Abstract
MCP-1-induced protein 1 (MCPIP1) plays an important role in the downregulation of the LPS-induced immune response by acting as an RNase targeting IL-6 and IL-12b mRNAs. A conserved domain located in the N-terminal part of MCPIP1 is thought to be responsible for its RNase activity, but its catalytic mechanism is not well understood due to the lack of an atomic resolution structure. We determined the 3D crystal structure of this MCPIP1 N-terminal conserved RNase domain at a resolution of 2.0 Å. The overall structure of MCPIP1 N-terminal conserved domain shares high structural homology with PilT N-terminal domain. We show that the RNase catalytic center is composed of several acidic residues, verifying their importance by site-specific mutagenesis. A positively charged arm close to the catalytic center may act as an RNA substrate-binding site, since exchange of critical positively charged residues on this arm with alanine partially abolish the RNase activity of MCPIP1 in vivo. Our structure of the MCPIP1 N-terminal conserved domain reveals the details of the catalytic center and provides a greater understanding of the RNA degradation mechanism.
INTRODUCTION
Pathogen invasion triggers complex immunological host responses, including recognition of pathogens by numerous cell surface and cytosolic receptors, subsequent signal transduction and expression of a series of inflammatory cytokines (1,2). Inflammatory and immune responses are precisely controlled to prevent the development of autoimmune disease which is caused by unregulated overreaction of the immune system. MCPIP1 (MCP-1-induced protein 1, encoded by gene zc3h12a) is a newly-identified immune response regulator whose expression is directly induced by TLR signaling (3–5). Recent studies have shown that MCPIP1 is a multifunctional protein involved in various distinct signaling pathways. The MCPIP family (including MCPIP1, MCPIP2, MCPIP3 and MCPIP4) was first reported to participate in the negative regulation of LPS-induced macrophage activation (6,7). Suppression of TNF-α and the inducible nitric-oxide synthase (iNOS) promoter, and blocking of LPS-induced NF-κB activation by MCPIP1 was originally thought to be the underlying mechanism of the negative regulation of macrophage activation (7). Akira et al., however, showed that MCPIP1 plays an essential role in downregulating the LPS-induced immune response by acting as an RNase (8). Overexpression of MCPIP1 decreases the production of LPS-induced inflammatory cytokines such as IL-1β, IL-12β and IL-6 by accelerating the degradation of their mRNAs (8,9). MCPIP1 significantly shortens the half-life of IL6 and IL-12β mRNA by directly targeting their 3′-untranslated terminal regions (3′-UTR), the intactness of which is crucial for preventing the degradation of these mRNAs in vivo (8). More recently, MCPIP1 was found to inhibit miRNA biogenesis by targeting a terminal loop of certain pre-miRNAs with its RNase domain (10). Sequence analysis and functional studies have identified several domains and residues essential for the RNase function of MCPIP1. All MCPIP family members harbor an N-terminal conserved domain (NCD, residues 100–300) which shares remote sequence homology with PilT N-terminus (PIN) domains (8). A CCCH type zinc-finger motif is located close to the C-terminal region of the RNase domain as shown in Figure 1A. This type of zinc finger has been found in many RNA-binding proteins, such as tristetraprolin (TTP) and has been shown to bind directly with the 3′-UTR regions of different mRNAs (11–13). In addition, two proline-rich regions have been found in both the N-terminal region (residues 100–126) and the C-terminal region (residues 458–536) of MCPIP1 (14). Mutagenesis studies have been performed to determine which residues are involved in the RNase functions of MCPIP1. A single mutation, D141N, completely abolishes the RNase activity of MCPIP1 (8). A computational study of the structure of the MCPIP1 N-terminal conserved domain has been reported. A conserved negatively charged pocket was predicted and several residues thought to be important for RNase activity were identified. However, to elucidate the details of MCPIP1 function, more detailed and accurate structural information is required.
Figure 1.
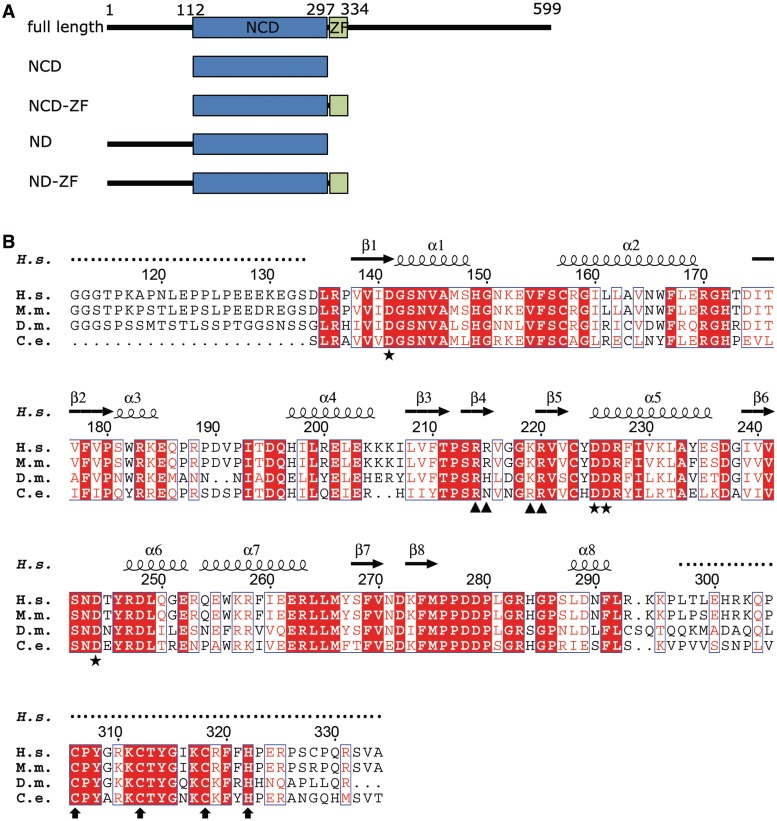
Sequence information of MCPIP1 and its N-terminal conserved domain. (A) Schematic domain structure of MCPIP1. Constructs used for structural determination are indicated. (B) Amino acid sequences of MCPIP1 NCD-ZF from Homo sapiens, Mus musculus, Drosophila melanogaster and Caenorhabditis elegans were aligned with the program CLUSTALW2 (28), and the alignment result was graphically displayed with the program ESPript (29). Secondary structure elements are indicated according to the structure of human MCPIP1 NCD-ZF. Residues involved in the catalytic center are marked with asterisks. Positively charged residues in the putative nucleotide-binding arm are marked with triangle. The conserved CCCH zinc-finger motif is marked with arrows.
