Abstract
Genome-wide DNA elimination accompanies development of the somatic macronucleus from the germ-line micronucleus during the sexual process of conjugation in the ciliated protozoan Tetrahymena thermophila. Small RNAs, referred to as “scan RNAs” (scnRNAs), that accumulate only during conjugation are highly enriched in the eliminated sequences, and mutations that prevent DNA elimination also affect the accumulation of scnRNAs, suggesting that an RNA interference (RNAi)-like mechanism is involved in DNA elimination. Histone H3 that is methylated at lysine 9 (K9) is a hallmark of heterochromatin and, in Tetrahymena, is found only in developing macronuclei (anlagen) in association with chromatin containing the sequences undergoing elimination. In this article, we demonstrate that a mutation in the TWI1 gene that eliminates the accumulation of scnRNAs also abolishes H3 methylation at K9. We created mutant strains of Tetrahymena in which the only major H3 contained a K9Q mutation. These mutants accumulated scnRNAs normally during conjugation but showed dramatically reduced efficiency of DNA elimination. These results provide strong genetic evidence linking an RNAi-like pathway, H3 K9 methylation, and DNA elimination in Tetrahymena.
Histone H3 lysine 9 (K9) methylation is an epigenetic mark of heterochromatin (1–3) that is recognized by some chromodomain-containing proteins, including HP1, a major heterochromatin component (4–6). Histone H3 methylated at K9 has been implicated in the formation and maintenance of heterochromatin and gene repression in centromeric regions in Schizosaccharomyces pombe (7), Drosophila (8), and mammalian cells (9–11).
Small interfering RNAs (siRNAs) produced by degradation of double-stranded or hairpin RNAs can regulate gene expression by degrading homologous mRNAs specifically by using a conserved mechanism known in various systems as posttranscriptional gene silencing, RNAi, or RNA silencing (12). Similar mechanisms have been implicated also in transcriptional gene silencing in plants (13) and Drosophila (14), as well as in cosuppression of endogenous genes by transgenes in the Caenorhabditis elegans germ line (15). In S. pombe, genes for siRNA production are required for heterochromatin formation and gene repression both in the centromere regions (16) and at the silent mating-type locus (17), and siRNAs homologous to centromeric repeats have been detected in wild-type cells (16). Elimination of siRNA production abolishes H3 K9 methylation, suggesting that siRNAs and K9 methylation are in the same pathway in S. pombe. Formation of heterochromatin in S. pombe also requires participation of Swi6p (18), a chromodomain protein homologous to HP1 that also binds K9-methylated H3.
A correlation among siRNAs, H3 K9 methylation, chromodomain proteins, and heterochromatin formation has been observed also in the developing macronuclei (anlagen) of the ciliated protozoan Tetrahymena thermophila. T. thermophila has two morphologically and functionally distinct nuclei, a small germ-line micronucleus, and a large somatic macronucleus (19). During conjugation, the sexual phase of the Tetrahymena life cycle, old macronuclei are destroyed and new macronuclei are derived from the micronuclei. By deletion and ligation, ≈6,000 interstitial DNA elements are removed from the developing macronuclei (20). These excised DNAs, referred to as internal eliminated sequences (IESs), consist mostly of moderately repeated sequences. No conserved cis-acting elements have been found among Tetrahymena IESs characterized so far. When an IES is transformed into vegetative macronuclei of Tetrahymena cells, that IES specifically fails to be eliminated from newly formed macronuclei when those cells conjugate (21), indicating that there is a sequence-specific epigenetic mechanism whereby sequences in the old macronucleus can affect elimination of IESs in the developing new macronucleus.
Both small RNAs and histone H3 K9 methylation have been implicated in the epigenetic mechanism of DNA elimination in Tetrahymena (22, 23). Heterogeneous, bidirectional transcripts of IESs from micronuclei have been detected during early conjugation (24), suggesting the existence of double-stranded RNAs that could be processed by an RNAi-like mechanism. Very recently, it has been shown that microinjection of double-stranded RNAs into conjugating Tetrahymena can result in specific elimination of the homologous sequences from the anlagen (25). Small RNAs, referred to as “scan RNAs” (scnRNAs), similar to siRNAs in other systems also accumulate during conjugation. They are enriched in micronuclear-limited sequences (22). These small RNAs are referred to as “scan RNAs” because they likely play a role in scanning the Tetrahymena genome and discriminating between sequences that should be retained and eliminated during macronuclear development (22). In Tetrahymena, histone H3 K9 methylation occurs only during conjugation, and it colocalizes (26) and is associated (23) with IESs in a heterochromatic structure.
