Abstract
Cardiomyocyte development in mammals is characterized by a transition from hyperplastic to hypertrophic growth soon after birth. The rise of cardiomyocyte cell mass in postnatal life goes along with a proportionally bigger increase in the mitochondrial mass in response to growing energy requirements. Relatively little is known about the molecular processes regulating mitochondrial biogenesis and mitochondrial DNA (mtDNA) maintenance during developmental cardiac hypertrophy. Genome-wide transcriptional profiling revealed the activation of transcriptional regulatory circuits controlling mitochondrial biogenesis in growing rat hearts. In particular, we detected a specific upregulation of factors involved in mtDNA expression and translation. More surprisingly, we found a specific upregulation of DNA repair proteins directly linked to increased oxidative damage during heart mitochondrial biogenesis, but only relatively minor changes in the mtDNA replication machinery. Our study paves the way for improved understanding of mitochondrial biogenesis, mtDNA maintenance and physiological adaptation processes in the heart and provides the first evidence for the recruitment of nucleotide excision repair proteins to mtDNA in cardiomyocytes upon DNA damage.
INTRODUCTION
Mammalian heart development is characterized by a transition from hyperplastic to hypertrophic growth associated with a marked increase in myofibril density, the appearance of mature intercalated discs and the formation of binucleated cardiomyocytes. In rodents, this transition occurs shortly before birth and during the first weeks of postnatal life. In humans, the switch from hyperplastic to hypertrophic growth is delayed as indicated by continued proliferation of human cardiomyocytes during the first few months after birth, after which it slows down (1–3). After completion of cardiomyocyte cell division, postnatal growth is mostly due to hypertrophic increase in cardiomyocyte size resulting in uniform growth of the ventricular wall and septum to match the increase in chamber dimension (4). In order to supply sufficient energy for the contractile machinery, ATP producing mitochondria multiply during physiological hypertrophy, causing a sharp increase in mitochondrial mass concomitant with a switch in energy metabolism from glucose and lactate to more energy-efficient fatty acid oxidation (5).
Proliferation of mitochondria occurs by extension of the existing organellar network. Mitochondria in mature adult rat heart have a half-life of about 6 days requiring continuous replacement of existing mitochondria, which is in contrast to the cardiomyocytes themselves that are only rarely renewed (5). The mechanisms to increase the mitochondrial mass are highly complex and require orchestration of two genomes since only 13 subunits of respiratory chain (RC) complexes are encoded by the mitochondrial DNA (mtDNA). The vast majority, over 1000 proteins, are encoded by nuclear genes including constituents of biochemical pathways of energy metabolism and RC complexes, assembly factors, carrier proteins as well as factors required for maintenance, expression and translation of mtDNA (6).
Several transcriptional activators and regulators control expression of nuclear-encoded mitochondrial (NEM) proteins. For example, ubiquitous transcriptional regulators such as SP1 provide basic expression of NEMs while nuclear respiratory factors (NRFs) are necessary for dynamic control of most NEM genes (5). Moreover, NEM gene promoters frequently contain cAMP-responsive elements (CRE) enabling regulation via CRE-binding protein (CREB) and its functional relatives (5). The metabolic switch to β-oxidation of fatty acids is mostly accomplished by regulation of genes involved in fatty acid metabolism and import (7). Key regulators of these genes are the nuclear receptor-type transcription factors called peroxisome proliferator-activated receptors (PPARs) and their coactivators, such as PGC-1α (known as PPARGC1A in rats and mice) (8). While PGC-1α activity itself is CREB dependent and sensitive to physiological signals such as fasting, cold and exercise, the related PGC-1β (PPARGC1B in rodents) is less inducible. The third member of the PGC family, the PGC-1 related coactivator or PRC (PPRC1) seems to be modulated by the cell cycle. In contrast, control of mitochondrial RC complex assembly, fission, fusion, membrane biogenesis as well as replication and expression of mtDNA are also controlled by estrogen-related receptors (ERRα, β and γ) and NRFs, and not purely by PGC-1α/β-activated PPARs (8).
The closed circular 16.5 kb mtDNA is packaged with proteins, forming nucleoprotein complexes called nucleoids, which exist in multiple copies inside mammalian mitochondria (9). mtDNA copy number is strictly controlled during embryonic development (10) and its levels can increase 5-fold per nuclear gene copy during developmental postnatal hypertrophy of the human heart (11). Surprisingly little is known about maintenance and control of steady-state levels of mtDNA in tissues and how this is related to the overall respiratory capacity. For example, the increase of mitochondrial mass in adult human skeletal muscle (12,13), adult rat heart (14) or in insect flight muscles (15) does not correlate directly with an increase in mtDNA while upregulation of mitochondrial functions in transgenic mice over-expressing the cytosolic form of phosphoenolpyruvate carboxykinase (PEPCK-C) results in stable mtDNA copy number increase (16).
It is generally assumed that NEM proteins regulating mtDNA replication and expression are controlled by PGC-1α coactivated NRF-1 and NRF-2 (GABP in rodents) (8,17). One of the many targets of these factors is mitochondrial transcription factor A (TFAM), a ubiquitous mtDNA-binding protein required for transcription of mtDNA. TFAM is believed to act also as a major regulator of mtDNA copy number (18,19) since depletion of TFAM results in loss of mtDNA (20,21). However, TFAM seems to act together with other factors in a context-dependent manner since over-expression of TFAM in cultured cells is not sufficient to increase mtDNA copy number (21–23) although mice expressing human TFAM have elevated mtDNA levels (24). A similar situation holds true for TWINKLE (PEO1), a mtDNA helicase, which elevates mtDNA levels in transgenic mice (25), but not in cultured cells (26). Interestingly, simultaneous over-expression of TWINKLE and TFAM increases mtDNA copy numbers additively in mouse, indicating that the proteins affect copy number control independently (27).
To identify factors contributing to mitochondrial biogenesis and mtDNA maintenance in a relevant physiological context, we performed a genome-wide transcriptome analysis of rat hearts during postnatal development and correlated the results with changes in mtDNA copy number, organization and replication. Rather surprisingly, we found that developmental upregulation of mitochondrial capacity did not result in an increase of mtDNA copy number or in a change in its topological organization. Instead, the increase in mitochondrial biogenesis during early developmental cardiac hypertrophy coincided with increase of oxidative mtDNA damage and upregulation of DNA damage response genes, among them members of the nucleotide excision repair (NER) XPA and RAD23A. We provide the first evidence of NER in mitochondria and its relation to oxidative DNA damage. Our results suggest major differences in the mtDNA maintenance between human and rat hearts and provide novel insights into the regulatory network controlling mitochondrial biogenesis during postnatal heart development.