In order to understand the molecular mechanism underlying the function of MCPIP1 in the negative regulation of immune responses, we determined high resolution 3D structures of its conserved RNase domain with and without the CCCH motif [amino acids 112–297 (NCD) and 112–334 (NCD-ZF)] by crystallographic methods. Our structures reveal a conserved RNase catalytic center, and a positively charged arm that may participate in substrate RNA binding. These findings provide useful information for further study of MCPIP1 function and for understanding the substrate mRNA-binding mechanism.
MATERIALS AND METHODS
Protein expression, purification and crystallization
Sequences encoding the MCPIP1 ND (residues 1–297) and ND-ZF (residues 1–334) were respectively inserted into pGEX-6 p-1 vectors (GE Healthcare), and Escherichia coli BL21 (DE3) cells were transformed with these vectors for expression. For Se-derived ND protein expression, E. coli B834 cells were transformed with the cloned vector. All E. coli cells were cultured at 16°C after adding Isopropyl β-d-1-thiogalactopyranoside (IPTG) to induce gene expression. Cells were harvested and then lysed by ultrasonication. After centrifugation at 30 700g, supernatants were applied to a Glutathione Sepharose 4B (GE Healthcare) column in order to remove other proteins. PreScission protease (GE Healthcare) was used to cleave the fusion proteins to obtain the ND and ND-ZF protein constructs after elution from the column by Glutathione. Amino acids 108–113 of MCPIP1 (L-V-P-R-G-G) are similar to those of the thrombin recognition cleavage site (L-V-P-R-G-S). Thus, thrombin protease (Novagen) was used to obtain the NCD (resides 112–297) and NCD-ZF (residues 112–334) protein, which were then further purified separately by Hitrap Q and Hitrap S before crystallization.
Crystallization was performed by the hanging drop vapor diffusion method. Proteins NCD, Se-NCD and NCD-ZF were concentrated to 7 mg/ml (in 50 mM NaCl, 20 mM Tris–HCl pH 8.0 buffer), 5 mg/ml (in 50 mM NaCl, 20 mM Tris–HCl pH 8.0 buffer) and 6 mg/ml (in 300 mM NaCl, 20 mM Tris–HCl pH 8.3 buffer), respectively, for crystallization. After optimization, an NCD crystal of size 200 × 10 × 10 µm3 was obtained in 1 μl of the above protein solution mixed with 1 μl of reservoir solution (0.1 M Bis–Tris pH 5.5, 7% PEG 3350). The Se-NCD crystal used for SAD data collection was obtained using the same condition. The single NCD-ZF crystal of size 200 × 200 × 50 µm3 used for data collection was grown in 4% Tacsimate pH 5, 6% PEG 3350. The NCD-ZF crystal was dehydrated in a solution of 30% ethanol, 4% Tacsimate pH 5, 15% PEG 3350 for a few seconds before mounting. All crystals were flash-frozen in liquid nitrogen before data collection.
Data collection and structure determination
Native NCD and Se-derived crystal diffraction data sets were collected at BL-17 A, Photon factory, KEK, Japan. The wavelength used for anomalous diffraction data collection was 0.9790 Å. Native NCD-ZF crystal diffraction data was collected at BL-17I, SSRF, China. All data sets were processed and scaled using the HKL2000 package (15). Due to anisotropic diffraction of NCD-ZF crystal, the ellipsoidal truncation and anisotropic scaling was applied to NCD-ZF data using the online Diffraction Anisotropy Server (16). The phasing of the NCD structure was solved using the Single-wavelength Anomalous Diffraction (SAD) method with the hkl2map program (17–20). The NCD-ZF structure was solved by the Molecular Replacement (MR) method using the NCD structure as a model (21,22). Mg2+-derived NCD crystals were obtained by soaking the NCD crystal in a reservoir containing 15 mM MgCl2. Mg2+-derived crystal data was collected at BL-17A, Photon factory, KEK, Japan. The Mg2+ ion-containing structure was solved by the MR method using the NCD structure as a model. All refinements were carried out using the Phenix.refine and Coot programs (23,24). Table 1 shows the crystallographic data and refinement statistics for the two structures. Figures were prepared using PyMol (25).
Table 1.