Two genes, TWI1 and PDD1, are required for both accumulation of scnRNAs and DNA elimination (22, 23, 26, 27). TWI1 is a member of the PPD (PAZ and Piwi domains) gene family, which includes homologues of Piwi and Ago1, which are involved in RNAi pathways in other systems (22). Knocking out TWI1 eliminates the accumulation of scnRNAs and prevents IES elimination and completion of conjugation (22). PDD1 is an abundant chromodomain protein of unknown function that can bind K9-methylated histone H3 in vitro (23). It is present only during conjugation and associates with IESs in a heterochromatic structure (26, 27). Eliminating parental PDD1 expression abolishes scnRNA accumulation during early conjugation (22), greatly reduces K9 methylation (23), and causes conjugation failure (23). Histone hypoacetylation also is characteristic of heterochromatin in many systems (1, 2), and lysine must be deacetylated for methylation to occur. A histone deacetylase inhibitor severely inhibits IES elimination in Tetrahymena (28).
Taken together, the above observations are consistent with the hypothesis that small RNAs enriched in micronuclear-limited sequences target histone H3 K9 methylation to IESs in developing macronuclei, leading to formation of a heterochromatic structure required for IES excision. Here, we provide genetic evidence that scnRNAs are required for H3 K9 methylation, which, in turn, is required for efficient IES elimination.
Materials and Methods
Tetrahymena Strains and Culture Conditions. Wild-type CU428 and B2086 strains of T. thermophila, provided by Peter J. Bruns (Cornell University, Ithaca, NY) were grown in super proteose peptone medium (29) at 30°C. To initiate conjugation, log-phase cells of different mating types were starved in 10 mM Tris (pH 7.4) for 16–24 h at 30°C and mixed at a concentration of 2 × 105 cells per ml.
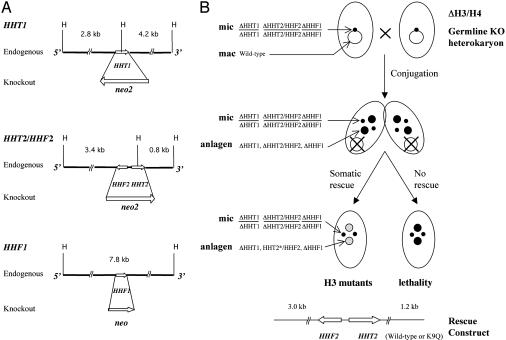
Construction of Germ-Line Knockout Heterokaryon and Somatic Rescue Strains. The knockout constructs for HHT1 and HHT2/HHF2 loci (divergently transcribed genes separated by >400 bp that were both disrupted in a single construct and treated as a single locus) were made by replacing the coding region of the corresponding gene with the neo2 cassette (30) that confers paromomycin resistance in Tetrahymena. The neo2 cassette was flanked by 1–4 kb of noncoding sequence on both ends to facilitate homologous recombination. The HHF1 knockout construct was made by replacing the HHF1 coding region with the neo coding region (31). To obtain germ-line transformants in which a single locus had been knocked out, the individual constructs were introduced into 2.5-h conjugating CU428 and B2086 cells by using the Biolistic PDS-1000/He particle-delivery system (Bio-Rad) as described (32). Germ-line knockout homozygous heterokaryons with a single disrupted locus (ΔHHT1, ΔHHT2/HHF2, or ΔHHF1) in both alleles in their micronuclei and wild-type genes in their macronuclei were created as described (30). Strains with germ-line knockouts of multiple loci were constructed by crossing the single gene knockout strains and further genetic manipulation (30). The ΔHHT1 strain was first crossed with the ΔHHT2/HHF2 strain. Progeny from this cross were then crossed with the ΔHHF1 strain to give rise to the ΔH3/H4 strain (ΔHHT1, ΔHHT2/HHF2, and ΔHHF1).
To demonstrate that the initial single-locus knockout heterokaryons had the intended genotypes, two germ-line knockout homozygous heterokaryon strains with the same genotype were crossed and the progeny were selected by paromomycin resistance. The derived homozygous homokaryon strains, which should contain the disrupted genes in both their micronuclear and macronuclear genomes, were then examined by Southern and Northern blot analyses to determine that they had inherited the expected disrupted gene from their knockout heterokaryon parents. The genotypes of all single-locus germ-line knockout strains were confirmed by this method.
As expected because they lacked genes for both the major H3 and H4, the progeny of mating between the two ΔH3/H4 triple knockout heterokaryon strains (ΔHHT1, ΔHHT2/HHF2, and ΔHHF1) were not viable. However, they could be rescued by transformation at late conjugation (24 h) with a construct containing the wild-type HHT2/HHF2 locus (see Fig. 2B). A construct with a null mutation in the HHT2 gene (K4Z mutation) could not rescue. Progeny cells of mating between two ΔH3/H4 germ-line knockout strains were transformed with DNA containing either a wild-type HHT2/HHF2 locus or a locus in which the codon for K9 had been mutated to glutamine to create the wild-type and K9Q rescued strains.
Fig. 2.