MATERIALS AND METHODS
Animals and transcriptome analysis
The animal experiments were performed in accordance with institutional and national ethical regulations and with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Embryonic day 18 (E18), neonatal (Neo), 10-day old (P10), young adult (6 months) Sprague-Dawley (SD) and young adult (6 months) and old (>18 months) Wistar (WS) rat hearts were collected and left ventricle RNA was immediately isolated with Trizol (Invitrogen) for the transcriptome analyses on Affymetrix Rat Genome 230 2.0 Array (28). As aged SD rats were not available, WS rats were chosen to represent aged heart samples and young adult WS rats were included to evaluate potential strain-dependent differences in gene expression between the two strains. Protein and DNA analyses were performed using neonatal (newborn to 3-day old), adolescent (10–21 days old), young adult (4–8 months) and old (>18 months) rat hearts. The microarray data has been submitted to ArrayExpress Archive (http://www.ebi.ac.uk/arrayexpress/) with access number E-MEXP-3590.
mtDNA analyses
Total heart DNA and mtDNA were isolated as previously described (29). Relative rat heart mtDNA copy number was determined by Southern blot using ClaI digested (cuts rat mtDNA once at 634 nt) total DNA separated on 0.7% 1× TBE agarose gels and quantifying the mtDNA signal using 18S rDNA as an internal loading control. 2D-AGEs, mtDNA topology analysis and Southern blotting were performed following standard procedures (21). Oxidative mtDNA damage was estimated using Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg, New England Biolabs). One microgram of total rat heart DNA was incubated with 8 U Fpg for 30 min under conditions recommended by the manufacturer. Cleavage of oxidized nucleotides by Fpg results in single-strand DNA breaks, which are normally visualized using alkaline gel conditions. As mtDNA completely disintegrates in alkaline gels due to its high ribonucleotide content (30), Fpg digested DNA was run in 0.7% 1× TBE agarose gels. The amount of linear mtDNA released from circular forms was used as an indicator of Fpg activity after blotting and hybridization with an mtDNA probe.
Human probes used in Southern hybridization have been described elsewhere (21). Rat probes were designed as follows (nucleotide coordinates indicated in the probe names, F—forward, R—reverse):
Rat mtDNA (GenBank AC_000022.2) probes
MT-10618F: CCACTCTTAATTGCCCTCA
MT-11167R: AGTTCGGCTGTGAATTCGTT
MT-15586F (7S F): TCTGTGTTATTAGACATGCACCATTA
MT-16180R (7S R): AGAATGGAGAAGCCTTTTAAGAATTA
Rat 18S gene (GenBank V01270.1) probes
18S-845F: CCGCAGCTAGGAATAATGGA
18S-1351R: AACTAAGAACGGCCATGCAC
South-Western blotting
To confirm data obtained using the Fpg-assay, 8-oxoguanine modifications (8-oxoG) of rat mtDNA were also analyzed using a South-Western approach and an antibody against 8-oxoG (Abcam mouse anti-8oxoG; ab64548). The XhoI digested rat heart mtDNA was run on gel and blotted on Hybond-N+ (Amersham) membrane as described above. After cross-linking, a 1.5-cm wide strip of the membrane containing the mtDNA was removed, cross-linked with UV and blocked in 5% non-fat milk in TBS-T for 1 h, washed once with TBS-T and incubated with primary antibody (1:1000) o/n. After incubation, the membrane was washed 2 × 30 s, 1 × 15 min, 2 × 5 min, followed by 2 h incubation with the peroxidase-coupled anti-mouse secondary antibody (1:10 000). Unbound secondary antibody was washed off as above and chemiluminescense (SuperSignal West Femto Chemiluminescent, Pierce) was detected using a VersaDoc imager (BioRad).
SDS–PAGE and Western blotting
A total of 7.5 and 12% SDS–PAGE (Laemmli) gels or precast NuPAGE 3–12% Bis–Tris gels (Invitrogen) were employed using standard conditions. Heart muscle samples were homogenized in 4% SDS, 10 mM Tris, pH 7.4, using an Ultra-Turrax and subsequently sonicated to reduce viscosity of the samples. DTT was added to a final sample concentration of 1 mM and samples boiled for 5 min after measurement of protein content. Western blotting and immunodetection were carried out using standard procedures and peroxidase-coupled secondary antibodies. Chemiluminescence detection and quantification was performed as above.
The following primary antibodies were used: Rabbit anti-TFB2M from Abcam (ab66014), goat anti-mouse/rat TFAM (mtTFA A-17) from Santa Cruz (sc-23588), rabbit anti-SOD2 from Abcam (ab13533), rabbit anti-HIF-1α from Novus Biologicals (NB100-449), rabbit IGF-1R from Santa Cruz (SC-712), mouse anti-STAT3 from BD Transduction Laboratories (612356), mouse anti-hHR23A (RAD23A) from Abcam (ab55725), chicken anti-PPARγ from Sigma (GW21258) and rabbit anti-GAPDH from Cell Signaling (14C10). The rabbit anti-PGC-1 antibody from Santa Cruz (sc-13067) raised against 300 amino acids N-terminal fragment of human PGC-1 is reported by the manufacturer to detect also the mouse and rat PGC-1. The rat PPARGC1A and PPARGC1B share 40% sequence similarity in their N-terminus and in our hands, the antibody detects a 90 and ∼109 kDa bands corresponding to the expected sizes of PPARGC1A and PPARGC1B, respectively.
Cardiomyocyte culture, immunocytochemistry and DNA-damage induction
Adult mouse cardiomyocytes were isolated using standard procedures (31) and grown on laminin-coated 3 cm plates or two-well chamber. For immunocytochemistry, cells were fixed with 3% PFA in PBS for 15 min at room temperature, washed twice with PBS, permeabilized in 0.5% Triton X-100 in PBS for 20 min, again washed twice with PBS and blocked using 3% BSA in PBS. After removal of the blocking solution, the cells were incubated with primary antibody (1:100 for anti-RAD23A and 1:200 for Santa-Cruz rabbit anti-TOM20, sc-11415) overnight at 4°C. Cells were washed twice with PBS, incubated at room temperature for 1 h in PBS with 1:500 dilution of cy2 (Dianova) or cy5 (Millipore) labeled secondary antibody and 2 ng/µl DAPI (Pierce). The washed coverslip was mounted using Mowiol 4-88 (Roth). For EdU incorporation the Invitrogen Click-iT® EdU Imaging Kit was used (Alexa Fluor® 594, C10339). Labeling of cells was achieved by overnight culture in 10 µM EdU. Signals were visualized following the manufacturers’ recommendations. Oxidative DNA damage was induced by incubating the cells in the presence of 1 µM doxorubicin (Sigma) for 16 h.
It should be noted that intercalating dyes such as DAPI and Picogreen are DNA conformation sensitive, making them suboptimal for staining supercoiled mtDNA. For example, altering mtDNA conformation by DNA-binding proteins can result in the loss of mitochondrial Picogreen signal without effects on mtDNA copy number (32). However, we noticed that Invitrogen’s Click-It® EdU detection kit buffer strongly enhances the DAPI signal from mtDNA. The kit is based on direct detection of EdU using click chemistry and assumable the component that enables reaction between the incorporated EdU and Alexa Fluor® azide also opens up mtDNA for DAPI.
RESULTS
In order to obtain a comprehensive view of postnatal transcriptional changes involving mitochondrial biogenesis and mtDNA maintenance, we performed a quantitative genome-wide transcriptomics analysis of embryonic Day 18 (E18), neonatal, 10-day old (P10), young adult (>6 months) and old (>18 months) rat hearts. We used two different rat strains, SD and WS rats, to identify strain-dependent variations in mitochondrial biogenesis and mtDNA maintenance and to bypass the limited availability of old SD rats. No major differences in the expression patterns between adult SD and WS hearts were monitored although SD rats expressed slightly higher levels of genes involved in immune system function, regeneration and collagen biogenesis whereas WS rats showed a barely significant enrichment of cuticular plate components (data not shown). No differences between SD and WS rats were detected in subsequent western and Southern blot analyses indicating that the regulation of mitochondrial biogenesis and mtDNA maintenance is subject to tight regulation and well-conserved within a species.