Data collection and refinement statistics
| Se-NCD | Native NCD | NCD-ZF | NCD-Mg | |
|---|---|---|---|---|
| Wavelength (Å) | 0.9790 | |||
| Temperature (K) | 100 | 100 | 100 | 100 |
| Resolution (Å) | 49.8–2.4 (2.49–2.40) | 39.9–2.00 (2.07–2.00) | 36.3–2.0 (2.07–2.00) | 25.4–2.0 (2.07–2.00) |
| Space group | P41 | P41 | P3121 | P41 |
| Unit cell parameters | a = b = 55.4, c = 113.8 | a = b = 56.4, c = 113.2 | a = b = 113.5 c = 78.0 | a = b = 56.4, c = 113.6 |
| Solvent content (%, v/v) | 41.5 | 43.1 | 57.7 | 43.3 |
| Unique reflections | 12 096 | 21 578 | 30 613a | 23 299 |
| Multiplicity | 13.2 (7.6) | 6.2 (2.9) | 5.8 (2.1) | 3.6 (3.0) |
| Completeness (%) | 90.3 (55.4) | 90.6 (51.5) | 77.8 (33.4) | 97.9 (92.2) |
| Rmergeb | 0.11 (0.25) | 0.08 (0.27) | 0.09 (0.57) | 0.09 (0.48) |
| I/σ(I) | 18.3 (3.4) | 17.5 (2.0) | 26.2 (1.4) | 16.1 (2.6) |
| FOM | 0.46 | |||
| FOM (after solvent flattening) | 0.68 | |||
| Refinement | ||||
| Rfreec | 0.244 | 0.226 | 0.240 | |
| Rworkd | 0.202 | 0.190 | 0.198 | |
| RMSD angles | 1.075 | 1.083 | 1.066 | |
| RMSD bonds | 0.008 | 0.008 | 0.008 | |
| Average B factor (Å2) | 24.03 | 47.62 | 28.37 | |
| Ramachandran plot regions (%) | ||||
| Most favored (%) | 97.04 | 97.48 | 97.39 | |
| Additionally allowed (%) | 2.96 | 2.52 | 2.61 | |
| Outlier (%) | 0 | 0 | 0 | |
| PDB code | 3V32 | 3V33 | 3V34 |
Values in parentheses are for highest-resolution shell.
aThe number of reflection is 29 309 after ellipsoidal truncation and anisotropic scaling.
bRmerge = Σhkl Σi|I(hkl)i − <I(hkl)>|/ Σhkl Σi I(hkl)i, where I(hkl) is the intensity of reflection hkl, and <I(hkl)> is the average intensity over all equivalent reflections.
cRfree was calculated for a test set of reflections (5%) omitted from the refinement.
dRwork = Σhkl |Fo(hkl) − Fc(hkl)|/Σhkl Fo (hkl).
Synthesis of [32P]-labeled RNAs and in vitro RNase activity determination
In vitro transcription was carried out using a MEGAshortscriptTM Kit (Ambion, Inc). UTP was replaced by 90% α-[32P]-UTP (PerkinElmer) to produce isotope-labeled mRNAs. A T7 RNA polymerase promoter was inserted upstream of the mouse IL-6 3′-UTR by PCR using cDNA from mouse endothelial cells. Amplified IL-6 3′-UTR and a control template in the kit were used as templates to synthesize different mRNAs. TURBO DNase was added to the reaction system after transcription to remove the DNA template. Enzymes and free nucleotides were removed by phenol:chloroform extraction and alcohol precipitation.
To measure RNase activity, purified [32P]-labeled RNA was incubated with purified MCPIP1 constructs at 37°C. Denaturing acrylamide gels were run and autoradiography was used to measure the amount of RNA left after digestion by different proteins.
In vivo RNase activity determination
Luciferase reporter assays were used according to a previously reported method (26,27). The MCPIP1 cDNA was inserted into pcDNA4TM/TO/myc-His A (Invitrogen) to express myc-tagged full-length MCPIP1 protein. Mutations were introduced into the vector directly using a quick-change single mutation method. The PGL3-IL-6 3′-UTR vector was generated by inserting the IL-6 3′-UTR sequence into a PGL-3 Luciferase Reporter Vector (Promega). 293A cells were transfected with 1 μg plasmid of each construct and 0.1 μg IL-6 3′-UTR PGL3 plasmid using LipofectamineTM 2000 (Invitrogen) according to the manufacturer’s instructions. A Renilla luciferase reporter pRL-TK (Promega), which is not sensitive to MCPIP1, was included for normalization of transfection efficiencies. Cells were harvested 48 h later and lysed in NP40 lysis buffer. After centrifuging for 20 min at 12 000g, 4°C, the supernatant was tested for luciferase activity using a luciferase assay system (Promega).