Generation of H3 K9Q mutants. (A) Restriction maps of the endogenous H3 and H4 loci in Tetrahymena and their corresponding knockout constructs. (B) Creation of strains with mutant histone H3 by somatic rescue of the two germ-line knockout heterokaryon strains ΔH3/H4. The strains were generated by crossing the appropriate single germ-line knockout strains and further genetic manipulation. The conjugation progeny of the ΔH3/H4 strains were not viable unless rescued with a construct carrying functional H3 and H4 genes.
Indirect Immunofluorescence Analysis. Cells were fixed in Lavdowsky's fixative (ethanol/formaldehyde/acetic acid/water; 50:10:1:39 dilution) overnight at 4°C and dried onto poly-l-lysine-coated cover slips. They were then incubated with the appropriate primary antibody [1:50 dilution for anti-dimethyl K9 H3 (Upstate Biotechnology, Lake Placid, NY); 1:200 dilution for anti-general H3; and 1:200 dilution for anti-Pdd1p] in blocking solution (10% normal goat serum/3% BSA/0.1% Tween 20/PBS) overnight at 4°C, followed by incubation with FITC-conjugated goat anti-rabbit secondary antibody (1:200 dilution). The samples were incubated with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Roche Diagnostics) in PBS, mounted, and observed by confocal microscopy (Leica, Deerfield, IL).
Analyses of M-Element Processing. For Southern blot analysis, genomic DNA was isolated from Tetrahymena by concentrating 1 × 107 cells to 0.5 ml of Tris buffer (10 mM; pH 7.4) and then adding 4 ml of lysis buffer (0.7 M NaCl/20 mM Tris, pH 7.4/20 mM EDTA/2% SDS) and incubating at room temperature for 15 min, followed by phenol–chloroform extraction.
For PCR analysis, cells were lysed by incubation for 1 h at 55°C, followed by 15 min at 98°C in 1× Taq polymerase buffer (Promega)/2 μg/μl proteinase K at a concentration of 1 × 102 cells per μl. The lysate was used directly for the PCR. The following primers were used for the PCR analysis: M5′-1 (nucleotide position 2–25, sense) and M3′-1 (nucleotide position 1172–1194, antisense). The GenBank accession number of the M element is M21936. The M-short PCR product (2–238 and 1147–1194) was used as the probe for Southern blot analysis.
Analysis of Small RNAs. Total RNA was extracted with TRIzol reagent (Invitrogen) and dissolved in formamide. RNA from 5 × 104 cells was separated on 12% polyacrylamide–urea DNA sequencing gels and visualized by soaking the gels in 1.5 μg/ml ethidium bromide (22).
Results
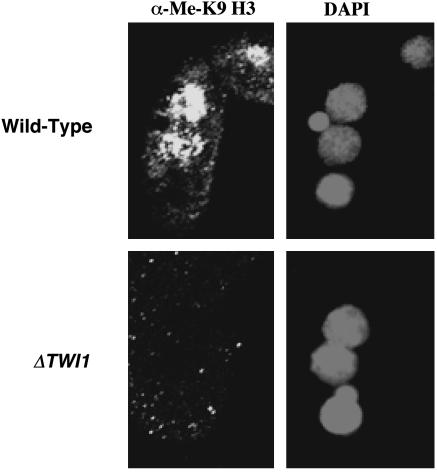
H3 K9 Methylation Is Eliminated in TWI1 Knockout Strains. TWI1 is required for accumulation of scnRNAs enriched in IES sequences and for IES elimination (22). If scnRNAs are required for H3 K9 methylation, then preventing their accumulation should affect H3 K9 methylation. To test this hypothesis, we examined conjugating cells of TWI1 knockout strains by immunofluorescence analysis using an antibody specific for K9-methylated H3. No signal was detected in TWI1 knockout cells at a stage when the anlagen of wild-type cells stained strongly (Fig. 1). Anlagen, micronuclei, and old macronuclei in knockout cells stained strongly with a control antibody that recognizes unmodified H3 (data not shown). Thus, TWI1 is required for H3 K9 methylation in anlagen. Somatic knockout of the PDD1 gene encoding the chromodomain-containing Pdd1 protein also greatly reduces scnRNA accumulation (22) and K9 methylation (23). These results suggest that the TWI1 and PDD1 genes, scnRNAs, and K9 methylation participate in the same pathway leading to IES excision. However, to prove this suggestion, it is necessary to demonstrate that K9 methylation is required for IES elimination.
Fig. 1.
TWI1 is required for H3 K9 methylation. TWI1 knockout cells (ΔTWI1-S2 ×ΔTWI1-S4) or wild-type cells (CU428 × B2086) were mated. Cells collected 14 h after mixing were fixed, processed for immunofluorescence staining with anti-dimethyl K9 H3 antibody, and stained with DAPI.