Massive upregulation of genes related to mitochondrial biogenesis during postnatal development
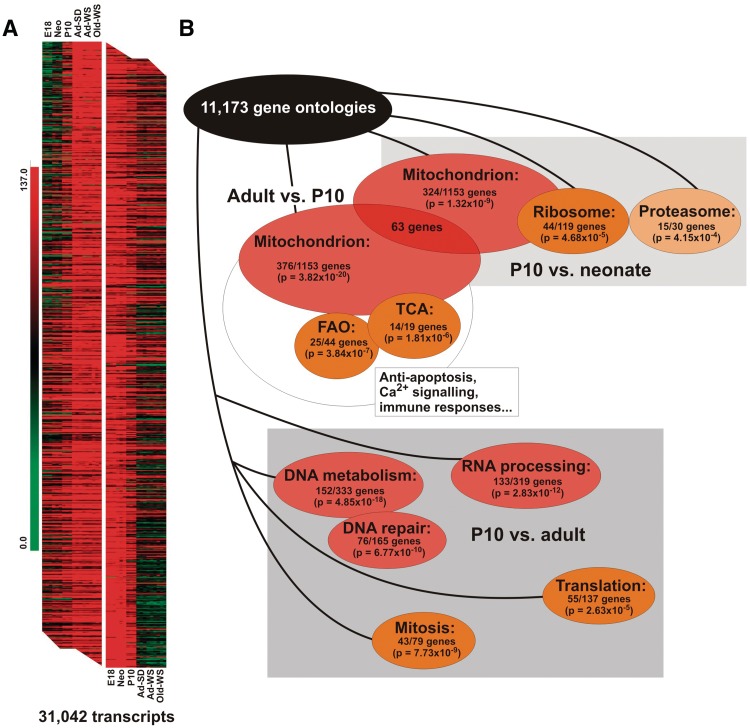
The gene expression patterns in rat heart changed dramatically during the postnatal period (Figure 1A). A total of 22% of all genes showed >1.5-fold expression differences between neonatal and adult hearts (Student’s t-test, P < 0.01 used for the initial sorting). A large fraction of these changes was due to differences in the expression of genes involved in mitochondrial biogenesis, oxidative phosphorylation and β-oxidation (Figure 1A and B; Supplementary Figure S1 and Supplementary Table S1). Almost one-third of all genes annotated to mitochondria (324 of 1153) were upregulated during the first 10 days of postnatal life. A total of 63 of these genes showed an even higher expression in adult hearts. Further 313 mitochondrially annotated genes were upregulated in adult compared to the P10 stage (Supplementary Table S1). In P10 rats, mitoribosomal proteins and other components of the translation apparatus dominated compared to other age groups, whereas induction of components of the OXPHOS chain and β-oxidation occurred only at later stages (Supplementary Figure S1 and Supplementary Table S1). Moreover, P10 rat heart, which is in transition from fetal- to adult-gene expression, displayed characteristics of both gene programs. The switch from fetal to adult genes, albeit evident upon closer inspection of expression data, was not immediately obvious from the gene ontology (GO) analysis since fetal and adult isoforms of the cardiac muscle contractile apparatus fall into the same cellular and functional category. Interestingly, P10 hearts showed a striking upregulation of DNA repair protein-gene expression (Figure 1B) constituting a unique property of P10 hearts.
Figure 1.
Pronounced upregulation of genes involved in mitochondrial and oxidative metabolism in adult rat hearts. (A) Heat-map clustering of embryonic Day 18 (E18), neonatal (Neo), 10-day old (P10) SD, adult SD (Ad-SD) (>4 months), adult WS (Ad-WS) and old (>18 months) WS rats. All three independent samples in each cohort cluster together and are separable from samples in other cohorts. (B) 11 173 out of 31 042 genes included in the Affymetrix microarray possess an annotated GO term. Mitochondrial and oxidative metabolism genes show significant upregulation at P10 compared to neonatal heart (324 out of 1153 annotated genes) and in adults compared to P10 heart (376 genes out of 1153). A total of 63 NEM genes upregulated at P10 are further upregulated in adults (see also Supplementary Table S1). P10 rat hearts show significant enrichment of genes involved in DNA repair when compared to any other age group (see also Supplementary Table S2). See (33) for the details of the GO term analysis. P-values are based on the hypergeometric score analysis of target (upregulated) and background (all GO terms) data sets (34).
Dynamic regulation of mitochondrial and NEM-gene expression during developmental cardiac hypertrophy
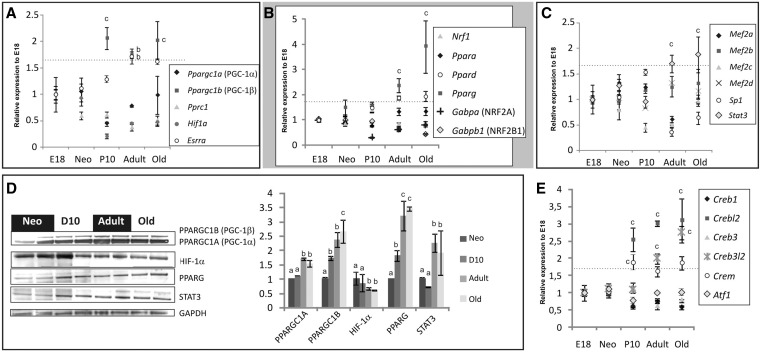
During postnatal development, the growing heart shows an impressive increase in both mitochondrial mass as well as in oxidative capacity. As PGC-1 transcriptional coactivators are important regulators of mitochondrial biogenesis and activity, we decided to have a closer look at the expression of the different isoforms of PGC-1, PGC-1α (PPARGC1A) and PGC-1β PPARGC1B). In contrast to other tissues (8), we found a striking correlation of PGC-1β PPARGC1B) expression to heart growth (Figure 2A and D) while the upregulation of PGC-1α (PPARGC1A) was less pronounced. PPARs, the main targets of PPARGC1s, as well as the key regulator of β-oxidation, ERRα (encoded by Esrra in rat), were strongly upregulated in adult animals, which reflects increased PGC-1β PPARGC1B) expression and enhanced requirement for fatty acids oxidation (Figure 2A and B; Supplementary Figure S1). Conversely, other key nuclear factors involved in the regulation of mitochondrial biogenesis such as Nrf1, Gabpa (and Gabpb in rat) (35) maintained rather constant expression levels during heart growth questioning a decisive role in the upregulation of mitochondrial mass (Figure 2B). Similarly, MEF2s, which control muscle specific COX subunit genes (35), showed little or no regulation (Figure 2C). However, the basal transcription regulator SP1 exhibited an expression peak at P10 rats, and the signal transducer and activator of transcription STAT3 showed a clear upregulation in aging animals (Figure 2C and D), which might be particularly relevant since STAT3 has been postulated to play an additional regulatory role inside mitochondria (36). Many NEM gene promoters possess CREB-binding sites (5,35) and the cytochrome c gene promoter is activated by CREB during muscle differentiation (37). Interestingly, three genes encoding proteins of the CREB-family, namely Crebl2, Creb3l2 and Crem, were upregulated during developmental cardiac hypertrophy indicating a role in the mitochondrial biogenesis in this tissue (Figure 2E).
Figure 2.
Transcriptional networks controlling heart mitochondrial biogenesis. (A) PPARGC1B ([Ppargc1b] PGC-1β in humans) is the only member of the PGC-1 family of transcriptional coactivators highly upregulated in P10 rat hearts. PPARGC1A ([Ppargc1a] or PGC-1α) maintains relatively stable expression while PPRC1 ([Pprc1] PRC in humans) as well as HIF-1α (Hif1a) are downregulated. (B) Expression of many known targets of PGC-1 s, such as NRFs (Nrf1, Gabps) is unchanged in growing rat hearts with exception of PPARG (Pparg), an important regulator of β-oxidation genes. (C) Stat3 expression increases concomitantly with postnatal heart growth. (D) Western blot analysis of the expression of some transcriptional regulators during postnatal heart development to validate transcriptome data. (E) Crem and Crebl2 but no other members of the Creb gene family are significantly upregulated during postnatal heart growth (a: P = n.s., b: P < 0.05 and c: P < 0.01, one-way ANOVA with Tukey’s multiple comparison test).
