these metabolic demands and the ability of mTOR to regulate function and metabolism.
Thymic development and selection
The process of thymic development and TCR selection is a period of dynamic proliferative and metabolic activity for a developing T cell. After migrating from the bone marrow, an immature thymocyte attempts TCR rearrangement during the double negative (DN) and double positive (DP) stages of development (154). Immature thymocytes upon passing the DN3 to DN4 transition also dramatically increase in size and metabolic rate (155). This process is accompanied by a burst of mTOR activity as well as an increase in expression of several mTOR-dependent transmembrane nutrient transporters, including the transferrin receptor (CD71) and surface neutral amino acid transporter (CD98) (155, 156). While rapamycin can reduce thymocyte proliferation and the expression of CD71 and CD98, it does not inhibit the process of differentiation. Interestingly, numerous extracellular cues can facilitate mTOR activity in a developing thymocyte including the Notch pathway. Notch signaling is a hallmark of thymocyte development and is critical for inducing the proliferation and glycolytic metabolic activity of pre-T cells (157). While Notch has previously been shown to facilitate thymocyte development via PDK-1 activation of AKT and subsequent activation of S6-kinase (155), Notch has recently been shown to also signal via mTORC2 (158). Thymocytes lacking the critical mTORC2 scaffolding protein Rictor exhibit a significant defect in thymocyte proliferation as well as differentiation. This was accompanied by a significant decrease in NF-κB activity and could be reversed by expression of a constituently active mutant of the mTORC2 target AKT.
Resting T cells in the periphery
Following thymic egress, a naive T cell must persist and circulate in the periphery for an extended period of time in a quiescent state. During this time, T cells express low levels of mTOR activity and rely primarily upon fatty acid oxidation as a source of energy (80). Alternatively, increasing mTOR activity by the genetic deletion of the mTORC1 inhibitory protein TSC-1 leads to an inability to maintain a large pool of resting immunologically naive cells (159-161). For these resting T cells, autophagy is indispensable for their survival and energetic homeostasis (162). Deletion of the critical autophagy gene Atg7 in mature circulating T cells results in a significant survival defect in part due to an inability to clear the large number of mitochondria generated during thymic development. This results in an increase in the generation of oxygen free radicals as well and an imbalance in the expression of pro/anti-apoptotic gene expression (162).
TCR dependent activation following antigen exposure
Within the first 24 h of activation, a naive T cell undergoes a radical shift in its metabolic profile, downregulating the expression of genes involved in fatty acid oxidation and shifting to a glycolytic profile. This process appears to be dependent on the transcription factor Myc (80). As mentioned previously, mTOR activity is modulated by a range of immunological factors during T-cell activation, including TCR ligation and the presence of costimulation and various cytokines. The inhibition or genetic deletion of mTOR in T cells leads to a decreased rate of proliferation and a dramatic shift in the path of differentiation. Metabolically, this lack of differentiation is associated with a failure to upregulate the glycolytic machinery. Interestingly, while TCR ligation and costimulation potently facilitate mTOR kinase activity, T-cell activation is also accompanied by a dramatic increase in the accumulation of LC3B coated autophagic vesicles (163). High levels of mTOR activity are normally associated with the inhibition of autophagy. Along these lines, autophagy appears to also modulate T-cell death following nutrient withdrawal. Knocking down components of the autophagic pathway , such as Atg7 and Beclin 1, leads to a higher rate of survival following serum starvation (163).
However, while the inhibition of autophagy can increase T-cell survival in vitro following growth factor withdrawal, it appears that T-cell survival, proliferation, and effector function in vivo is significantly impaired in the absence of autophagy (164, 165). T cells lacking the critical macroautophagy gene Atg5 develop normally in the thymus but show significantly higher rates of cell death upon entry to the periphery. Furthermore, upon TCR stimulation, these Atg5-deficient T cells fail to proliferate and instead undergo rapid apoptosis. The inability of T cells to proliferate in the absence of autophagy is due to a dysregulation of ER integrity and calcium homeostasis (166).Therefore to some degree, autophagy is critical for the initiation and persistence of the highly energetic process of T-cell activation and proliferation.
While effector T cells display an essentially glycolytic metabolic phenotype during the acute phase of an immune response, there is increasing evidence that chronically activated T cells depend more on mitochondrial oxidative phosphorylation to fulfill their bioenergetic needs. Chronically stimulated alloreactive murine splenocytes display a significant increase in mitochondrial oxidative phosphorylation in experimental models of both lupus and graft-vs-host disease (GVHD) compared to normal controls (167, 168). Furthermore, T cells isolated from human patients with systemic lupus erythematosus (SLE) have been observed not only to possess abnormally high mitochondrial mass and mitochondrial transmembrane potential but also to exhibit profound ATP depletion (169, 170).
CD4+ T-cell differentiation
Upon antigen recognition, the effector fate of a naive CD4+ T cell is dictated by the integration of multiple cues from the immune microenvironment. While these T-cell subsets have classically been characterized by their cytokine secretion profile and transcription factor expression, it has recently become apparent that each subset also possesses a unique metabolic profile and corresponding set of mTOR signal requirements.