Creation of Tetrahymena Strains with H3 K9Q Mutation. To eliminate K9 methylation, we created strains of Tetrahymena in which a HHT2 gene with a K9Q mutation was the only gene encoding the major H3 protein (Fig. 2). We constructed a pair of germ-line knockout heterokaryon strains (30) in which the micronuclear copies of the two genes (HHT1 and HHT2) that encode (the same) major H3 histone are among the genes disrupted (as well as the two H4 genes) but in which the macronuclei contain wild-type copies of these genes. During conjugation, the old macronuclei are destroyed and new macronuclei are derived from the micronuclei. Because histones H3 and H4 are essential, the progeny of these germ-line knockout heterokaryons cannot survive unless they are rescued by transformation with a gene encoding a functional H3 (and H4). Using this strategy, we produced strains whose only macronuclear gene encoding a major H3 was either a wild-type HHT2 gene or a gene encoding a K9Q mutation.
Southern and Northern blot analyses showed that the rescue constructs integrated into the homologous locus and that the rescuing wild-type or mutated HHT2 gene was expressed normally (data not shown). The genotypes of the mutants and wild-type strains also were confirmed by PCR amplification and sequencing of the genomic DNA. Histone H3 isolated from the K9Q mutants showed a reduced migration in acid–urea gels, consistent with a reduction in its overall positive charge by one (data not shown). The quantitatively minor H3 variant hv2 (encoded by HHT3) was not changed in these experiments. However, this factor did not affect our results and interpretations significantly because it contributes only ≈10% of the total H3 protein (33).
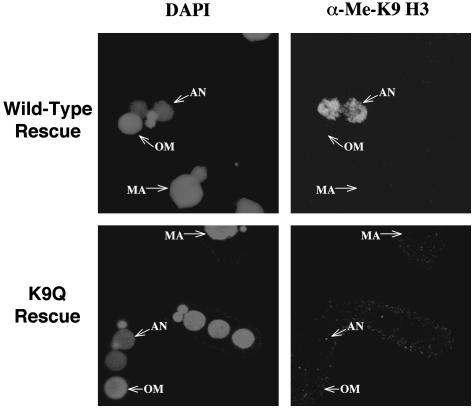
The H3 K9Q Mutation Eliminates K9 Methylation. There was no detectable phenotype in growing cells of the K9Q mutants, which is consistent with the failure to detect H3 K9 methylation in vegetative Tetrahymena cells (23, 34). They had a doubling time (2.6 h) that was indistinguishable from either true wild-type cells or cells that had been rescued with a wild-type HHT2 gene. No differences were detected in the mRNA levels of several growth-related, cadmium-induced, starvation-induced, or conjugation-induced genes (data not shown). The overall pairing efficiency was high (≈80%), and the progression of cells through different stages of conjugation was similar for K9Q and wild-type rescued cells (data not shown). However, conjugating K9Q and wild-type rescued cells both differed from true wild-type cells because they failed to produce any viable progeny and arrested before resorbing one of the two new micronuclei, which is presumably because of the lack of sufficient newly synthesized H3 and H4 proteins to enable chromatin replication in these germ-line knockout heterokaryons. However, because development of the progeny of rescued cells proceeded past the stage where K9 methylation normally occurs (23), we could analyze K9 methylation in matings of rescued cells by immunofluorescence analysis using an antibody specific for dimethyl K9 histone H3. When rescued cells containing the wild-type HHT2 gene were mated, the anlagen were specifically stained (Fig. 3) in a pattern that was indistinguishable from true wild-type cells, demonstrating that this modification could occur normally, even in the absence of expression by the new macronucleus of the genes encoding major H3. In conjugating cells containing the K9Q mutation, no staining was detected, indicating that this mutation abolished H3 K9 methylation. A control antibody against general histone H3 stained both the macronuclei and micronuclei of both strains (data not shown).
Fig. 3.
H3 K9 methylation is not detectable in H3 K9Q mutants. The cells from 14 h after mixing were fixed and processed for immunofluorescence analysis using anti-dimethyl K9 H3 primary antibody and FITC-conjugated secondary antibody. Cells were also stained with DAPI. AN, anlagen; OM, old macronuclei; MA, macronuclei in nonmating cells.