RESULTS
The N-terminal conserved domain degrades IL-6 mRNA 3′-UTR in vitro
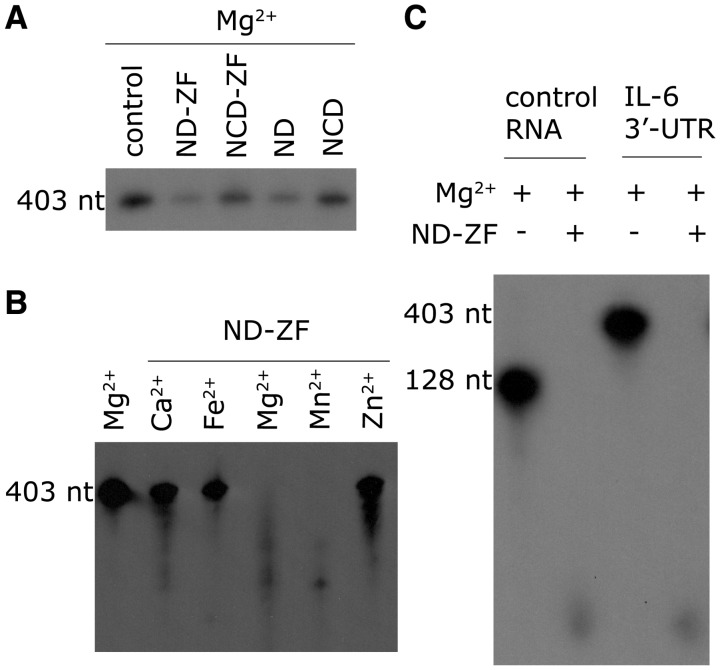
To determine the minimal part of MCPIP1 that is required for its RNase activity, we constructed four truncations, spanning residues 1–297, 112–297, 1–334 and 112–334 (referred to hereafter as ND, NCD, ND-ZF and NCD-ZF, Figure 1A). These four recombinant protein truncations were incubated respectively (50 ng each) with [32P]-labeled IL-6 mRNA 3′-UTR (50 ng) for 20 min in the presence of 5 mM Mg2+ (the reaction was stopped before the RNA was completely digested to reveal differences between the different truncations). All truncations were able to digest [32P]-labeled IL-6 mRNA 3′-UTR, but their RNase activities were different (Figure 2A). ND-ZF had the highest RNase activity while that of NCD was the lowest, suggesting that both the first 111 N terminal residues and the zinc-finger motif are required for efficient digestion of substrate mRNA. In addition, ND had comparable activity to ND-ZF, and both constructs had much higher activity than constructs which lacked the N-terminal region (NCD and NCD-ZF), implying that the N-terminal region is more important for RNase activity than the zinc-finger domain. From the above results, we conclude that while the conserved domain of MCPIP1 (112–297) is sufficient for RNase activity in vitro, the zinc-finger domain and the N-terminal region may also play important roles in RNA digestion in vitro and in vivo.
Figure 2.
MCPIP1 in vitro RNase activity assay. (A) The MCPIP1 conserved domain alone is sufficient for RNase activity. RNase activity, from highest to lowest, measured by the remaining RNA after a 20 min incubation, was as follows; ND-ZF (1–334), ND (1–297), NCD-ZF (112–334) and NCD (112–297). (B) Effect of bivalent cations on MCPIP1 RNase activity. MCPIP1 required either Mg2+ or Mn2+ to function as an RNase. MCPIP1 in the presence of Mg2+ or Mn2+ was able to digest RNA efficiently, but RNase activity was almost completely abolished in the presence of Ca2+, Fe2+ and Zn2+. (C) The ND-ZF construct showed no RNA selectivity in vitro. Both control RNA and the IL-6 3′-UTR were digested completely after a 1-h incubation with MCPIP1 ND-ZF protein in the presence of Mg2+.
We also tested the in vitro metal ion selectivity and the RNA specificity of the RNase activity. Ca2+, Fe2+, Mg2+, Mn2+ and Zn2+ (at a final concentration of 5 mM) were added separately to a system containing 50 ng [32P]-labeled IL-6 mRNA 3′-UTR and 50 ng ND-ZF to test their influence on RNase activity. High RNase activity was observed both in the presence of Mg2+ and Mn2+, while in the presence of Ca2+, Fe2+ and Zn2+, ND-ZF lost almost all its RNase activity (Figure 2B). Since the zinc finger might participate in RNA recognition, ND-ZF was also incubated respectively with IL-6 mRNA 3′-UTR and control mRNA in the presence of 5 mM Mg2+ to test for RNA specificity. No RNA was left after 1 h incubation in the presence of ND-ZF (Figure 2C). These results indicate that MCPIP1 is a Mn2+ or Mg2+ dependent RNase and that substrate specificity is lost in vitro in the absence of the C-terminal region of MCPIP1.