Th1 CD4+ T cells
Th1 CD4+ T cells are characterized by their production of IFNγ and their expression of the master transcription factor Tbet (171). The development of Th1 CD4+ T cells is dependent on IL-12-mediated signal transducer and activator of transcription 4 (STAT4) activation, while the addition of IFNγ to the culture conditions serves to enhance differentiation. CD4+ T cells skewed to a Th1 phenotype exhibit a robustly glycolytic phenotype and express high surface levels of the Glut1 glucose transporter (90). Inhibition of glycolysis by the addition of the competitive glucose analog 2-DG profoundly inhibits the secretion of IFNγ by Th1 CD4+ T cells (29). The development of Th1 T cells is dependent on the presence of the mTORC1 signaling complex (44). CD4+ T cells lacking the critical mTORC1 activating GTPase Rheb fail to differentiate towards a Th1 phenotype and show reduced STAT4 phosphorylation in response to IL-12. As might be anticipated the inhibition of mTOR signaling in CD4+ T cells by the genetic deletion of mTOR (15), the addition of rapamycin or the AMP-kinase activating compound AICAR (29), as well as the pharmacological inhibition of glycolysis or amino acid transport all serve to mitigate Th1 differentiation (29, 90). While the presence of glucose is absolutely required for the activation and differentiation of Th1 T cells, the addition of fatty acids to skewing cultures can dramatically inhibit IFNγ production (90). Furthermore, deletion of the intracellular fatty acid sensing receptor PPARα in CD4+ T cells results in enhanced IFNγ secretion and an increase in the severity of the experimental autoimmune model EAE (172). Interestingly, this phenomenon of increased IFNγ secretion in PPARα knockout cells is only observed in male animals. This observation is potentially explained by the fact that male animals also express higher levels of PPARα. Likewise, activation of PPARγ in CD4+ T cells significantly decreases IL-2 and IFNγ secretion (173, 174). This anti-inflammatory effect of PPARγ ligation has shown great promise in relieving the severity of many models of autoimmunity, including autoimmune diabetes, inflammatory bowel disease, allergic asthma, and experimental autoimmune encephalomyelitis (175-179). Interestingly, despite the many anti-inflammatory effects attributed to PPARγ ligation, CD4+ T cells lacking PPARγ expression lack the ability to induce systemic autoimmunity following adoptive transfer into a lymphopenic host (180).
While the process of glycolysis is undoubtedly necessary for the generation of IFNγ secreting Th1 T cells (29, 90), the exact role if any that HIF-1 plays in this process is slightly convoluted. Early studies utilizing T-cell specific HIF-1a isoform knockdowns or chimeric animals demonstrated enhanced IFNγ secretion by both CD4+ and CD8+ T cells with reduced HIF-1α activity (181, 182). However, more recent studies utilizing CD4cre-driven excision of the gene has shown no difference in IFNγ secretion under Th1-skewing conditions by CD4+ T cells lacking HIF-1α expression (183). Therefore, the importance of HIF-1 in the generation of IFNγ-producing CD4+ T cells remains to be clearly defined.
Th2 CD4+ T cells
Th2 T cells express high levels of the transcription factor GATA3, express the cytokines IL-4, IL-5, and IL-13, and play a role in immunity against extracellular parasitic infections (184, 185). Like their Th1 counterparts, Th2 T cells express high surface levels of the Glut1 glucose transporter and exhibit a high rate of glucose uptake and lactate production following stimulation (90, 186). In addition, stimulation of the fatty acid sensing protein PPARγ inhibits the production of this cytokine (90, 187).
While the development of Th1 T cells is dependent on mTORC1 activity, Th2 cells readily develop in the absence of mTORC1 but fail to differentiate in the absence of the mTORC2 signaling complex. Murine CD4+ T cells lacking the critical mTORC2 scaffolding factor Rictor fail to produce IL-4 under appropriate skewing conditions (44, 45). The observation that the generation of Th2 cells does not require mTORC1 activity is very interesting in light of the fact that Th2 cells differentiated from wildtype naive CD4+ T cells exhibit a robustly glycolytic metabolic phenotype (90). This raises the interesting possibility that although Th2 cells exhibit a high rate of glycolysis when differentiated from wildtype cells, perhaps they do not necessarily depend on the same high rate of glycolysis as their Th1 counterparts. Along these lines, in vitro generated Th2 cells exhibit a significantly higher rate of autophagy than Th1 cells (163). Since autophagy is inhibited by high levels of mTORC1 activity, the ability of Rheb-deficient CD4+ T cells to skew towards a Th2 phenotype might be promoted by their ability to utilize autophagy.
Th17 CD4+ T cells
The Th17 effector linage is one of the more recently define CD4+ T-cell subsets that appears to play a critical role in the clearance of some bacterial infections as well as the induction of tissue inflammation and a number of experimental models of autoimmunity (188). Th17 cells are generated in the presence of TGFβ and IL-6 and are defined by the expression of the master transcription factor RORγt. Like Th1 and Th2 CD4+ T cells, Th17 cells are highly glycolytic and perhaps rely even more on this metabolic program than other T-cell subsets (90, 186). The development of IL-17-secreting CD4+ T cells is severely diminished in T cells lacking mTOR, T cells lacking the mTORC1 complex and T cells treated with rapamycin (15, 44, 186). However, as in the case of Th1 cells, the absence of mTORC2 does not appear to inhibit the development of Th17 cells. The transcription factor HIF-1 has recently been shown to play a critical role in regulating the differentiation and metabolic activity of Th17 cells (183, 186). HIF-1α expression is dramatically induced in Th17 cells following activation, facilitating the expression of many critical components of the glycolytic pathway (186). Inhibition of glycolysis by 2-DG treatment strongly inhibits the development of Th17 cells. The development of Th17 cells is also inhibited by the absence of HIF-1α. In addition to facilitating glycolytic metabolism, HIF-1 also appears to directly stabilize RORγt expression (183). Since Th17 cells are glycolytic, the addition of exogenous fatty acids to skewing cultures, as well as the direct ligation of the fatty acid sensing receptor PPARγ, serves to suppress Th17 development (90, 189). Interestingly, the activation of the transcription factor SREBP, which facilitates the expression of many genes involved in lipogenesis and cholesterol synthesis also impairs the development of Th17 cells (190). The precise significance of this finding from a metabolic perspective is unclear.