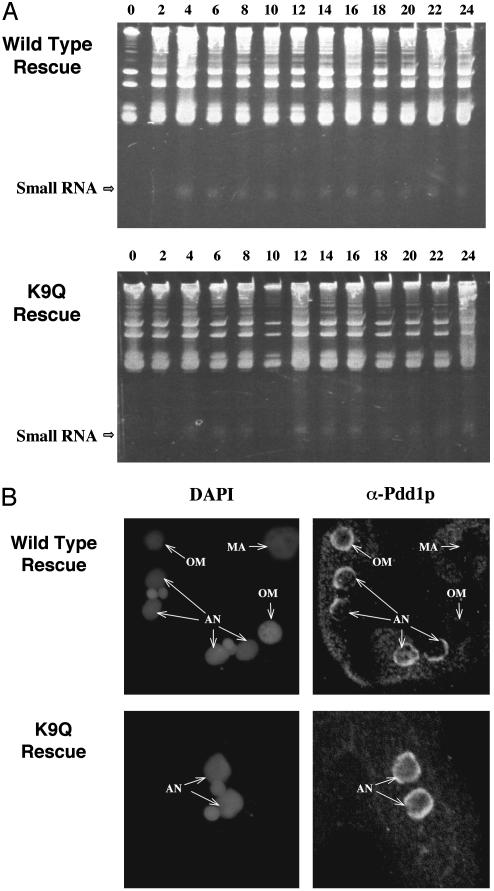
scnRNAs and Pdd1p Accumulate Normally in K9Q Mutants. Disruption of TWI1 prevents both scnRNA accumulation (22) and K9 methylation (see above), and disruption of PDD1 has similar effects (22, 23). Although K9Q and wild-type rescued heterokaryons failed to complete conjugation, wild-type rescued cells did specifically methylate K9 at the correct stage of conjugation, and the K9Q mutation prevented this modification. Thus, these rescued cells could be used to determine whether K9 methylation occurred upstream or downstream of Twi1p and Pdd1p expression. We, therefore, compared the accumulation of small RNAs and of Pdd1p in both K9Q and wild-type rescued strains. Small RNAs were detected as early as 2–4 h after mixing in both strains (Fig. 4A), as described in ref. 22 for true wild-type cells. This finding indicates that K9 methylation of H3 has little or no effect on the initiation of small RNA accumulation during conjugation, indicating that K9 methylation is downstream of the RNAi pathway. In true wild-type cells, the level of small RNA diminishes after 16 h into conjugation when the conjugation process is nearly complete (22). However, in both the K9Q and wild-type rescued cells, the level of small RNAs remained high, even 24 h after mixing, probably because these cells arrest in late conjugation.
Fig. 4.
Absence of K9 methylation does not affect accumulation of scnRNAs or Pdd1p. (A) scnRNA accumulation in conjugating K9Q or wild-type rescued cells. Total RNA was extracted from mating cells (at the indicated hours after mixing) and separated in 12% acrylamide–urea gels and stained by ethidium bromide. Position of the scnRNAs (Small RNA) is indicated by the arrow. (B) Localization of Pdd1p in conjugating K9Q or wild-type rescued cells. Cells were fixed 14 h after mixing, processed for immunofluorescence analysis by using primary anti-Pdd1p antibody and FITC-conjugated secondary antibody, and stained with DAPI. AN, anlagen; OM, old macronuclei; MA, macronuclei in nonmating cells.
We also examined the expression of Pdd1p in both K9Q and wild-type rescued strains. Immunofluorescence analysis using an antibody against Pdd1p showed that the nuclear localization pattern of Pdd1p was not distinguishably different in conjugating cells of both strains (Fig. 4B) and was similar to nuclear localization pattern described for true wild-type cells (26, 27). Pdd1p was not detected in the macronuclei or micronuclei of nonmating cells. It could be detected in old macronuclei and in anlagen at later conjugation stages (Fig. 4B). This finding suggests that the accumulation and migration of Pdd1p are not affected either by the K9Q mutation or by the lack of newly synthesized H3 in the knockout heterokaryon progeny.
The observations that knockout of TWI1 or PDD1 inhibits scnRNA accumulation and H3 K9 methylation and that scnRNAs appear normally in conjugating K9Q cells lacking histone H3 K9 methylation argue strongly that Twi1p, Pdd1p, scnRNAs, and K9 methylation are in the same pathway and suggest, but do not prove, that K9 methylation is required for IES elimination.
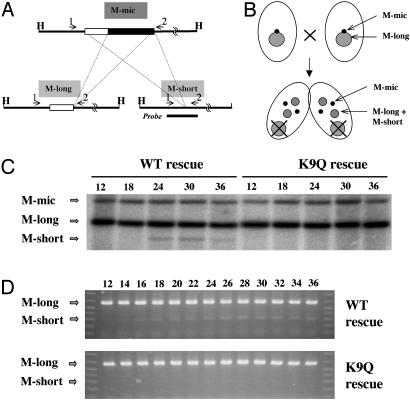
H3 K9 Methylation Is Required for Efficient IES Processing. To address whether IES elimination was affected by the conjugation of K9Q and wild-type rescued strains, a well studied IES, the M element (35), was examined. This element hybridizes like a single-copy sequence and is processed with equal frequencies into two stable macronuclear-specific forms, referred to as M-long and M-short (Fig. 5A). Because macronuclei divide amitotically, all allelic sequences in them are segregated randomly into daughter cells, a process referred to as phenotypic assortment. With repeated divisions, this process generates cells with homozygous macronuclei from heterozygous progenitors (36). Using this approach, we obtained wild-type and K9Q rescued strains whose macronuclei contained only the M-long form of the rearranged M region. When these cells are allowed to conjugate, the only way that the M-short form can be produced is by processing the intact, micronuclear-specific M-IES (Fig. 5B). Thus, the appearance of the M-short form in conjugating rescued cell is an assay for IES rearrangement (28).