Aging has no effect on rat heart mtDNA topological organization
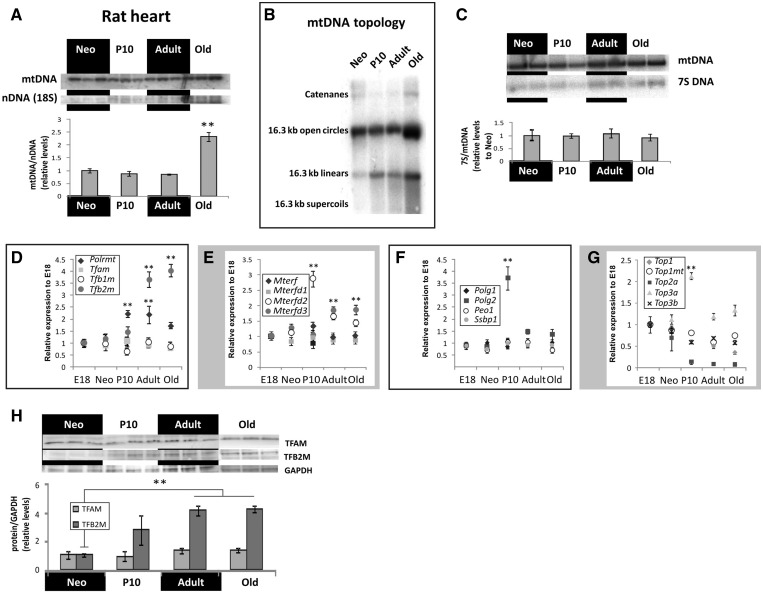
In humans, mtDNA copy number increases during childhood concomitant with profound changes in topological organization and replication mode of mtDNA (11). Surprisingly, we found that the rat heart followed a different path. Despite the massive upregulation of nuclear mitochondrial genes and enhanced mitochondrial biogenesis in postnatal rat hearts, no changes in mtDNA copy number, topological organization or in 7S DNA quantity occurred (Figure 3A–C). Higher levels of mtDNA were detected only in aging hearts, which is in line with earlier measurements of old rat hearts (38). In addition, we detected increased quantities of mtDNA replication intermediates in aged rat hearts compared to younger individuals (Supplementary Figure S2), although no significant changes in replication mode of mtDNA was observed.
Figure 3.
mtDNA maintenance and expression in growing rat heart. No significant differences in mtDNA copy number (A), topological organization (B) or 7S DNA quantity (C) were detected between neonatal and adult rat hearts. Aged rats had about 2-fold higher mtDNA levels than other age groups. Of the known factors involved in the expression of mtDNA, mitochondrial RNA polymerase POLRMT, TFB2M (D), MTERFD2 and MTERFD3 (E) show a significant positive response to increasing mitochondrial function in postnatal rat heart. The accessory subunit of POLG, POLG2 (F) and the topoisomerase TOP3A (G) were the only proteins involved in mtDNA maintenance that were induced during increased mitochondrial biogenesis. (H) TFAM and TFB2M were used to confirm the transcriptome results on protein level. Western blots were quantified and normalized against neonatal rats as loading control. Results are presented as mean ± SD. (**P < 0.01, one-way ANOVA with Tukey’s multiple comparison test).
The regulation of mtDNA expression and maintenance factors during postnatal rat heart development
Among the classical mtDNA maintenance factors, TFAM levels have been suggested to correlate with the amount of mtDNA in cells (19). In the rat heart, TFAM RNA and protein levels remained rather constant (Figure 3D and H) throughout life, which corresponded to the mtDNA copy number in young rats. In contrast, the expression of the mitochondrial transcription factor (TFB2M) and the mitochondrial RNA polymerase (POLRMT) increased in growing rat hearts (Figure 3D and H), correlating with the increased mitochondrial biogenesis. The mitochondrial transcription terminating factor mTERF (or MTERF1), belonging to the MTERF family of mitochondrial regulators, seemed to maintain a steady expression level throughout postnatal heart growth (Figure 3E). However, the expression of MTERFD2 protein [or MTERF4 in (39)] rose almost 3-fold in P10 rats, and was also maintained at significantly higher expression level in aged animals compared to neonates. Similarly, MTERFD3 [MTERF2 in (39)] showed higher expression levels in adult and aged animals.
Relatively few factors involved in mtDNA replication are known. The minimal replisome can be reconstituted in vitro using DNA Polymerase γ (POLG1), the DNA helicase TWINKLE (PEO1 in rat) and the mitochondrial single-stranded DNA-binding protein (SSBP1) (40). All of these factors displayed constant expression levels during postnatal heart development (Figure 3F). Only POLG2, the accessory subunit of the polymerase γ rose by almost 4-fold in 10-day old rats compared to neonates and maintained a statistically significant higher expression level in aged rats, too. A similar expression profile was found for topoisomerase IIIα (Top3a; Figure 2G). Top3a is a decatenase, capable of resolving catenated DNA as well as replication and recombination junctions in the nucleus (41). An alternative translation product of Topoisomerase IIIα is required for mtDNA maintenance in Drosophila (42). Expression of all other nuclear topoisomerases declined sharply during cardiac growth as expected for post-mitotic tissue whereas expression of the mitochondrial paralogue of Top1 (43), Top1mt, was only mildly reduced in aging hearts.
Increased oxidative mtDNA damage and specific upregulation of the DNA repair mechanisms in growing rat heart
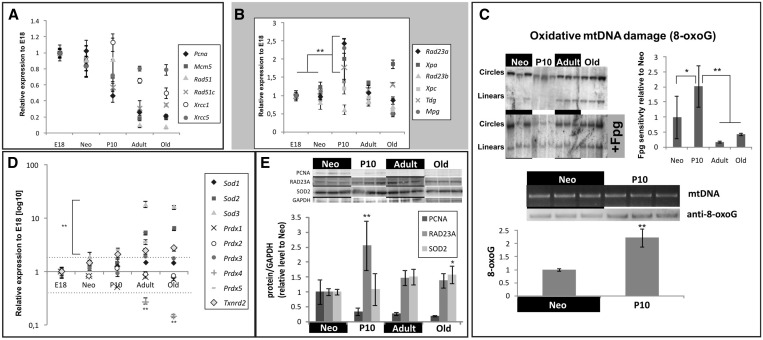
Despite a significant downregulation of factors involved in nuclear DNA (nDNA) maintenance, we found a significant induction of DNA repair genes in 10-day old rat hearts (Figures 1B and 4A and B). While many of these genes are either involved in direct repair of alkylated nucleotides or in base excision repair (BER), we also detected an upregulation of several genes directing NER (Figure 4B, Supplementary Table S2). It seems likely that the relative downregulation of proteins involved in nDNA maintenance during developmental heart hypertrophy results from the post-mitotic state of adult cardiomyocytes as well as the smaller nucleus/cytoplasm ratio compared to fetal and newborn cardiomyocytes. Since cardiomyocytes are effectively post-mitotic and thus no longer exposed to genetic stress caused by cell division, a major cause of DNA damage in cells (44), we reasoned that any induction of repair proteins is related to mitochondrial biogenesis. As the main natural source of DNA damage in a post-mitotic internal organ is reactive oxygen species (ROS), we measured the relative amounts of 8-oxoG substitutions in rat heart mtDNA by E. coli Fpg assay and by direct antibody-mediated detection of 8-oxoG (Figure 4C). mtDNA from 10-day old rat hearts exhibited significantly increased amounts of 8-oxoG compared to all other age groups indicating increased oxidative DNA damage in postnatal growing rat hearts. So far, it has been assumed that cellular antioxidant defenses should effectively detoxify ROS originating from physiological processes and thus protect young animals from oxidative DNA damage (45). Yet, the bulk of antioxidant enzymes are upregulated only at later stages (Figure 4D) permitting increased oxidative DNA damage during the postnatal transition period when mitochondria have increased their activity and thereby also ROS production. Delayed activation of the antioxidant defense system was particularly evident for the mitochondrial superoxide dismutase SOD2, which was expressed at high levels only in adult and aging but not in adolescent heart muscle. Interestingly, the increase of SOD2 mRNA in aging hearts was not matched by a concomitant increase at the protein level, which is contrast to other DNA maintenance proteins such as PCNA and RAD23A that showed comparable effects both at mRNA and protein levels (Figure 4E). Most likely, SOD2 is post-transcriptionally regulated in aged hearts by regulation of its translation or by changes in protein stability.