Regulatory T cells
Regulatory T cells are generated naturally in the thymus (nTregs), as well as in the periphery as a result of TGFβ (iTregs) or IL-35 (Tr35) exposure following TCR engagement (191, 192). In contrast to the highly glycolytic metabolic phenotype of effector CD4+ T-cell subsets, regulatory T cells display a much more oxidative metabolic profile utilizing fatty acid oxidation and mitochondrial respiration to derive energy (90). Furthermore, the treatment of naive CD4+ T cells with compounds that inhibit the process of glycolysis, such as 2-DG or rapamycin, profoundly enhance the development of FoxP3+ regulatory T cells (90, 186). Conversely, inhibiting the process of fatty acid oxidation with the irreversible carnitine palmitoyltransferase I inhibitor etomoxir antagonizes the development of regulatory T cells, while not effecting the development of IFNγ–secreting Th1 cells (90). Inclusion of supplemental fatty acids to culture conditions facilitates the development of regulatory T cells. Furthermore, ligation of the fatty acid sensing proteins PPARα and PPARγ can facilitate the conversion of CD4+CD25− T cells into FoxP3+ regulatory T cells in the presence of TGFβ (193). PPARγ activity in particular appears to play a critical role in the development and stability of FoxP3+ regulatory T cells (194, 195). As mentioned previously, the expression of HIF-1α is critical for the development of Th17 cells. HIF-1 directly antagonizes the development of FoxP3+ regulatory T cells and deletion of HIF-1α in CD4+ T cells facilitates the development of FoxP3+ regulatory cells (183, 186).
In contrast to their effector counterparts, regulatory T cells exhibit low levels of mTOR activity throughout their lifespan, and their development is facilitated by culture conditions conducive to low mTOR signaling (196-199). While it does appear that regulatory T cells transiently up regulate mTOR activity during their initial activation phase (200), mTOR-deficient CD4+ T cells readily differentiate towards a regulatory T-cell phenotype at the expense of effector differentiation (15). Pharmacologic inhibition of mTOR activity also facilitates the development of regulatory T cells and enhances TGFβ sensitivity (196, 201). Furthermore, stimulation of naive CD4+ T cells under conditions that facilitate low mTOR activity, such as sub-optimal antigen, the presence of low-affinity peptide, short periods of TCR stimulation, or the premature termination of TCR stimulation all facilitates the development of FoxP3+ regulatory T cells (199, 201-203).
Anergy
When Th1 CD4+ T cells are stimulated through the TCR in the absence of costimulation, this leads to a state of tolerance known as anergy (204, 205). Anergic cells fail to proliferate or secrete IL-2 upon rechallenge; however, they proliferate in response to exogenous IL-2 which reverses the anergic state. Full activation (or conversely stated, the prevention of anergy) has been linked to TCR engagement in the presence of robust mTOR activity. Indeed, the PI3K-mediated mTOR activity induced by TCR stimulation alone is insufficient to prevent anergy (206). Likewise, robust stimulation of Th1 cells in the presence of the mTOR inhibitor rapamycin will lead to anergy even in the presence of full costimulation (29, 207). Along these lines, anergy can be induced by activating Th1 cells in the presence of the metabolic inhibitors AICAR, 2-DG, and NALA, all of which, by different mechanisms, inhibit mTOR activity. Interestingly, anergic T cells demonstrate a failure to upregulate the metabolic machinery necessary for full activation. That is, upon full stimulation anergic T cells demonstrate mitigated mTOR activity, a failure to upregulate the transferrin receptor, a failure to upregulate the extracellular amino acid transport pathway (CD98) and a decrease in glycolysis. This state of metabolic anergy is also reversed by stimulation with exogenous IL-2. These observations suggest that the state of non-responsiveness characteristic of anergy may in fact be maintained in part by the failure to upregulate the necessary metabolic machinery.
CD8+ memory T-cell development
Inasmuch as the initiation phase of a T-cell-mediated immune response is accompanied by a dramatic upregulation in mTOR signaling, metabolic activity and glycolysis, the generation of a stable pool of memory T cells is reliant on a corresponding decrease in mTOR signaling and a transition to a non-glycolytic metabolic profile. The pharmacological inhibition of mTOR signaling by the administration of low-dose rapamycin during an active LCMV infection can markedly increase the number of antigen specific memory CD8+ T cells without inhibiting the ability of the infected animal to clear the pathogen (16). Furthermore, long-lived antigen-specific CD8+ T cells can be generated in vitro by the addition of rapamycin resulting in a stable pool of memory cells that persist when adoptively transferred into a recipient animal (17). The inclusion of rapamycin during T-cell stimulation facilitates the development of memory CD8+ T cells in part by inhibiting the expression of Tbet, while simultaneously stabilizing the expression of the related transcription factor Eomesodermin (18).
mTOR’s regulation of Tbet and eomesodermin expression has also been show to play a critical role in the process of homeostatic proliferation (HP)-induced memory CD8+ T-cell development (208). CD8+ T cells adoptively transferred into lymphopenic hosts rapidly acquire a memory cell phenotype in a process dependent on major histocompatibility complex (MHC) class I expressed self-peptide and the cytokines IL-7 and IL-15 (209, 210). Although not as potent as classically induced memory CD8+ T cells, these HP-induced memory CD8+ T cells are capable of rapid proliferation upon antigenic rechallenge, and possess potent anti-tumor capability (211, 212). Li et al. (208) have recently shown that the development of HP-derived memory CD8+ T cells is dependent on mTOR.