Fig. 5.
Analysis of M-element processing in the progeny of K9Q and wild-type rescued cells. (A) Alternative processing of the M element. The micronuclear-specific form of the M element can be processed into either of two macronuclear-specific forms, M-long and M-short, which occur in about equal frequency (35). Both elements have the same 3′ break site. In the M-long region, 0.6 kb of sequence 5′ of the break point is eliminated. In the M-short region, 0.9 kb of sequence 5′ of the break point is eliminated. The M element is flanked by HindIII sites. Primers 1 (M5′-1) and 2 (M3′-1) were used in PCR analysis of M processing. Dashed lines indicate the regions that are eliminated in each of the two processing alternatives. (B) Schematic representation of the rationale for detecting M-element processing in conjugating rescued cells. The parental rescued cells were assorted, and clones containing only the M-long form of the M element were used for conjugation. The appearance of the M-short form in conjugant progeny is indicative of the processing of the M element during macronuclear development. (C) Southern blot analysis of M-element processing in conjugating K9Q and wild-type rescued cells. Genomic DNA was isolated from K9Q and wild-type rescued cells at different times of conjugation and digested with HindIII. The blot was probed with the M-short PCR product (see A and D), which detects both the M-long and M-short fragments with equal affinity. (D) PCR analysis of M-element processing in conjugating K9Q and wild-type rescued cells. PCR was conducted on conjugating whole-cell lysates collected at 2-h intervals 12–36 h after mixing. Primers 1 and 2 were used to examine the relative abundance of the two alternative macronuclear-specific forms, M-long and M-short.
Fig. 5C shows a Southern blot analysis of DNA isolated at different times during conjugation. In wild-type rescued cells, the M-short form was observed 18 h after conjugation was initiated, demonstrating that mating progeny of these cells underwent IES elimination, consistent with earlier observations showing that IES elimination is independent of the endoreplication in anlagen that occurs in the late stages of conjugation (37). We conclude that, even though these strains cannot produce viable conjugation progeny, they can be used to study the mechanism of IES elimination.
We next analyzed the appearance of M-short in K9Q rescued cells and could detect only trace (<10% that in wild-type rescued cell) amounts, even after 36 h of conjugation. A more sensitive PCR assay of the time course of M-element elimination (Fig. 5D) showed that, in wild-type rescued cells, M-short was detected at 14–16 h of conjugation and the amount plateaued at ≈24 h after mixing. In K9Q rescued cells, there were again only trace amounts of M-short, even 36 h after mixing. Thus, both Southern blot and PCR analyses demonstrated that elimination of the M element was reduced greatly in K9Q compared with wild-type rescued strains. It is not clear whether the small amount of IES elimination that remains in the K9Q rescued cells reflects the presence of small amounts of K9 methylation on the replacement histone variant hv2 or indicates that the elimination machinery can associate inefficiently with IESs in the absence of K9 methylation. In either case, these results argue that K9 methylation is required for efficient IES elimination, allowing the IES elimination pathway to be ordered as follows: Twi1p/Pdd1p →scnRNAs→K9 methylation→IES elimination.
Discussion
Small RNAs Likely Target H3 K9 Methylation to Heterochromatin. Recent studies have shown that, in S. pombe, the mutation of genes required for RNAi in other systems leads to deficiencies in K9 methylation and heterochromatin formation (16, 17). Also, it has been established in Tetrahymena that TWI1 and PDD1 are required for scnRNA accumulation (22) and for H3 K9 methylation (ref. 23 and this article). These findings make it likely that the function of these small RNAs in heterochromatin formation is to target the K9-specific histone methyltransferases (and perhaps other chromatin-modifying proteins as well) to the sequences to which they are homologous.
There is precedence for RNA-mediated chromatin modifications, notably the marking of the X chromosome for dosage compensation in Drosophila and mammals (38, 39). Sequence-specific histone modifications targeted by homologous RNAs are involved in the initiation phase of male X chromosome activation in Drosophila (40). The RNA transcripts, roX1 and roX2, are stabilized by a protein complex containing MOF and MSL3 (40, 41) whose chromodomains can interact directly with RNA in vitro (42). MOF also has H4 K16-specific histone acetyltransferase activity (43). Based on these observations, it seems likely that a protein complex that can stabilize small RNAs and facilitate homology-based targeting is involved in the scnRNA–DNA interactions in IES elimination. Alternatively, in keeping with the known ability of siRNAs to interact with other RNAs in the RNAi pathway, the scnRNAs could interact with nascent transcripts of the IES regions. It is tempting to speculate that this proposed complex contains at least one chromodomain protein, probably Pdd1p, which concentrates in the heterochromatic structure specialized for IES processing and is required for accumulation of scnRNAs. Pdd1p also can bind K9-methylated H3 in vitro (23), suggesting that it acts as a bridge between scnRNAs and K9-methylated H3. It could function to strengthen the interactions between the small RNA complexes and chromatin and to establish a feedback loop that could spread K9 methylation in a manner analogous to Swi6p in S. pombe. Twi1p, which also is required for the accumulation of scnRNAs, is another likely member of this complex. The similarity between the localization of Pdd1p (27, 44) and Twi1p (22), first in the old macronucleus and then in the developing macronucleus, and the proposed flow of information from micronuclei to old macronuclei and then to anlagen (22) also suggests that scnRNAs may reside in a complex with these two proteins.