Overall structure of the conserved domain of MCPIP1
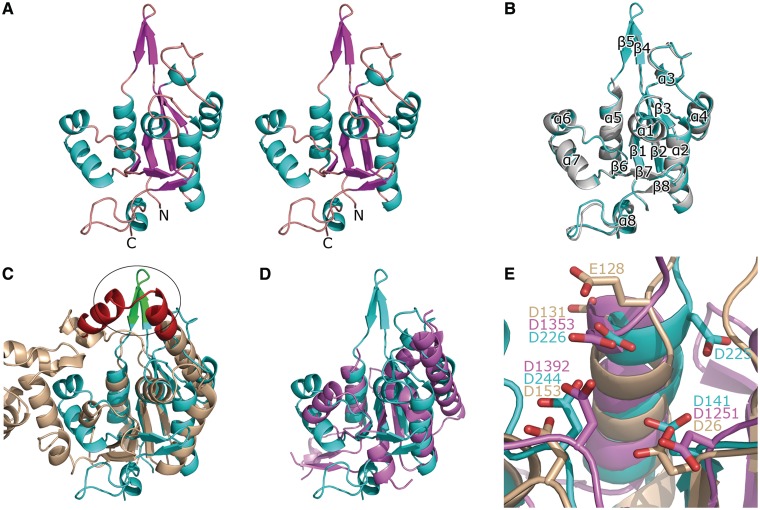
To obtain an atomic resolution structure for further analysis, we crystallized NCD and NCD-ZF and solved their structures. The NCD crystal belongs to space group P41 with unit cell parameters a = b = 56.4 Å, c = 113.2 Å. Matthews coefficient analysis (30) suggested that there are two molecules in one asymmetric unit at a solvent content of 43.08% (VM = 2.16 Å3/Da). The initial phases were obtained using the multiwavelength anomalous diffraction method. The N-terminal 22 residues (i.e. 112–133) and residues 216–222 are missing in the final model due to the absence of interpretable electron density. Residues 134–215 and 223–296 can be well traced in the density map, while residues 216–222 extended into the solvent region without interacting with neighboring molecules in the crystal and thus tended to be flexible. The NCD-ZF crystal belongs to space group P3121 and has a unit cell parameter a = b = 113.5 Å, c = 78.0 Å. The initial phases were obtained by MR using the NCD structure as a search model. As was the case for NCD, there are two molecules in one asymmetric unit, and loop 216–222 was well defined in the structure of NCD-ZF. This loop forms a positively charged arm whose significance will be discussed below. The CCCH zinc-finger motif (297–334) was totally disordered in our structure of NCD-ZF, although SDS–PAGE analysis confirmed that the CCCH zinc finger was not degraded in the crystal. Residues 134–296 were well build in chain A, while residues 188–190 were not able to be traced in chain B of the NCD-ZF structure due to the weak density. A stereo view of chain A of NCD-ZF structure was shown in Figure 3A. The structures of NCD and NCD-ZF are quite similar, and the RMSD between the two structures is 0.479 Å (Figure 3B). Secondary structures were labeled according to NCD-ZF structure in Figures 1B and 3B. The reliably modeled range of NCD-ZF structure was also indicated in Figure 1B as untraceable residues are marked with dot above them.
Figure 3.
Overall structure of MCPIP1 NCD and its comparison with homology proteins. (A) Stereo view of the crystal structure of MCPIP1 NCD-ZF. The structure of MCPIP1 NCD-ZF adopts an α-β-α sandwich-like fold. The residues 112–223 and the zinc-finger motif (residues 297–334) are not included in the model due to absence of interpretable electron density. (B) Superposition of the NCD-ZF structure on the NCD structure. The NCD structure is in violet and NCD-ZF in cyan. Eight α-helices and eight β-strands in the structure are numbered from the N-terminal to C-terminal respectively. There are only slight differences in the loop region between these two structures. (C) Superposition of the MCPIP1 NCD-ZF structure and the T5 D15 5′-exonuclease structure (pdb code: 1exn). The 1exn structure is colored in wheat and NCD-ZF in cyan. The oval indicates the positively charged arm of MCPIP1 (green) in the same position with the nuclear acid-binding arch of 1exn (red). (D) Superposition of the MCPIP1 NCD-ZF structure and the PIN domain of human SMG6 (pdb code: 2hww). The 2hww is colored in violet. A good fit in the central β-sheets and several α-helixes could be seen in this figure. (E) Conserved residues and similar arrangement of them in the catalytic center of MCPIP1 NCD-ZF (cyan), T5 D15 5′-exonuclease (wheat) and PIN domain of human SMG6 (violet). Residues involved in the catalytic center are shown in stick representation and labeled.
NCD and NCD-ZF were packed in an identical dimer arrangement in their respective crystals. However, analytical ultracentrifugation indicated that the monomer was the predominant form of MCPIP1 NCD in solution (data not shown). The dimer we observed may be a crystallographic artifact which has no physiological significance.
The NCD structure adopts a Rossmann-like α/β/α sandwich fold (Figure 3A and B). β-strands 1, 2, 3 and 6 form a β-sheet surrounded by sets of α-helices at both sides. Dali results show that MCPIP1 NCD shares highest structural homology with T5 D15 5′-exonuclease (PDB: 1exn) (31,32) (RMSD 2.4 Å for 92 Cα of aligned residues), suggesting a previous assumption that the conserved domain of MCPIP1 is a nuclease. Alignment of the NCD-ZF structure with that of T5 D15 5′-exonuclease showed that the central four-stranded β-sheet of the NCD-ZF structure is very similar to that of T5 D15 5′-exonuclease (Figure 3C). There was a good fit for the three α-helices (α 1, α 2 and α 5) surrounding the β-sheet in these two structures. While the arm in the T5 D15 5′-exonuclease structure that is responsible for nucleotide binding was not observed in our structure, a flexible positively charged region (residues 214–222) was found near the location of the nucleotide-binding arm. Since MCPIP1 N-terminal conserved domain shared remote homology with PIN domain as suggested in an early study (8), we also compared the NCD-ZF structure with PIN domain of SMG6 (PDB: 2hww) (33). A good fit in the central part of these structures was shown in Figure 3D. Although the sequence identity was <10%, the three structures share highly conserved catalytic center (shown in Figure 3E).