Figure 4.
Induction of DNA repair genes corresponds to increased oxidative damage during postnatal heart development. (A) Factors involved in nDNA replication (Pcna, Mcm5), single- (Xrcc1) or double-strand break repair (Rad51, Rad51c and Xrcc5) are either downregulated or unchanged in growing heart. (B) Factors either involved in NER (Rad23a, Xpa) or BER (Tdg, Mpg) are transiently upregulated in P10 rat hearts (Supplementary Table S2). (C) Upregulation of NER and BER genes coincides with increased sensitivity of mtDNA to Fpg, an enzyme capable of cleaving a wide range of oxidized purine-bases. Neonatal and P10 rat heart mtDNAs were probed for the presence of 8-oxoG modifications by South-Western blot analysis. (D) Cellular antioxidant defenses are upregulated only in the adult animals although increased levels of mtDNA damage are evident only in P10 rats. (E) Changes in the expression of nDNA maintenance genes (PCNA), NER (RAD23A) and antioxidant defenses (SOD2) were validated on protein level. Western blots were quantified and normalized against neonatal rats as loading control (**P < 0.01, *P < 0.05, one-way ANOVA with Tukey’s multiple comparison test).
Localization of a NER protein to cardiac mitochondria upon oxidative DNA damage
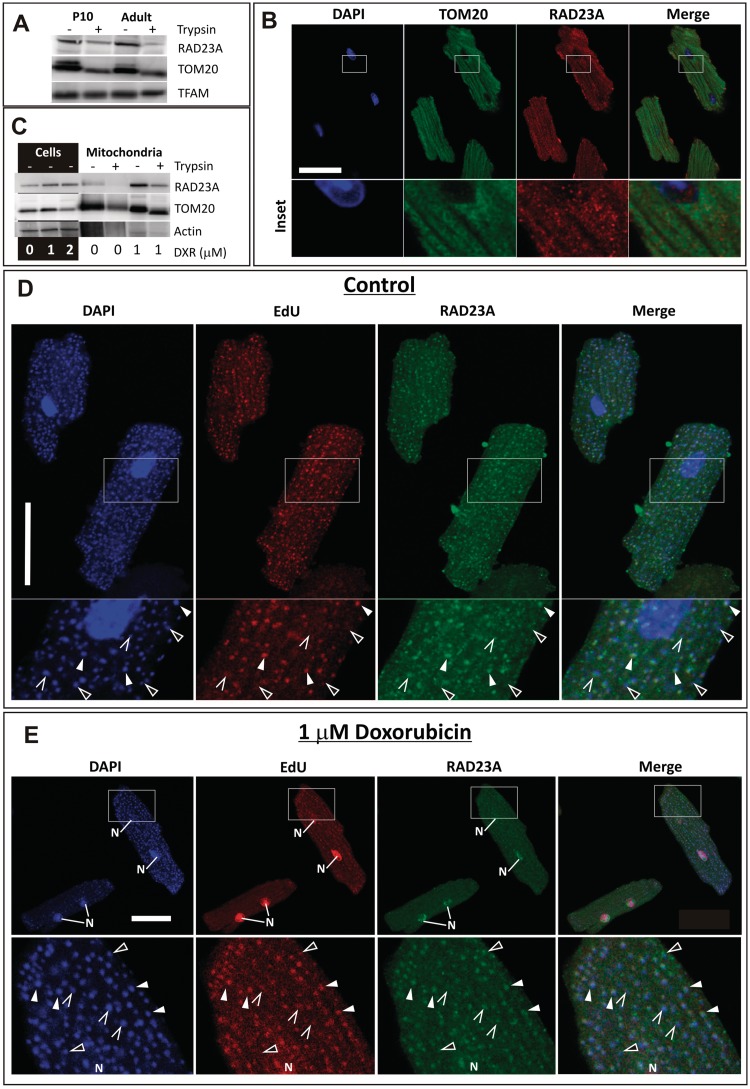
So far, no clear evidence exists for NER in mitochondria, which seem to rely mostly on direct repair of DNA damage or BER (46). However, as the expression of some NER proteins correlated with increased mitochondrial biogenesis and oxidative mtDNA damage, we decided to investigate the possibility of their mitochondrial function. RAD23B and XPC are the classical partners for DNA damage recognition for NER in the nucleus (47) and their levels are stable or decreasing during heart growth (Figure 4B). However, RAD23B can be substituted by its homolog RAD23A (48), which is strongly upregulated during increased mitochondrial biogenesis. Interestingly, RAD23A was also detected in sucrose gradient purified rat heart mitochondria and was partially resistant to trypsin digestion of these preparations, indicating a mitochondrial localization (Figure 5A). Immunocytochemical analysis of cardiomyocytes revealed a partial co-localization of RAD23A to the mitochondrial network, sometimes presenting as distinct foci (Figure 5B).
Figure 5.
RAD23A, a key NER protein is localized to cardiomyocyte mitochondria. (A) Western blot analysis of RAD23A presence in sucrose-gradient purified rat heart mitochondrial preparations subjected to trypsin digestion. TOM20 a mitochondrial outer membrane protein, partially accessible to trypsin, and the mitochondrial matrix protein TFAM were used as positive and negative controls, respectively. (B) Immunofluorescent analysis of the localization of RAD23A in the mitochondrial network (TOM20 and DAPI were used to label mitochondria and nucleus, respectively). (C) DXR for 16 h causes a marked increase in RAD23A protein levels in whole cell as well as in mitochondrial preparations. Sarcomeric actin demonstrates the relative enrichment of mitochondria in the preparations. (D) RAD23A co-localizes with mtDNA partially with nucleoids (cytoplasmic DAPI foci) under basal conditions without DNA damage induction. All nucleoids that had incorporated EdU stained positive for RAD23A (white arrows, N = 3 cells, 300 nucleoids). Open arrows indicate nucleoids that were positive for RAD23A but not obviously for EdU. Closed arrows show nucleoids negative for both EdU and RAD23A. (E) Exposure of cells to 1 µM DXR for 16 h results in the uptake of RAD23A into nuclei (N), together with more contrasted mitochondrial nucleoid staining. The nuclear signal in the EdU panels is due to auto-fluorescence of DXR when bound to nDNA, although some EdU incorporation due to nDNA repair cannot be excluded. The nucleus in the lower panels is mostly out of the confocal plane. Arrows as above. All scale bars 25 µm.