Small RNA-Targeted H3 K9 Methylation as a General Mechanism for Heterochromatin Formation. Eliminating a significant fraction of the germ-line genome from the somatic genome is a highly specialized feature of ciliate biology. However, a related process is probably involved in the formation of centromeric heterochromatin in other eukaryotes. A small RNA component and H3 K9 methylation are involved in centromere function and silencing in S. pombe (16, 17). K9 methylation and an RNA component also have been implicated in pericentric heterochromatin of mammalian cells (10, 11). In all of these cases, the challenge is to understand the mechanism that specifically recognizes and silences (or eliminates) mostly repeated DNA sequences that constitute a relatively small percentage of the genomic DNA. This mechanism is particularly difficult to understand in the case of centromeric heterochromatin in which highly conserved proteins (e.g., a centromere-specific H3 or chromodomain proteins) must associate with highly divergent and rapidly evolving DNA elements (45). The RNAi-like mechanism that appears to be operating in gene silencing in S. pombe heterochromatin and in IES elimination in Tetrahymena heterochromatin seems ideally suited to this role. In both cases, the silencing/elimination likely depends on the ability of the targeted sequences to generate double-stranded RNAs, which activate the RNAi pathway of the system to produce the small RNAs. Dispersed repeated sequences, likely to be present in both orientations relative to endogenous promoters, could produce transcripts of both sense and antisense strands and, therefore, be subject to heterochromatin formation and gene repression (12). Clearly, although considerable progress has been made in understanding the mechanisms underlying the formation and maintenance of heterochromatin, much remains to be explored. The studies described here firmly establish IES processing in Tetrahymena as an accessible model for studying the general mechanisms by which small RNAs and histone-modifying activities interact in this process.
Acknowledgments
We thank Josephine Bowen for assistance with confocal imaging. The anti-Pdd1p antibody was kindly provided by Dr. C. David Allis. This work was supported by National Institutes of Health Grant GM21793.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: scnRNA, scan RNA; siRNA, small interfering RNA; IES, internal eliminated sequence; Kn, lysine n; RNAi, RNA interference; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- 2.Grewal, S. I. & Elgin, S. C. (2002) Curr. Opin. Genet. Dev. 12, 178-187. [DOI] [PubMed] [Google Scholar]
- 3.Lachner, M. & Jenuwein, T. (2002) Curr. Opin. Cell Biol. 14, 286-298. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C. & Kouzarides, T. (2001) Nature 410, 120-124. [DOI] [PubMed] [Google Scholar]
- 5.Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. (2001) Nature 410, 116-120. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs, S. A., Taverna, S. D., Zhang, Y., Briggs, S. D., Li, J., Eissenberg, J. C., Allis, C. D. & Khorasanizadeh, S. (2001) EMBO J. 20, 5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. (2001) Science 292, 110-113. [DOI] [PubMed] [Google Scholar]
- 8.Schotta, G., Ebert, A., Krauss, V., Fischer, A., Hoffmann, J., Rea, S., Jenuwein, T., Dorn, R. & Reuter, G. (2002) EMBO J. 21, 1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., et al. (2001) Cell 107, 323-337. [DOI] [PubMed] [Google Scholar]
- 10.Maison, C., Bailly, D., Peters, A. H., Quivy, J. P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T. & Almouzni, G. (2002) Nat. Genet. 30, 329-334. [DOI] [PubMed] [Google Scholar]
- 11.Muchardt, C., Guilleme, M., Seeler, J. S., Trouche, D., Dejean, A. & Yaniv, M. (2002) EMBO Rep. 3, 975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plasterk, R. H. (2002) Science 296, 1263-1265. [DOI] [PubMed] [Google Scholar]