Recent work from the laboratory of Erika Pearce (30, 31) has highlighted the significant metabolic changes that a CD8+ T cell must undergo during the effector to memory cell transition. It was found that tumor necrosis factor receptor-associated factor 6 (TRAF6)-null T cells could become effector cells but failed to develop into memory T cells. Upon further analysis it was observed that the TRAF6-deficient CD8+ T cells exhibit a profound defect in their ability to up regulate genes associated with fatty acid oxidation. This was accompanied by an inability to induce mitochondrial fatty acid oxidation following growth factor withdrawal. Treatment with rapamycin or metformin rescued the memory defect in TRAF6-deficient CD8+ T cells and further enhanced the development of memory CD8+ T cells wildtype animals. Subsequently, this group went on to demonstrate that the effector to memory transition of CD8+ T cells is accompanied by a dramatic increase in the number of mitochondria in the cells. This results in an increase in the oxygen consumption of memory cells and the development of increased spare respiratory capacity (31).
Conclusion
In this review, we highlight the role of mTOR in regulating T-cell differentiation and function as well as the role of mTOR in regulating cellular metabolic function. Clearly, the metabolic demands of different subsets of T cells are quite diverse. To this end, the metabolic machinery plays an important role in regulating the outcome of T-cell activation. In this regard, we tried to accentuate the potential intersection between mTOR signaling and metabolic regulation and the ability of mTOR to guide the outcome of antigen recognition. In many cases, the details of this intersection have yet to be defined. However, the elucidation of these precise signaling pathways will provide the opportunity to specifically target mediators downstream of mTOR to selectively regulate T-cell responses. Likewise, pharmacologic inhibition of specific metabolic pathways might greatly facilitate the enhancement and/or inhibition of selective T-cell function.
Acknowledgments
We thank Drs. Fan Pan, Thi Bui, and Michael Wolfgang for their invaluable editorial assistance. This work was supported by NIAID grants, R01AI077610 and R01 AI091481-01. The authors have no conflicts of interest to declare.
References
- 1.Lafferty KJ, Cunningham AJ. A new analysis of allogeneic interactions. Aust J Exp Biol Med Sci. 1975;53:27–42. doi: 10.1038/icb.1975.3. [DOI] [PubMed] [Google Scholar]
- 2.Bretscher P, Cohn M. A theory of self-nonself discrimination. Science. 1970;169:1042–1049. doi: 10.1126/science.169.3950.1042. [DOI] [PubMed] [Google Scholar]
- 3.Powell JD, Ragheb JA, Kitagawa-Sakakida S, Schwartz RH. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol Rev. 1998;165:287–300. doi: 10.1111/j.1600-065x.1998.tb01246.x. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins MK, Taylor PS, Norton SD, Urdahl KB. CD28 delivers a costimulatory signal involved in antigen-specific IL-2 production by human T cells. J Immunol. 1991;147:2461–2466. [PubMed] [Google Scholar]
- 5.Colombetti S, Basso V, Mueller DL, Mondino A. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J Immunol. 2006;176:2730–2738. doi: 10.4049/jimmunol.176.5.2730. [DOI] [PubMed] [Google Scholar]
- 6.O’Garra A. Cytokines induce the development of functionally heterogeneous T helper cell subsets. Immunity. 1998;8:275–283. doi: 10.1016/s1074-7613(00)80533-6. [DOI] [PubMed] [Google Scholar]
- 7.Soroosh P, Doherty TA. Th9 and allergic disease. Immunology. 2009;127:450–458. doi: 10.1111/j.1365-2567.2009.03114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;18:349–356. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Rudensky AY. Regulatory T cells and Foxp3. Immunol Rev. 2011;241:260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powell JD, Delgoffe GM. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 2010;33:301–311. doi: 10.1016/j.immuni.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgoffe GM, Powell JD. mTOR: taking cues from the immune microenvironment. Immunology. 2009;127:459–465. doi: 10.1111/j.1365-2567.2009.03125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. J Immunol. 1999;162:2775–2784. [PubMed] [Google Scholar]
- 15.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He S, et al. Characterization of the metabolic phenotype of rapamycin-treated CD8 T cells with augmented ability to generate long-lasting memory cells. PLoS One. 2011;6:e20107. doi: 10.1371/journal.pone.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, et al. Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood. 2011;117:1228–1238. doi: 10.1182/blood-2010-05-287821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazorchak AS, et al. Sin1-mTORC2 suppresses rag and il7r gene expression through Akt2 in B cells. Mol Cell. 2010;39:433–443. doi: 10.1016/j.molcel.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sathaliyawala T, et al. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity. 2010;33:597–606. doi: 10.1016/j.immuni.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turnquist HR, Raimondi G, Zahorchak AF, Fischer RT, Wang Z, Thomson AW. Rapamycin-conditioned dendritic cells are poor stimulators of allogeneic CD4+ T cells, but enrich for antigen-specific Foxp3+ T regulatory cells and promote organ transplant tolerance. J Immunol. 2007;178:7018–7031. doi: 10.4049/jimmunol.178.11.7018. [DOI] [PubMed] [Google Scholar]