The catalytic center has several residues important for RNase function
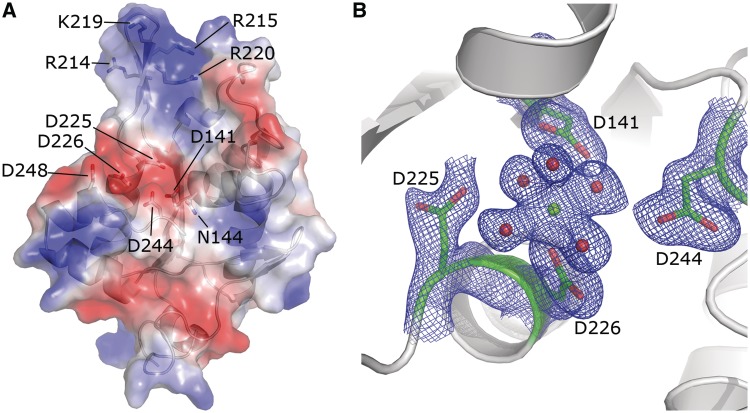
Analysis of the electrostatic surface potential of the NCD-ZF structure revealed a negatively-charged pocket which is presumably the catalytic center of this RNase domain (Figure 4A). Consistent with a previous prediction (8), this pocket is composed of several conserved acidic residues including Asp141, Asn144, Asp225, Asp226, Asp244 and Asp248.
Figure 4.
The detailed catalytic center and putative RNA-binding arm. (A) Electrostatic surface potential analysis of the NCD-ZF structure. A negatively-charged pocket formed by residues 141 Asp, 144 Asn, 225 Asp, 226 Asp, 244 Asp and 248 Asp was found in the structure of NCD-ZF. A positively charged arm was found close to the negatively-charged pocket. Four positively charged residues, 214 Arg, 215 Arg, 219 Lys and 220 Arg, are located in this region. (B) Density map of the NCD structure after Mg2+ soaking. A magnesium ion was found in the pocket, interacting directly with 226 Asp and five hydrogen molecules. Residues 141 Asp, 225 Asp and 226 Asp are thought to be important in stabilization of these water molecules.
To obtain an accurate structure of the catalytic center and identify which residues are important for processing RNA, we solved the structure after soaking magnesium into the NCD crystal. Although many nucleases require two metal ions for complete enzymatic activity, only one magnesium ion was found in the catalytic center of MCPIP1. The magnesium ion forms six coordination bonds with surrounding atoms including an oxygen from residue Asp 226 and five water molecules, forming an octahedron (Figure 4B). The length of these bonds ranged from 2.0 to 2.3 Å, consistent with the empirical value of 2.07 Å (34). Asp141, Asp225 and Asp244 form hydrogen bonds with these water molecules which help to maintain the magnesium ion in the correct position, suggesting that they may be important for the RNase activity of MCPIP1. While Asn144 and Asp248, which were previously predicted to be involved in the catalytic center (8), were 6.5 and 8.4 Å away from the magnesium ion, they had no interaction with either the magnesium ion or the water molecules around the magnesium ion, suggesting that they probably do not contribute to the RNase activity. Taken together, the structure of the MCPIP1 conserved domain containing a magnesium ion shows that Asp 141, Asp 225, Asp 226 and Asp 244 should be important for its RNase activity.
In vivo studies confirm which residues of the catalytic center are essential
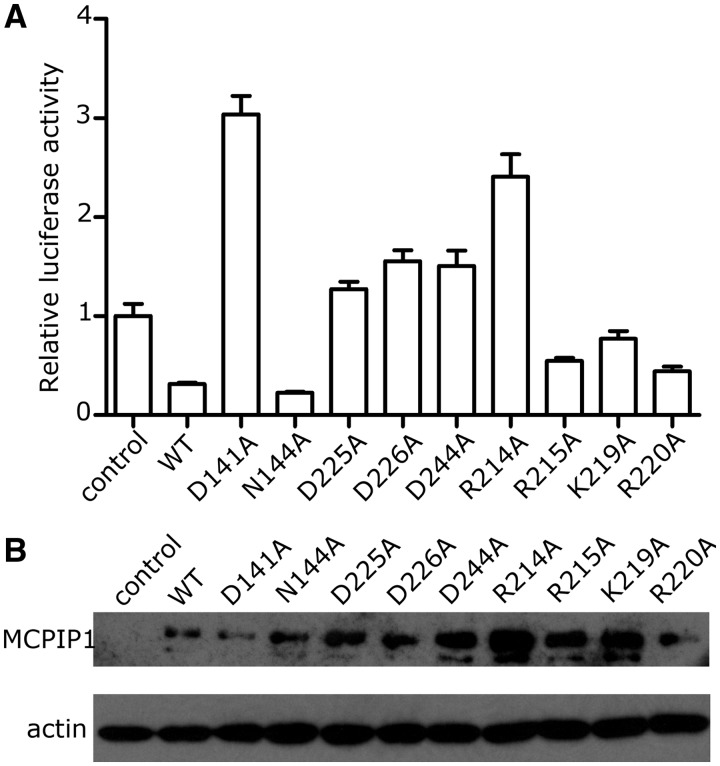
Luciferase assays were used to determine whether specific residues in the catalytic center could affect RNase activity in vivo. An IL-6 3′-UTR sequence was inserted upstream of the luciferase gene in a PGL3 vector and a full-length MCPIP1 cDNA was inserted into a pcDNA4 vector to be overexpressed. The PGL3-IL-6 3′-UTR and pcDNA4–MCPIP1 plasmids were cotransfected into 293A cells. Luciferase activity was measured 48 h after transfection. MCPIP1 protein expression was confirmed by western blotting (Figure 5B). In general, expression levels of MCPIP1 mutants were higher than those of the native protein, but some showed lower activity or even no activity at all. Results showed complete loss of RNase activity in single mutants D141A, D225A, D226A, D244A (Figure 5A), even though these mutations enhanced the level of protein expression. This result is easy to interpret since residue Asp 226 interacts directly with the magnesium ion while residues Asp 141, Asp 225 and Asp 244 hold the magnesium ion in position by coordination bond bridges mediated by water molecules. Mutation of any of these residues would disrupt the elaborate coordination bonds and hydrogen bond network which holds the magnesium ion in the correct place allowing native RNase activity. Also these four residues lie in the conserved catalytic center as suggested by superposition of NCD-ZF with 1exn and 2hww (Figure 3E). In contrast to these mutants, mutant N144A had the same RNase activity as native MCPIP1; this result is in contrast to previous studies but is consistent with the assumptions based on the crystal structure that Asn 144 may not participate in digesting RNA. Asn 144, which previous computational studies have suggested to be important, appears to play no significant role in RNase activity as measured by luciferase assays. These findings are consistent with findings from the structure that the magnesium ion is important in RNA digestion and that residues interacting with the magnesium ion directly or indirectly are all crucial for RNA decay.