- 13.Mette, M. F., Aufsatz, W., van der Winden, J., Matzke, M. A. & Matzke, A. J. (2000) EMBO J. 19, 5194-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal-Bhadra, M., Bhadra, U. & Birchler, J. A. (2002) Mol. Cell 9, 315-327. [DOI] [PubMed] [Google Scholar]
- 15.Ketting, R. F., Haverkamp, T. H., van Luenen, H. G. & Plasterk, R. H. (1999) Cell 99, 133-141. [DOI] [PubMed] [Google Scholar]
- 16.Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I. & Martienssen, R. A. (2002) Science 297, 1833-1837. [DOI] [PubMed] [Google Scholar]
- 17.Hall, I. M., Shankaranarayana, G. D., Noma, K., Ayoub, N., Cohen, A. & Grewal, S. I. (2002) Science 297, 2232-2237. [DOI] [PubMed] [Google Scholar]
- 18.Ekwall, K., Javerzat, J. P., Lorentz, A., Schmidt, H., Cranston, G. & Allshire, R. (1995) Science 269, 1429-1431. [DOI] [PubMed] [Google Scholar]
- 19.Karrer, K. M. (2000) Methods Cell Biol. 62, 127-186. [DOI] [PubMed] [Google Scholar]
- 20.Jahn, C. L. & Klobutcher, L. A. (2002) Annu. Rev. Microbiol. 56, 489-520. [DOI] [PubMed] [Google Scholar]
- 21.Chalker, D. L. & Yao, M. C. (1996) Mol. Cell. Biol. 16, 3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mochizuki, K., Fine, N. A., Fujisawa, T. & Gorovsky, M. A. (2002) Cell 110, 689-699. [DOI] [PubMed] [Google Scholar]
- 23.Taverna, S. D., Coyne, R. S. & Allis, C. D. (2002) Cell 110, 701-711. [DOI] [PubMed] [Google Scholar]
- 24.Chalker, D. L. & Yao, M. C. (2001) Genes Dev. 15, 1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao, M. C., Fuller, P. & Xi, X. (2003) Science 300, 1581-1584. [DOI] [PubMed] [Google Scholar]
- 26.Madireddi, M. T., Coyne, R. S., Smothers, J. F., Mickey, K. M., Yao, M. C. & Allis, C. D. (1996) Cell 87, 75-84. [DOI] [PubMed] [Google Scholar]
- 27.Coyne, R. S., Nikiforov, M. A., Smothers, J. F., Allis, C. D. & Yao, M. C. (1999) Mol. Cell 4, 865-872. [DOI] [PubMed] [Google Scholar]
- 28.Duharcourt, S. & Yao, M. C. (2002) Eukaryot. Cell 1, 293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorovsky, M. A., Yao, M.-C., Keevert, J. B. & Pleger, G. L. (1975) Methods Cell Biol. 9, 311-327. [DOI] [PubMed] [Google Scholar]
- 30.Hai, B. & Gorovsky, M. A. (1997) Proc. Natl. Acad. Sci. USA 94, 1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahn, R. W., Andersen, B. H. & Brunk, C. F. (1993) Proc. Natl. Acad. Sci. USA 90, 9295-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cassidy-Hanley, D., Bowen, J., Lee, J., Cole, E. S., VerPlank, L. A., Gaertig, J., Gorovsky, M. A. & Bruns, P. J. (1997) Genetics 146, 135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allis, C. D., Glover, C. V. D., Bowen, J. K. & Gorovsky, M. A. (1980) Cell 20, 609-617. [DOI] [PubMed] [Google Scholar]
- 34.Strahl, B. D., Ohba, R., Cook, R. G. & Allis, C. D. (1999) Proc. Natl. Acad. Sci. USA 96, 14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Austerberry, C. F., Allis, C. D. & Yao, M.-C. (1984) Proc. Natl. Acad. Sci. USA 81, 7383-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orias, E. & Flacks, M. (1975) Genetics 79, 187-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikiforov, M. A., Smothers, J. F., Gorovsky, M. A. & Allis, C. D. (1999) Genes Dev. 13, 2852-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, Y. & Kuroda, M. I. (2001) Science 293, 1083-1085. [DOI] [PubMed] [Google Scholar]
- 39.Kelley, R. L. & Kuroda, M. I. (2000) Curr. Opin. Genet. Dev. 10, 555-561. [DOI] [PubMed] [Google Scholar]
- 40.Meller, V. H. & Rattner, B. P. (2002) EMBO J. 21, 1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meller, V. H., Gordadze, P. R., Park, Y., Chu, X., Stuckenholz, C., Kelley, R. L. & Kuroda, M. I. (2000) Curr. Biol. 10, 136-143. [DOI] [PubMed] [Google Scholar]
- 42.Akhtar, A., Zink, D. & Becker, P. B. (2000) Nature 407, 405-409. [DOI] [PubMed] [Google Scholar]
- 43.Akhtar, A. & Becker, P. B. (2000) Mol. Cell 5, 367-375. [DOI] [PubMed] [Google Scholar]
- 44.Janetopoulos, C., Cole, E., Smothers, J. F., Allis, C. D. & Aufderheide, K. J. (1999) J. Cell Sci. 112, 1003-1011. [DOI] [PubMed] [Google Scholar]
- 45.Henikoff, S., Ahmad, K. & Malik, H. S. (2001) Science 293, 1098-1102. [DOI] [PubMed] [Google Scholar]