- 23.Weichhart T, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 25.Bauer DE, et al. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J. 2004;18:1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frauwirth KA, Thompson CB. Regulation of T lymphocyte metabolism. J Immunol. 2004;172:4661–4665. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- 27.Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 28.Carr EL, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. J Immunol. 2009;183:6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamagata K, et al. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- 33.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 34.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 35.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 37.Beugnet A, Tee AR, Taylor PM, Proud CG. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J. 2003;372:555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 41.Chen CH, et al. ER stress inhibits mTORC2 and Akt signaling through GSK-3beta-mediated phosphorylation of rictor. Sci Signal. 2011;4:ra10. doi: 10.1126/scisignal.2001731. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 43.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Exley M, Varticovski L, Peter M, Sancho J, Terhorst C. Association of phosphatidylinositol 3-kinase with a specific sequence of the T cell receptor zeta chain is dependent on T cell activation. J Biol Chem. 1994;269:15140–15146. [PubMed] [Google Scholar]
- 47.Harada Y, et al. Critical requirement for the membrane-proximal cytosolic tyrosine residue for CD28-mediated costimulation in vivo. J Immunol. 2001;166:3797–3803. doi: 10.4049/jimmunol.166.6.3797. [DOI] [PubMed] [Google Scholar]
- 48.Kane LP, Andres PG, Howland KC, Abbas AK, Weiss A. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-gamma but not TH2 cytokines. Nat Immunol. 2001;2:37–44. doi: 10.1038/83144. [DOI] [PubMed] [Google Scholar]
- 49.Fos C, et al. ICOS ligation recruits the p50alpha PI3K regulatory subunit to the immunological synapse. J Immunol. 2008;181:1969–1977. doi: 10.4049/jimmunol.181.3.1969. [DOI] [PubMed] [Google Scholar]
- 50.Xiao X, et al. New Insights on OX40 in the Control of T Cell Immunity and Immune Tolerance In Vivo. J Immunol. 2012;188:892–901. doi: 10.4049/jimmunol.1101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.So T, Choi H, Croft M. OX40 complexes with phosphoinositide 3-kinase and protein kinase B (PKB) to augment TCR-dependent PKB signaling. J Immunol. 2011;186:3547–3555. doi: 10.4049/jimmunol.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruby CE, Redmond WL, Haley D, Weinberg AD. Anti-OX40 stimulation in vivo enhances CD8+ memory T cell survival and significantly increases recall responses. Eur J Immunol. 2007;37:157–166. doi: 10.1002/eji.200636428. [DOI] [PubMed] [Google Scholar]
- 53.Parry RV, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell BIol. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuang E, et al. The CD28 and CTLA-4 receptors associate with the serine/threonine phosphatase PP2A. Immunity. 2000;13:313–322. doi: 10.1016/s1074-7613(00)00031-5. [DOI] [PubMed] [Google Scholar]
- 55.Francisco LM, et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abraham RT, Wiederrecht GJ. Immunopharmacology of rapamycin. Annu Rev Immunol. 1996;14:483–510. doi: 10.1146/annurev.immunol.14.1.483. [DOI] [PubMed] [Google Scholar]
- 57.Stephenson LM, Park DS, Mora AL, Goenka S, Boothby M. Sequence motifs in IL-4R alpha mediating cell-cycle progression of primary lymphocytes. J Immunol. 2005;175:5178–5185. doi: 10.4049/jimmunol.175.8.5178. [DOI] [PubMed] [Google Scholar]
- 58.Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004;200:659–669. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curnock AP, Ward SG. Development and characterisation of tetracycline-regulated phosphoinositide 3-kinase mutants: assessing the role of multiple phosphoinositide 3-kinases in chemokine signaling. J Immunol Methods. 2003;273:29–41. doi: 10.1016/s0022-1759(02)00416-7. [DOI] [PubMed] [Google Scholar]
- 60.Sasaki T, et al. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 61.Liu G, et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10:769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat Immunol. 2010;11:1047–1056. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uddin S, Yenush L, Sun XJ, Sweet ME, White MF, Platanias LC. Interferon-alpha engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3′-kinase. J Biol Chem. 1995;270:15938–15941. doi: 10.1074/jbc.270.27.15938. [DOI] [PubMed] [Google Scholar]
- 64.Platanias LC, Uddin S, Yetter A, Sun XJ, White MF. The type I interferon receptor mediates tyrosine phosphorylation of insulin receptor substrate 2. J Biol Chem. 1996;271:278–282. doi: 10.1074/jbc.271.1.278. [DOI] [PubMed] [Google Scholar]
- 65.Navarro A, Anand-Apte B, Tanabe Y, Feldman G, Larner AC. A PI-3 kinase-dependent, Stat1-independent signaling pathway regulates interferon-stimulated monocyte adhesion. J Leukoc Biol. 2003;73:540–545. doi: 10.1189/jlb.1002508. [DOI] [PubMed] [Google Scholar]
- 66.Myers MG., Jr Leptin receptor signaling and the regulation of mammalian physiology. Rec Prog Hormone Res. 2004;59:287–304. doi: 10.1210/rp.59.1.287. [DOI] [PubMed] [Google Scholar]
- 67.Kellerer M, Koch M, Metzinger E, Mushack J, Capp E, Haring HU. Leptin activates PI-3 kinase in C2C12 myotubes via janus kinase-2 (JAK-2) and insulin receptor substrate-2 (IRS-2) dependent pathways. Diabetologia. 1997;40:1358–1362. doi: 10.1007/s001250050832. [DOI] [PubMed] [Google Scholar]
- 68.Galgani M, et al. Leptin modulates the survival of autoreactive CD4+ T cells through the nutrient/energy-sensing mammalian target of rapamycin signaling pathway. J Immunol. 2010;185:7474–7479. doi: 10.4049/jimmunol.1001674. [DOI] [PubMed] [Google Scholar]
- 69.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 74.Feng Z, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 75.Ellisen LW, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 76.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 77.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun Q, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci USA. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iyer NV, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tozzi MG, Camici M, Mascia L, Sgarrella F, Ipata PL. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 2006;273:1089–1101. doi: 10.1111/j.1742-4658.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 83.Christie GR, Hajduch E, Hundal HS, Proud CG, Taylor PM. Intracellular sensing of amino acids in Xenopus laevis oocytes stimulates p70 S6 kinase in a target of rapamycin-dependent manner. J Biol Chem. 2002;277:9952–9957. doi: 10.1074/jbc.M107694200. [DOI] [PubMed] [Google Scholar]