Since similar foci are formed by mtDNA that is organized as nucleoprotein complexes or nucleoids in mitochondria (49), we analyzed the presence of RAD23A in mtDNA nucleoids. In order to distinguish inactive from replicating nucleoids ones, we used DAPI in combination with EdU incorporation. Furthermore, the cardiomyocytes were treated with 1 µM doxorubicin (DXR), a potent chemical oxidizer of DNA (50,51), simultaneously with the addition of EdU to investigate the role of RAD23A in oxidative DNA lesion repair.
DXR treatment for 16 h resulted in increased RAD23A protein levels in both total cardiomyocyte lysates and mitochondrial preparations (Figure 5C). Although RAD23A was convincingly localized to cardiomyocyte mitochondrial nucleoids already under basal conditions (Figure 5D), the DXR treatment diminished the diffuse cytoplasmic RAD23A signal and induced recruitment of RAD23A to the nucleus (Figure 5E). In both cases, not all nucleoids were positive for EdU or RAD23A. Interestingly, although RAD23A was present also in inactive nucleoids, all nucleoids that had incorporated EdU stained positive also for RAD23A.
DISCUSSION
Surprisingly, little is known about the regulation of mitochondrial biogenesis during postnatal heart development although the switch from glycolytic to oxidative metabolism is one of the key features that distinguish fetal and adult heart gene programs (7,8,52). Moreover, we also know relatively little about the physiological regulation of mitochondrial biogenesis and mtDNA maintenance. Especially, the regulation of mtDNA copy number in cells and tissues is only poorly understood (10,53). In the present study, we assessed the physiological consequences of the developmental switch on mitochondrial biogenesis and maintenance. Our results provide novel insights into both mtDNA maintenance and potential regulatory circuits directing mitochondrial biogenesis.
Postnatal heart development is closely linked to mitochondrial biogenesis
Developmental cardiac hypertrophy is a gradual process, which makes it difficult to identify all relevant effectors based on expression data. However, the advent of genome-wide comparative transcriptomics facilitates identification of potential ‘switch-on’ type regulators and provides detailed insights in complex regulatory changes between different age groups. We found that one-third of all mitochondrial proteins increased their expression more than 1.5-fold during early postnatal life, which identifies this group of genes as one of the major constituents of the rapid transition of the fetal expression program to a more adult-type during the first 10 postnatal days (Figure 1). The need for a significant upregulation of genes involved in mitochondrial biogenesis is underscored by the fact that relative mitochondrial volume increases 2.6-fold from P1 to P10 while the volume of cardiomyocytes in the left ventricle increases only 1.5-fold in the same time period (54). We found that the gain of mitochondrial mass starts with increased expression of mitoribosomal proteins and other components of the translation apparatus required for setting up the mitochondrial compartment, they are only later joined by increased expression of components of the OXPHOS chain and β-oxidation (Supplementary Figure S1 and Supplementary Table S1), which argues for tightly controlled, distinct regulatory circuits.
POLRMT and TFB2M showed the highest upregulation during heart growth (Figure 3D) of known factors required for expression of mtDNA-encoded genes, which fits the role of TFB2M as an accessory subunit of POLRMT required for promoter recognition (55,56). Other putative regulators of mtDNA expression, which were strongly expressed during postnatal heart growth, belong to the mTERF family. mTERF (MTERF1) is a transcription terminator required for rRNA expression (57), but is also involved in mtDNA replication pausing where it is suggested to regulate the passage of replication fork and RNA polymerase in the highly transcribed rRNA region (58). mTERF (MTERF1) is rather stably expressed during heart growth indicating a constant requirement for mitochondrial function. The same is true for MTERFD1 [MTERF3, (59)], whereas MTERFD2 (MTERF4) and MTERFD3 (MTERF2) show an impressive upregulation during postnatal heart growth suggesting an important role in de novo mitochondrial biogenesis. Notably, the expression pattern of MTERFD2 correlates with many mitochondrial ribosomal proteins that are strongly upregulated in adolescent animals and reach an expression plateau in aging hearts (Figure 3E, Supplementary Figure S1) suggesting a role in mitochondrial translation (60). MTERFD3 has been suggested to modulate mitochondrial transcription and OXHPOS function (61), a role, which corresponds nicely to its co-expression with TFB2M (Figure 2D–H) and mitochondrial transcripts (Supplementary Figure S1). MTERFD2 and MTERFD3 proteins, together with POLRMT and TFB2M, are likely to be also important regulators of mitochondrial gene expression needed to meet the increased energy demand of the adult heart.
Transcriptional networks controlling mitochondrial biogenesis in postnatal heart
Under physiological conditions, PPARGC1A responds to stimuli such as exercise, fasting and cold (8). Overexpression of PPARGC1A (PGC-1α) in mice dramatically increases mitochondrial biogenesis in heart and skeletal muscle (8,62). In our rats, the expression level remained rather stable in P10 animals despite the general upregulation of mitochondrial biogenesis during heart growth (Figure 2A and D). In contrast, we found that PPARGC1B (PGC-1β) was dramatically upregulated in growing rat hearts both on transcript and protein level, suggesting that that PPARGC1B is the key developmental activator of mitochondrial biogenesis while PPARGC1A is responsible for fine-tuning of mitochondrial activity in response to physiological stimuli.
Unlike, in the case of PPARs, which control β-oxidation, we did not find a strong increase in NRFs (Figure 2B), postulated to act as key expression regulators of mtDNA gene expression, electron transport chain assembly, mitochondrial fission/fusion and membrane biogenesis. As CRE-binding proteins (CREBs) are known to control many mitochondrial genes, we propose that Crem, Crebl2 and Creb3l2 could be important for the heart mitochondrial biogenesis since their expression specifically increases during postnatal rat heart growth.
Another transcriptional factor strongly upregulated during postnatal heart development was STAT3, originally characterized as a nuclear transcription activator and known to be important for heart function (63). Recently, STAT3 has been shown to regulate mitochondrial electron transport chain activity; it decreases ROS production and protects cardiomyocytes from ischemic damage (36). Although the molecular details of this mechanism are yet unclear, the mitochondrial role of STAT3 is in agreement with our data indicating a strong STAT3 upregulation in adult hearts with high OXPHOS capacity.
Mitochondrial biogenesis and mtDNA maintenance in heart
During the postnatal period, human heart mtDNA experiences dramatic changes in copy number, organization and mode of replication (11). In contrast to adult human hearts, which seem to employ recombination-dependent replication, murine hearts and human neonates have a simple mtDNA organization and utilize standard θ-type replication (9,29). Interestingly, we did not observe any changes in the topological organization of rat mtDNA (Figure 3B) and detected only subtle changes in the replication (Supplementary Figure S2) during postnatal heart growth, which further emphasizes the differences in the biogenesis of heart mitochondria between human and rodents. In agreement with earlier reports (38), mtDNA levels remained unchanged during development while an ∼2-fold increase in the mtDNA copy number was observed in the aged rat heart (Figure 3A). Since cardiomyocytes grow in size resulting in a 15-fold larger volume in 6-month old adult rats compared to neonates (54), the relative amount of nDNA compared to total cellular mass decreases in adult rat compared to neonatal heart. While in neonatal rats nuclei constitute almost 9% of left ventricle cell volume, the proportion drops to <1% in 6-month old rats (54). However, since the ratio between nDNA and mtDNA remains nearly constant between neonatal and adult heart, obviously fewer mtDNA copies are sufficient to maintain the larger mitochondrial volume with higher oxidative capacity in larger adult rat cardiomyocytes. Unlike human newborns, it seems that neonatal rat hearts possess an excess of mtDNA compared to the size of their mitochondrial compartment. It seems likely that the functional parameters responsible for species-specific differences in heart mtDNA maintenance, are relative workload, reserve capacity, beat rate and duration of adult life. Similarly, the different body and heart size of young adults compared to aging animals (body weight 250 g in young versus up to 600 g in aged rats, heart weight 700 mg versus 1.5 g, respectively) might account for the modest increase in mtDNA copy number during the adult rat life.