Figure 5.
Mutagenesis studies of important residues of MCPIP1. (A) In vivo luciferase assay results. Luciferase activity is normalized. The exchange of residues with alanine in the catalytic center or those which interact with the magnesium ion resulted in the loss of RNase activity. Mutation of 144 Asn, which was located at some distance from the Mg2+ ion, did not affect RNase activity. Mutation of 214 Arg in the positively charged region also significantly affected RNase activity, while mutation of the other three positively-charged residues did not have such significant effect. Luciferase activity (relative) data are means + SEM from three independent experiments. (B) Western blotting of the cell lysate used in the luciferase assay. The protein expression level of mutant proteins was higher than that of the native protein.
A putative RNA-binding site is revealed by in vivo luciferase assays
The catalytic center of MCPIP1 can be clearly seen in our MCPIP1 structure. We noticed that four residues (Arg 214, Arg 215, Lys 219 and Arg 220) formed a positively charged arm located close to the catalytic center (Figure 4A). When we superposed our MCPIP1 structure with that of T5 5′-exonuclease [PDB: 1exn (31)], we found that this positively charged arm was in the same direction with a positively charged arch which is thought to be the nucleotide-binding region of T5 5′-exonuclease (oval region in Figure 3C). Since the positively charged arm was flexible, we hypothesized that this positively charged arm in MCPIP1 may also be involved in mRNA binding. In order to verify this speculation, a luciferase activity assay was used to test the RNase activity of different mutants in vivo (Figure 5A). The single mutation R214A yielded a high level of luciferase activity which was similar to the negative control in which there was no MCPIP1 protein expression, suggesting a complete loss of RNase activity. Mutation of Lys 219, Arg 215 and Arg 220 to alanine also yielded higher luciferase activity compared to wild-type MCPIP1, indicating that these residues may affect MCPIP1 RNase activity. Considering the location of these positively charged residues and their influence on RNase activity, we believe these residues may play a role in RNA binding. Our results indicate that these two positively charged antiparallel β-strands are likely to be a potential RNA-binding site.
DISCUSSION
MCPIP1 is a newly-identified CCCH type zinc-finger protein which plays an essential role in preventing immune disorders by negatively regulating the expression of cytokine IL-6 and IL-12 at the mRNA level. It targets the 3′-UTR region of IL-6 and IL-12 mRNAs and promotes their degradation. The NCD of MCPIP1 is thought to be responsible for its RNase activity, as it has been categorized into family RNase_Zc3h12a (PF11977) in Pfam database (35). In this study, we solved the 2.0 Å crystal structure of the MCPIP1 NCD. Dali results showed that the NCD of MCPIP1 shares highest structural similarity with a T5 exonuclease, confirming its RNase function. Using in vivo luciferase assays, we also identified four residues (Asp 141, Asp 225, Asp 226 and Asp 244) important for RNase activity. This finding provides significant insights into the mechanism of MCPIP1 RNase activity. The catalytic center was clearly visible in our structure since a Mg2+ ion was soaked into the crystal. A typical octahedron with a magnesium ion in the center was formed by five water molecules and Asp226. In vivo luciferase assays confirmed that Asp141, Asp225, Asp226 and Asp244 are the crucial residues in the catalytic center, consistent with their role in stabilizing the Mg2+ ion. Both our structure and in vivo luciferase assays show that Asn144, a residue which computational modeling had previously suggested is important, is probably not involved in RNase activity, highlighting the importance of atomic resolution 3D structures rather than computational models for understanding the details of enzyme function.
The first indication that MCPIP1 may have RNase activity was the discovery that the N-terminal sequence of MCPIP1 shares homology with the PilT N-terminus (PIN) domain which is present in flap endonuclease 1 (FEN1) superfamily proteins (36). The T5 5′-exonuclease (PDB: 1exn), which Dali analysis showed shares highest similarity with MCPIP1 NCD, is also a member of the FEN1 superfamily. Sequence alignment and structural superposition of the MCPIP1 NCD further suggests that it is a PIN nuclease. It has long been accepted that members of the FEN1 superfamily contain two metal ions in their catalytic center, both of which participate in their nuclease activity (36). However, in some members which only have a single metal ion in their catalytic center, the single metal ion is thought to play the same role in catalysis as the two metal ions (37). It has also been proposed that only one metal ion is essential for nuclease function (38). Here, only one Mg2+ ion was found in the structure of MCPIP1 NCD, and it is probable that MCPIP1 NCD represents a novel type of PIN domain which needs only one metal ion for its nuclease activity.