- 84.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cohen A, Hall MN. An amino acid shuffle activates mTORC1. Cell. 2009;136:399–400. doi: 10.1016/j.cell.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 87.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sipula IJ, Brown NF, Perdomo G. Rapamycin-mediated inhibition of mammalian target of rapamycin in skeletal muscle cells reduces glucose utilization and increases fatty acid oxidation. Metab Clin Exp. 2006;55:1637–1644. doi: 10.1016/j.metabol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 89.Brown NF, Stefanovic-Racic M, Sipula IJ, Perdomo G. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metab Clin Exp. 2007;56:1500–1507. doi: 10.1016/j.metabol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 90.Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schieke SM, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 92.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 93.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 94.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 95.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell SCi. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 96.Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 97.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 98.Ng S, Wu YT, Chen B, Zhou J, Shen HM. Impaired autophagy due to constitutive mTOR activation sensitizes TSC2-null cells to cell death under stress. Autophagy. 2011;7:1173–1186. doi: 10.4161/auto.7.10.16681. [DOI] [PubMed] [Google Scholar]
- 99.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 100.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 103.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 104.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 105.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 106.Seagroves TN, et al. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21:3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 108.Hudson CC, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Majumder PK, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 110.Nakamura H, et al. TCR engagement increases hypoxia-inducible factor-1 alpha protein synthesis via rapamycin-sensitive pathway under hypoxic conditions in human peripheral T cells. J Immunol. 2005;174:7592–7599. doi: 10.4049/jimmunol.174.12.7592. [DOI] [PubMed] [Google Scholar]
- 111.Treins C, Giorgetti-Peraldi S, Murdaca J, Monthouel-Kartmann MN, Van Obberghen E. Regulation of hypoxia-inducible factor (HIF)-1 activity and expression of HIF hydroxylases in response to insulin-like growth factor I. Mol Endocrinol. 2005;19:1304–1317. doi: 10.1210/me.2004-0239. [DOI] [PubMed] [Google Scholar]
- 112.Zhou J, et al. Regulation of hypoxia-inducible factor 1 by glucose availability under hypoxic conditions. Kobe J Med Sci. 2007;53:283–296. [PubMed] [Google Scholar]
- 113.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 115.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22:9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 117.Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 118.West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 119.Murooka TT, Rahbar R, Fish EN. CCL5 promotes proliferation of MCF-7 cells through mTOR-dependent mRNA translation. Biochem Biophys Res Commun. 2009;387:381–386. doi: 10.1016/j.bbrc.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 120.Shi Y, Sharma A, Wu H, Lichtenstein A, Gera J. Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J Biol Chem. 2005;280:10964–10973. doi: 10.1074/jbc.M407874200. [DOI] [PubMed] [Google Scholar]
- 121.Marderosian M, et al. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene. 2006;25:6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- 122.Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem. 2008;283:23274–23287. doi: 10.1074/jbc.M801185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vartanian R, et al. AP-1 regulates cyclin D1 and c-MYC transcription in an AKT-dependent manner in response to mTOR inhibition: role of AIP4/Itch-mediated JUNB degradation. Mol Cancer Res. 2011;9:115–130. doi: 10.1158/1541-7786.MCR-10-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmidt EV, Ravitz MJ, Chen L, Lynch M. Growth controls connect: interactions between c-myc and the tuberous sclerosis complex-mTOR pathway. Cell Cycle. 2009;8:1344–1351. doi: 10.4161/cc.8.9.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hosoi H, et al. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol Pharmacol. 1998;54:815–824. doi: 10.1124/mol.54.5.815. [DOI] [PubMed] [Google Scholar]
- 126.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kliewer SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci U A. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hostetler HA, Petrescu AD, Kier AB, Schroeder F. Peroxisome proliferator-activated receptor alpha interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem. 2005;280:18667–18682. doi: 10.1074/jbc.M412062200. [DOI] [PubMed] [Google Scholar]
- 130.Xu HE, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 131.Brandt JM, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J Biol CHem. 1998;273:23786–23792. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 132.Rodriguez JC, Gil-Gomez G, Hegardt FG, Haro D. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. J Biol Chem. 1994;269:18767–18772. [PubMed] [Google Scholar]
- 133.Motojima K, Passilly P, Peters JM, Gonzalez FJ, Latruffe N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor alpha and gamma activators in a tissue- and inducer-specific manner. J Biol Chem. 1998;273:16710–16714. doi: 10.1074/jbc.273.27.16710. [DOI] [PubMed] [Google Scholar]
- 134.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Delerive P, et al. Peroxisome proliferator-activated receptor activators inhibit thrombin-induced endothelin-1 production in human vascular endothelial cells by inhibiting the activator protein-1 signaling pathway. Circ Res. 1999;85:394–402. doi: 10.1161/01.res.85.5.394. [DOI] [PubMed] [Google Scholar]
- 136.Delerive P, Gervois P, Fruchart JC, Staels B. Induction of IkappaBalpha expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-alpha activators. J Biol Chem. 2000;275:36703–36707. doi: 10.1074/jbc.M004045200. [DOI] [PubMed] [Google Scholar]