Our observations clearly question the relevance of mtDNA copy number for mitochondrial biogenesis and oxidative capacity. Apparently, rats do not require an increase of mtDNA to cope with increased mitochondrial biogenesis during postnatal heart development. In fact, this unexpected conclusion corresponds well to other physiological states in which an increase in mitochondrial mass and oxidative capacity does not go along with increased mtDNA copy number (12,13,15,37). RRM2B knockout mice represent a striking example in this respect, showing severe mtDNA depletion but no obvious effect on OXPHOS capacity (64). Obviously, high OXPHOS capacity can be maintained with only limited amounts of mtDNA leaving the question why murines and humans employ such different strategies for maintenance of heart mtDNA. Most likely, the high mtDNA copy number and peculiar replication mode in adult human heart (29) is not related to the high ATP demand but caused by some other physiological requirements.
The expendable genome: oxidative stress and mtDNA maintenance
One of the physiological causes that might require high mtDNA copy numbers and a special mode of replication in human cardiomyocytes is the high and persistent exposure to ROS during the long adult life-time of human beings (9,29). mtDNA is located in a highly oxidative environment and several lines of evidence show that mtDNA is an immediate target for ROS (65). It has been suggested that replication and turnover of mtDNA in somatic cells is energetically more efficient than damage repair, unlike nDNA (66). Moreover, the high copy number of mtDNA might act as a reserve during damage, continuously supplying some undamaged templates for ‘good enough’ expression of mitochondrial genes.
In our study, we detected a dramatic upregulation of DNA repair proteins in adolescent rat hearts coinciding with increased oxidative mtDNA damage (Figure 4). Notably, upregulated DNA repair proteins included members of NER and to a lesser extent also BER and some double-strand break repair (DSBR) proteins but not mismatch repair proteins (MMR; Figure 4, Supplementary Table S2). While BER and MMR have been reported to be present in mitochondria, so far, no evidence has existed for NER proteins (46). Although NER has been traditionally associated with repair of UV-induced DNA lesions, there is increasing evidence of its importance in the repair of oxidative damage as well (67). In this study, we analyzed, in detail, the distribution of RAD23A as representative of NER proteins. We found an elevated expression of RAD23A during mitochondrial biogenesis and demonstrated that it co-localizes with mtDNA. RAD23A was recruited to both mitochondria and nucleus upon oxidative DNA damage (Figure 5), which makes it another DNA repair protein with two homes (68,69).
DXR is a DNA-intercalating anticancer drug, whose main mode of DNA damage is based on its ability to produce superoxide radicals from molecular oxygen while being bound to DNA (50,51). DXR has been shown to cause remodeling of mitochondrial nucleoids and mtDNA copy number depletion in proliferating cells, apparently due to inhibition of replication (70). Interestingly, in agreement with our data, the same study reported that some nucleoids did incorporate BrdU (analogous to EdU used in our study) despite the presence of DXR. It is plausible that this EdU incorporation is due to repair processes. Clearly, functional studies are needed to decipher the actual composition of the putative mitochondrial NER machinery as well as the mechanisms of their recruitment into mitochondria.
Besides mitochondria, monoamine and NADPH oxidases are the other known major sources of ROS in heart muscle (71). None of these were substantially upregulated in P10 rats (Supplementary Figure S3). This suggests that the main source for elevated ROS levels during early phases of mitochondrial biogenesis derives from sub-optimal function of the RC releasing high levels of ROS. It is tempting to speculate that increased ROS levels play a physiological role during heart muscle development since ROS at P10 stage are not neutralized by cellular antioxidant defenses. In fact, it is well-known that ROS act as important stimulators of cardiomyocyte differentiation and hypertrophic signaling pathways (72) and might induce adaptation responses that result in long-term reduction of oxidative stress in cells (73).
Our study provides the first comprehensive insight into mitochondrial biogenesis during postnatal heart development, and the complex machinery required for the maintenance and expression of mtDNA. We believe that our work lays the groundwork for future functional studies that will enable us to distinguish functionally important mechanisms from epiphenomena.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–3.
FUNDING
European Molecular Biology Organization (ALTF 1125-2009 to JLOP), Academy of Finland, the Max-Planck Society, the Excellence Cluster Cardiopulmonary System (ECCPS), the University of Giessen-Marburg Lung Center (UGMLC), the Cell and Gene Therapy Center (CGT) of the University of Frankfurt. Funding for open access charge: Max Planck Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Marion Wiesnet, Dr Matthew Wheeler, Jennifer Norris and Aaron Ruhs (all MPI, Bad Nauheim) for their excellent technical support. Dr Ian Holt (MRC Mitochondria Biology Unit, Cambridge, UK), Prof. Hans Spelbrink (University of Nijmegen, The Netherlands), Dr Eric Dufour and Prof. Howy Jacobs (University of Tampere, Finland) are thanked for their valuable comments.
REFERENCES
- 1.Tam SK, Gu W, Mahdavi V, Nadal-Ginard B. Cardiac myocyte terminal differentiation. Potential for cardiac regeneration. Ann. N Y Acad. Sci. 1995;752:72–79. doi: 10.1111/j.1749-6632.1995.tb17407.x. [DOI] [PubMed] [Google Scholar]
- 2.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ. Res. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- 4.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 5.Goffart S, von Kleist-Retzow JC, Wiesner RJ. Regulation of mitochondrial proliferation in the heart: power-plant failure contributes to cardiac failure in hypertrophy. Cardiovasc. Res. 2004;64:198–207. doi: 10.1016/j.cardiores.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 6.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ. Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 8.Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ. Res. 2010;107:825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pohjoismaki JL, Goffart S. Of circles, forks and humanity: topological organisation and replication of mammalian mitochondrial DNA. Bioessays. 2011;33:290–299. doi: 10.1002/bies.201000137. [DOI] [PubMed] [Google Scholar]
- 10.Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 11.Pohjoismaki JL, Goffart S, Taylor RW, Turnbull DM, Suomalainen A, Jacobs HT, Karhunen PJ. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS One. 2010;5:e10426. doi: 10.1371/journal.pone.0010426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes MA, Carraway MS, Piantadosi CA, Reynolds CM, Cherry AD, Wester TE, Natoli MJ, Massey EW, Moon RE, Suliman HB. Carbon monoxide, skeletal muscle oxidative stress, and mitochondrial biogenesis in humans. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H392–H399. doi: 10.1152/ajpheart.00164.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taivassalo T, Gardner JL, Taylor RW, Schaefer AM, Newman J, Barron MJ, Haller RG, Turnbull DM. Endurance training and detraining in mitochondrial myopathies due to single large-scale mtDNA deletions. Brain. 2006;129:3391–3401. doi: 10.1093/brain/awl282. [DOI] [PubMed] [Google Scholar]
- 14.Wiesner RJ, Hornung TV, Garman JD, Clayton DA, O'Gorman E, Wallimann T. Stimulation of mitochondrial gene expression and proliferation of mitochondria following impairment of cellular energy transfer by inhibition of the phosphocreatine circuit in rat hearts. J. Bioenerg. Biomembr. 1999;31:559–567. doi: 10.1023/a:1005417011436. [DOI] [PubMed] [Google Scholar]
- 15.Sogl B, Gellissen G, Wiesner RJ. Biogenesis of giant mitochondria during insect flight muscle development in the locust, Locusta migratoria (L.). Transcription, translation and copy number of mitochondrial DNA. Eur. J. Biochem. 2000;267:11–17. doi: 10.1046/j.1432-1327.2000.00936.x. [DOI] [PubMed] [Google Scholar]