Another important question to be considered is how MCPIP1 achieves the substrate specificity observed in previous studies. MCPIP1 targets the 3′-UTRs of IL-6 and IL-12 b but not the 3′-UTR of TNFα. The MCPIP1 recognition site was thought to be a conserved AU rich element in the IL-6 3′-UTR. However, a region in MCPIP1 that can specifically bind to the IL-6 3′-UTR conserved element has not yet been identified. Moreover, in vitro RNase assays demonstrated that both of the N-terminal conserved domains of MCPIP1, with or without the CCCH zinc-finger motif, do not have substrate specificity. This difference suggests that an elaborate regulation mechanism is needed to confer MCPIP1 substrate specificity in vivo. A possible mechanism is that the NCD may be regulated by the MCPIP1 C terminal region or other adaptor proteins which still remain to be identified.
Given the current structural information, it remains an open question whether the zinc-finger motif is involved in RNA binding. In vivo luciferase assays revealed that Arg214 is crucial for RNase activity, suggesting that this region may be responsible for RNA binding. A protein–RNA complex structure is needed to investigate the interaction between MCPIP1 and the IL-6 3′-UTR. However, from the superposition of the MCPIP1 conserved domain and T5 D15 5′-exonuclease structures, we note that while our structure lacks the nucleotide-binding arch, it has a positively charged arm located near the position of the arch. This, together with results from in vivo luciferase assays, leads us to believe that the positively charged arm is responsible for RNA binding. Our current structure does not provide insights into the mechanism underlying the newly discovered deubiquitinase function of MCPIP1 (39). Availability of a full-length structure of MCPIP1 would facilitate further investigations of the molecular mechanisms underlying substrate specificity and deubiquitinase activity. Since the deubiquitinase activity is located in the N-terminus of MCPIP1, we expect that MCPIP1 is a novel type of deubiquitinase.
ACCESSION NUMBERS
The coordinates and structure factors have been deposited in the Protein Data Bank. Accession codes: 3V32, 3V33 and 3V34.
FUNDING
The ‘973’ Project [2011CB915501, 2011CB910304]; [SIMMKF1106FK-02]. Funding for open access charge: The ‘973’ Project [2011CB910304].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr X. Cai Zhang and Dr Yingfang Liu for helpful discussions. The authors are grateful to Xiaoxia Yu, Ya Wang, Zhongnian Zhou and other members of staff at the Structural Biology Core Facility of the Institute of Biophysics, Chinese Academy of Sciences (CAS).
REFERENCES
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Liu F, Gu J. Retinoic acid inducible gene-I, more than a virus sensor. Protein Cell. 2011;2:351–357. doi: 10.1007/s13238-011-1045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-kappaB and MCP-1-induced protein 1 RNase. FEBS J. 2009;276:5892–5905. doi: 10.1111/j.1742-4658.2009.07273.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM, Younce C, Binkley PF, Kolattukudy PE. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ. Res. 2006;98:1177–1185. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jura J, Wegrzyn P, Korostynski M, Guzik K, Oczko-Wojciechowska M, Jarzab M, Kowalska M, Piechota M, Przewlocki R, Koj A. Identification of interleukin-1 and interleukin-6-responsive genes in human monocyte-derived macrophages using microarrays. Biochim. Biophys. Acta. 2008;1779:383–389. doi: 10.1016/j.bbagrm.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One. 2008;3:e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, Fu M. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J. Biol. Chem. 2008;283:6337–6346. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 8.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 9.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, Jarzab B. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 12.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 13.Hake LE, Mendez R, Richter JD. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell. Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bidzhekov K, Zernecke A, Weber C. MCP-1 induces a novel transcription factor with proapoptotic activity. Circ. Res. 2006;98:1107–1109. doi: 10.1161/01.RES.0000223483.12225.80. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pape TS, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 2004;37:843–844. [Google Scholar]
- 18.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 19.Sheldrick GM. Macromolecular phasing with SHELXE. Z Kristallogr. 2002;217:644–650. [Google Scholar]
- 20.Sheldrick GM. SHELXC. Goettingen University; 2003. [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Afonine PV, Grosse-Kunstleve RW, Adams PD. CCP4 Newsl. 2005;42 http://www.ccp4.ac.uk/newsletters/newsletter42/content.html. [Google Scholar]
- 24.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 26.Huang Z, Wang X, Gao G. Analyses of SELEX-derived ZAP-binding RNA aptamers suggest that the binding specificity is determined by both structure and sequence of the RNA. Protein Cell. 2010;1:752–759. doi: 10.1007/s13238-010-0096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jie LI, Yutao YANG, Keqian MA, Zhuqing YAN, Xiaoxiao LI, Chunli ZHAO, Zhiqing X. Cloning and preliminary functional analysis of human galanin receptor 2 promoter. Acta Biophys. Sin. 2011;27:417–423. [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 30.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 31.Ceska TA, Sayers JR, Stier G, Suck D. A helical arch allowing single-stranded DNA to thread through T5 5'-exonuclease. Nature. 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 32.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harding MM. The geometry of metal-ligand interactions relevant to proteins. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1432–1443. doi: 10.1107/s0907444999007374. [DOI] [PubMed] [Google Scholar]
- 35.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat. Struct. Mol. Biol. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupureur CM. One is enough: insights into the two-metal ion nuclease mechanism from global analysis and computational studies. Metallomics. 2010;2:609–620. doi: 10.1039/c0mt00013b. [DOI] [PubMed] [Google Scholar]
- 39.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 2010;207: 2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]