- 137.Delerive P, et al. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappaB and AP-1. J Biol Chem. 1999;274:32048–32054. doi: 10.1074/jbc.274.45.32048. [DOI] [PubMed] [Google Scholar]
- 138.Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 139.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 140.Fajas L, Fruchart JC, Auwerx J. PPARgamma3 mRNA: a distinct PPARgamma mRNA subtype transcribed from an independent promoter. FEBS Lett. 1998;438:55–60. doi: 10.1016/s0014-5793(98)01273-3. [DOI] [PubMed] [Google Scholar]
- 141.Spiegelman BM, Flier JS. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 142.Rosen ED, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 143.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 144.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 145.Kim JE, Chen J. regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 146.Zhang HH, et al. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One. 2009;4:e6189. doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 148.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bakan I, Laplante M. Connecting mTORC1 signaling to SREBP-1 activation. Curr Opin Lipidol. 2012;23:226–234. doi: 10.1097/MOL.0b013e328352dd03. [DOI] [PubMed] [Google Scholar]
- 150.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Amemiya-Kudo M, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 152.Peterson TR, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.De Boer RJ, Oprea M, Antia R, Murali-Krishna K, Ahmed R, Perelson AS. Recruitment times, proliferation, and apoptosis rates during the CD8(+) T-cell response to lymphocytic choriomeningitis virus. J Virol. 2001;75:10663–10669. doi: 10.1128/JVI.75.22.10663-10669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol. 2012;13:121–128. doi: 10.1038/ni.2190. [DOI] [PubMed] [Google Scholar]
- 155.Kelly AP, et al. Notch-induced T cell development requires phosphoinositide-dependent kinase 1. EMBO J. 2007;26:3441–3450. doi: 10.1038/sj.emboj.7601761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tamas P, et al. LKB1 is essential for the proliferation of T-cell progenitors and mature peripheral T cells. Eur J Immunol. 2010;40:242–253. doi: 10.1002/eji.200939677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 158.Lee K, et al. Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J Exp Med. 2012;209:713–728. doi: 10.1084/jem.20111470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wu Q, et al. The tuberous sclerosis complex-mammalian target of rapamycin pathway maintains the quiescence and survival of naive T cells. J Immunol. 2011;187:1106–1112. doi: 10.4049/jimmunol.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.O’Brien TF, et al. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. Eur J Immunol. 2011;41:3361–3370. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 163.Li C, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 164.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 166.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wahl DR, Petersen B, Warner R, Richardson BC, Glick GD, Opipari AW. Characterization of the metabolic phenotype of chronically activated lymphocytes. Lupus. 2010;19:1492–1501. doi: 10.1177/0961203310373109. [DOI] [PubMed] [Google Scholar]
- 168.Gatza E, et al. Manipulating the bioenergetics of alloreactive T cells causes their selective apoptosis and arrests graft-versus-host disease. Sci Transl Med. 2011;3:67ra68. doi: 10.1126/scitranslmed.3001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gergely P, Jr., Niland B, Gonchoroff N, Pullmann R, Jr., Phillips PE, Perl A. Persistent mitochondrial hyperpolarization, increased reactive oxygen intermediate production, and cytoplasmic alkalinization characterize altered IL-10 signaling in patients with systemic lupus erythematosus. J Immunol. 2002;169:1092–1101. doi: 10.4049/jimmunol.169.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gergely P, Jr., et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:175–190. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 172.Dunn SE, et al. Peroxisome proliferator-activated receptor (PPAR)alpha expression in T cells mediates gender differences in development of T cell-mediated autoimmunity. J Exp Med. 2007;204:321–330. doi: 10.1084/jem.20061839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Clark RB, Bishop-Bailey D, Estrada-Hernandez T, Hla T, Puddington L, Padula SJ. The nuclear receptor PPAR gamma and immunoregulation: PPAR gamma mediates inhibition of helper T cell responses. J Immunol. 2000;164:1364–1371. doi: 10.4049/jimmunol.164.3.1364. [DOI] [PubMed] [Google Scholar]
- 174.Cunard R, Eto Y, Muljadi JT, Glass CK, Kelly CJ, Ricote M. Repression of IFN-gamma expression by peroxisome proliferator-activated receptor gamma. J Immunol. 2004;172:7530–7536. doi: 10.4049/jimmunol.172.12.7530. [DOI] [PubMed] [Google Scholar]
- 175.Bassaganya-Riera J, et al. Activation of PPAR gamma and delta by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology. 2004;127:777–791. doi: 10.1053/j.gastro.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 176.Beales PE, et al. Troglitazone prevents insulin dependent diabetes in the non-obese diabetic mouse. Eur J Pharmacol. 1998;357:221–225. doi: 10.1016/s0014-2999(98)00574-3. [DOI] [PubMed] [Google Scholar]
- 177.Diab A, et al. Peroxisome proliferator-activated receptor-gamma agonist 15-deoxy-Delta(12,14)-prostaglandin J(2) ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2002;168:2508–2515. doi: 10.4049/jimmunol.168.5.2508. [DOI] [PubMed] [Google Scholar]
- 178.Feinstein DL, et al. Peroxisome proliferator-activated receptor-gamma agonists prevent experimental autoimmune encephalomyelitis. Ann Neurol. 2002;51:694–702. doi: 10.1002/ana.10206. [DOI] [PubMed] [Google Scholar]