- 16.Hakimi P, Yang J, Casadesus G, Massillon D, Tolentino-Silva F, Nye CK, Cabrera ME, Hagen DR, Utter CB, Baghdy Y, et al. Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J. Biol. Chem. 2007;282:32844–32855. doi: 10.1074/jbc.M706127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruni F, Polosa PL, Gadaleta MN, Cantatore P, Roberti M. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J. Biol. Chem. 2010;285:3939–3948. doi: 10.1074/jbc.M109.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushima Y, Goto Y, Kaguni LS. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proc. Natl Acad. Sci. USA. 2010;107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 21.Pohjoismaki JL, Wanrooij S, Hyvarinen AK, Goffart S, Holt IJ, Spelbrink JN, Jacobs HT. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noack H, Bednarek T, Heidler J, Ladig R, Holtz J, Szibor M. TFAM-dependent and independent dynamics of mtDNA levels in C2C12 myoblasts caused by redox stress. Biochim. Biophys. Acta. 2006;1760:141–150. doi: 10.1016/j.bbagen.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, et al. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 25.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 26.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 2010;19:2695–2705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- 28.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 29.Pohjoismaki JL, Goffart S, Tyynismaa H, Willcox S, Ide T, Kang D, Suomalainen A, Karhunen PJ, Griffith JD, Holt IJ, et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 2009;284:21446–21457. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 31.Schips TG, Wietelmann A, Hohn K, Schimanski S, Walther P, Braun T, Wirth T, Maier HJ. FoxO3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc. Res. 2011;91:587–597. doi: 10.1093/cvr/cvr144. [DOI] [PubMed] [Google Scholar]
- 32.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 36.Szczepanek K, Chen Q, Derecka M, Salloum FN, Zhang Q, Szelag M, Cichy J, Kukreja RC, Dulak J, Lesnefsky EJ, et al. Mitochondrial-targeted signal transducer and activator of transcription (STAT3) protects against ischemia-induced changes in the electron transport chain and the generation of reactive oxygen species. J. Biol. Chem. 2011;286:29610–29620. doi: 10.1074/jbc.M111.226209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franko A, Mayer S, Thiel G, Mercy L, Arnould T, Hornig-Do HT, Wiesner RJ, Goffart S. CREB-1alpha is recruited to and mediates upregulation of the cytochrome c promoter during enhanced mitochondrial biogenesis accompanying skeletal muscle differentiation. Mol. Cell Biol. 2008;28:2446–2459. doi: 10.1128/MCB.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gadaleta MN, Rainaldi G, Lezza AM, Milella F, Fracasso F, Cantatore P. Mitochondrial DNA copy number and mitochondrial DNA deletion in adult and senescent rats. Mutat. Res. 1992;275:181–193. doi: 10.1016/0921-8734(92)90022-h. [DOI] [PubMed] [Google Scholar]
- 39.Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005;48:265–269. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- 40.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Bachrati CZ, Ou J, Hickson ID, Brown GW. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J. Biol. Chem. 2010;285:21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Feng L, Hsieh TS. Drosophila topo IIIalpha is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proc. Natl Acad. Sci. USA. 2010;107:6228–6233. doi: 10.1073/pnas.1001855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalla Rosa I, Goffart S, Wurm M, Wiek C, Essmann F, Sobek S, Schroeder P, Zhang H, Krutmann J, Hanenberg H, et al. Adaptation of topoisomerase I paralogs to nuclear and mitochondrial DNA. Nucleic Acids Res. 2009;37:6414–6428. doi: 10.1093/nar/gkp708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 45.Jackson MJ, McArdle A. Age-related changes in skeletal muscle reactive oxygen species generation and adaptive responses to reactive oxygen species. J. Physiol. 2011;589:2139–2145. doi: 10.1113/jphysiol.2011.206623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta. 2011;1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Rechkunova NI, Lavrik OI. Nucleotide excision repair in higher eukaryotes: mechanism of primary damage recognition in global genome repair. Subcell Biochem. 2010;50:251–277. doi: 10.1007/978-90-481-3471-7_13. [DOI] [PubMed] [Google Scholar]
- 48.Sugasawa K, Ng JM, Masutani C, Maekawa T, Uchida A, van der Spek PJ, Eker AP, Rademakers S, Visser C, Aboussekhra A, et al. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell Biol. 1997;17:6924–6931. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spelbrink JN. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010;62:19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- 50.Taatjes DJ, Gaudiano G, Resing K, Koch TH. Redox pathway leading to the alkylation of DNA by the anthracycline, antitumor drugs adriamycin and daunomycin. J. Med. Chem. 1997;40:1276–1286. doi: 10.1021/jm960835d. [DOI] [PubMed] [Google Scholar]
- 51.Ferreira AL, Matsubara LS, Matsubara BB. Anthracycline-induced cardiotoxicity. Cardiovasc. Hematol. Agents Med. Chem. 2008;6:278–281. doi: 10.2174/187152508785909474. [DOI] [PubMed] [Google Scholar]
- 52.Lopaschuk GD, Jaswal JS. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 53.Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J. Genet. Genomics. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.David H, Bozner A, Meyer R, Wassilew G. Pre- and postnatal development and ageing of the heart. Ultrastructural results and quantitative data. Exp. Pathol. Suppl. 1981;7:1–176. [PubMed] [Google Scholar]
- 55.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 56.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 58.Hyvarinen AK, Pohjoismaki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 60.Camara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Wenz T, Luca C, Torraco A, Moraes CT. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Wenz T, Diaz F, Spiegelman BM, Moraes CT. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Hilfiker-Kleiner D, Hilfiker A, Drexler H. Many good reasons to have STAT3 in the heart. Pharmacol. Ther. 2005;107:131–137. doi: 10.1016/j.pharmthera.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 65.Liu P, Demple B. DNA repair in mammalian mitochondria: much more than we thought? Environ. Mol. Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 66.Bendich AJ. Mitochondrial DNA, chloroplast DNA and the origins of development in eukaryotic organisms. Biol. Direct. 2010;5:42. doi: 10.1186/1745-6150-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pascucci B, D'Errico M, Parlanti E, Giovannini S, Dogliotti E. Role of nucleotide excision repair proteins in oxidative DNA damage repair: an updating. Biochemistry (Mosc) 2011;76:4–15. doi: 10.1134/s0006297911010032. [DOI] [PubMed] [Google Scholar]
- 68.Dmitrieva NI, Malide D, Burg MB. Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;301:R632–R640. doi: 10.1152/ajpregu.00853.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sage JM, Gildemeister OS, Knight KL. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J. Biol. Chem. 2010;285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashley N, Poulton J. Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene. 2009;28:3880–3891. doi: 10.1038/onc.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaludercic N, Carpi A, Menabo R, Di Lisa F, Paolocci N. Monoamine oxidases (MAO) in the pathogenesis of heart failure and ischemia/reperfusion injury. Biochim. Biophys. Acta. 2011;1813:1323–1332. doi: 10.1016/j.bbamcr.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Santos CX, Anilkumar N, Zhang M, Brewer AC, Shah AM. Redox signaling in cardiac myocytes. Free Radic. Biol. Med. 2011;50:777–793. doi: 10.1016/j.freeradbiomed.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: the concept of mitochondrial hormesis (mitohormesis) Exp. Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.