- 179.Mueller C, Weaver V, Vanden Heuvel JP, August A, Cantorna MT. Peroxisome proliferator-activated receptor gamma ligands attenuate immunological symptoms of experimental allergic asthma. Arch Biochem Biophys. 2003;418:186–196. doi: 10.1016/j.abb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 180.Housley WJ, et al. Peroxisome proliferator-activated receptor gamma is required for CD4+ T cell-mediated lymphopenia-associated autoimmunity. J Immunol. 2011;187:4161–4169. doi: 10.4049/jimmunol.1101731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guo J, Lu W, Shimoda LA, Semenza GL, Georas SN. Enhanced interferon-gamma gene expression in T Cells and reduced ovalbumin-dependent lung eosinophilia in hypoxia-inducible factor-1-alpha-deficient mice. Int Arch Allergy Immunol. 2009;149:98–102. doi: 10.1159/000189191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lukashev D, et al. Cutting edge: hypoxia-inducible factor 1alpha and its activation-inducible short isoform I.1 negatively regulate functions of CD4+ and CD8+ T lymphocytes. J Immunol. 2006;177:4962–4965. doi: 10.4049/jimmunol.177.8.4962. [DOI] [PubMed] [Google Scholar]
- 183.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 185.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 186.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chung SW, Kang BY, Kim TS. Inhibition of interleukin-4 production in CD4+ T cells by peroxisome proliferator-activated receptor-gamma (PPAR-gamma) ligands: involvement of physical association between PPAR-gamma and the nuclear factor of activated T cells transcription factor. Mol Pharmacol. 2003;64:1169–1179. doi: 10.1124/mol.64.5.1169. [DOI] [PubMed] [Google Scholar]
- 188.Peters A, Lee Y, Kuchroo VK. The many faces of Th17 cells. Curr Opin Immunol. 2011;23:702–706. doi: 10.1016/j.coi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Klotz L, et al. The nuclear receptor PPAR gamma selectively inhibits Th17 differentiation in a T cell-intrinsic fashion and suppresses CNS autoimmunity. J Exp Med. 2009;206:2079–2089. doi: 10.1084/jem.20082771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cui G, et al. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J Clin Invest. 2011;121:658–670. doi: 10.1172/JCI42974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Collison LW, et al. IL-35-mediated induction of a potent regulatory T cell population. Nat Immunol. 2010;11:1093–1101. doi: 10.1038/ni.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lei J, Hasegawa H, Matsumoto T, Yasukawa M. Peroxisome proliferator-activated receptor alpha and gamma agonists together with TGF-beta convert human CD4+CD25-T cells into functional Foxp3+ regulatory T cells. J Immunol. 2010;185:7186–7198. doi: 10.4049/jimmunol.1001437. [DOI] [PubMed] [Google Scholar]
- 194.Wohlfert EA, Nichols FC, Nevius E, Clark RB. Peroxisome proliferator-activated receptor gamma (PPARgamma) and immunoregulation: enhancement of regulatory T cells through PPARgamma-dependent and -independent mechanisms. J Immunol. 2007;178:4129–4135. doi: 10.4049/jimmunol.178.7.4129. [DOI] [PubMed] [Google Scholar]
- 195.Hontecillas R, Bassaganya-Riera J. Peroxisome proliferator-activated receptor gamma is required for regulatory CD4+ T cell-mediated protection against colitis. J Immunol. 2007;178:2940–2949. doi: 10.4049/jimmunol.178.5.2940. [DOI] [PubMed] [Google Scholar]
- 196.Kang J, Huddleston SJ, Fraser JM, Khoruts A. De novo induction of antigen-specific CD4+CD25+Foxp3+ regulatory T cells in vivo following systemic antigen administration accompanied by blockade of mTOR. J Leukoc Biol. 2008;83:1230–1239. doi: 10.1189/jlb.1207851. [DOI] [PubMed] [Google Scholar]
- 197.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zeiser R, et al. Differential impact of mammalian target of rapamycin inhibition on CD4+CD25+Foxp3+ regulatory T cells compared with conventional CD4+ T cells. Blood. 2008;111:453–462. doi: 10.1182/blood-2007-06-094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sauer S, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Procaccini C, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gabrysova L, Christensen JR, Wu X, Kissenpfennig A, Malissen B, O’Garra A. Integrated T-cell receptor and costimulatory signals determine TGF-beta-dependent differentiation and maintenance of Foxp3+ regulatory T cells. Eur J Immunol. 2011;41:1242–1248. doi: 10.1002/eji.201041073. [DOI] [PubMed] [Google Scholar]
- 202.Katzman SD, et al. Duration of antigen receptor signaling determines T-cell tolerance or activation. Proc Natl Acad Sci USA. 2010;107:18085–18090. doi: 10.1073/pnas.1010560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gottschalk RA, Corse E, Allison JP. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J Exp Med. 2010;207:1701–1711. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chappert P, Schwartz RH. Induction of T cell anergy: integration of environmental cues and infectious tolerance. Curr Opin Immunol. 2010;22:552–559. doi: 10.1016/j.coi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 206.Zheng Y, et al. A role for mammalian target of rapamycin in regulating T cell activation versus anergy. J Immunol. 2007;178:2163–2170. doi: 10.4049/jimmunol.178.4.2163. [DOI] [PubMed] [Google Scholar]
- 207.Vanasek TL, Khoruts A, Zell T, Mueller DL. Antagonistic roles for CTLA-4 and the mammalian target of rapamycin in the regulation of clonal anergy: enhanced cell cycle progression promotes recall antigen responsiveness. J Immunol. 2001;167:5636–5644. doi: 10.4049/jimmunol.167.10.5636. [DOI] [PubMed] [Google Scholar]
- 208.Li Q, et al. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–553. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cheung KP, Yang E, Goldrath AW. Memory-like CD8+ T cells generated during homeostatic proliferation defer to antigen-experienced memory cells. J Immunol. 2009;183:3364–3372. doi: 10.4049/jimmunol.0900641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Cho BK, Rao VP, Ge Q, Eisen HN, Chen J. Homeostasis-stimulated proliferation drives naive T cells to differentiate directly into memory T cells. J Exp Med. 2000;192:549–556. doi: 10.1084/jem.192.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dummer W, et al. T cell homeostatic proliferation elicits effective antitumor autoimmunity. J Clin Invest. 2002;110:185–192. doi: 10.1172/JCI15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ma J, Urba WJ, Si L, Wang Y, Fox BA, Hu HM. Anti-tumor T cell response and protective immunity in mice that received sublethal irradiation and immune reconstitution. Eur J Immunol. 2003;33:2123–2132. doi: 10.1002/eji.200324034. [DOI] [PubMed] [Google Scholar]