Abstract
UDP-glucose (UDPG) and derivatives are naturally-occurring agonists of the Gi protein-coupled P2Y14 receptor, which occurs in the immune system. We synthesized and characterized pharmacologically novel analogues of UDPG modified on the nucleobase, ribose, and glucose moieties, as the basis for designing novel ligands in conjunction with modeling. The recombinant human P2Y14 receptor expressed in COS-7 cells was coupled to phospholipase C through an engineered Gα-q/i protein. Most modifications of the uracil or ribose moieties abolished activity; this is among the least permissive P2Y receptors. However, a 2-thiouracil modification in 15 (EC50 49 ± 2 nM) enhanced the potency of UDPG (but not UDP-glucuronic acid) by 7-fold. 4-Thio analogue 13 was equipotent to UDPG, but S-alkylation was detrimental. Compound 15 was docked in a rhodposin-based receptor homology model, which correctly predicted potent agonism of UDP-fructose, UDP-mannose, and UDP-inositol. The hexose moiety of UDPG interacts with multiple H-bonding and charged resides and provides a fertile region for agonist modification.
Keywords: G protein-coupled receptor, nucleotides, pyrimidines, phospholipase C, carbohydrates, uracil
Introduction
The nucleotide-activated P2 receptors occur in two subfamilies of plasma membrane receptors. P2Y receptor subtypes include eight members, the P2Y1, P2Y2, P2Y4, P2Y6, P2Y11, P2Y12, P2Y13, and P2Y14 receptors, which couple to G proteins. The seven subtypes of ligand-gated cation channels that comprise the P2X receptor subfamily are denoted P2X1-P2X7. All of these subtypes have been cloned and functionally characterized.1–10 The family of P2Y receptors can be divided into two subgroups. A P2Y1-like subgroup (P2Y1, P2Y2, P2Y4, P2Y6, P2Y11) is Gq-coupled and stimulates phospholipase C (PLC), and a P2Y12-like subgroup (P2Y12, P2Y13, P2Y14) is Gi coupled and inhibits adenylyl cyclase.11 P2Y receptors are distributed in a broad range of tissues and are of interest as therapeutic targets, including antithrombotic therapy, modulation of the immune system and cardiovascular system, inflammation, pain, diabetes, and treatment of cystic fibrosis and other pulmonary diseases.12–15 P2X receptors are activated principally by adenine nucleotides, and P2Y receptors are activated by adenine and/or uracil nucleotides. For example, the P2Y2 receptor is activated by both uridine 5′-triphosphate (UTP) and adenosine 5′-triphosphate (ATP), the P2Y4 receptor by UTP, and the P2Y6 receptor by uridine 5′-diphosphate (UDP).16
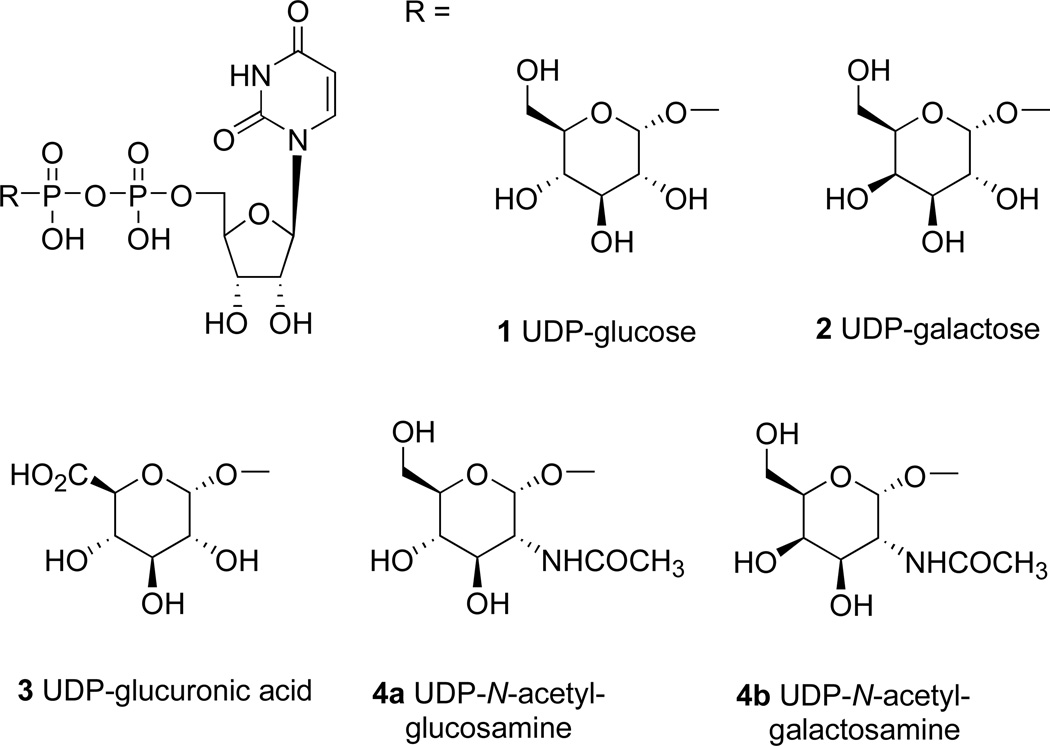
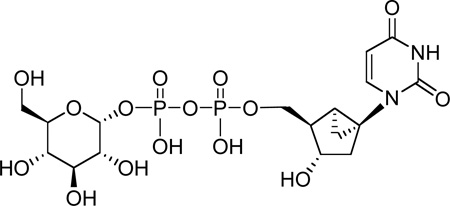
The P2Y14 receptor is distributed in various tissues including placenta, adipose, stomach, intestine, brain, spleen, thymus, lung, and heart.3 Potent and selective ligands, currently lacking, are needed for pharmacological studies. UDP-glucose (1, UDPG), UDP-galactose (2), and UDP-N-acetylglucosamine (4a) are naturally-occurring agonists of the P2Y14 receptor (Chart 1). The precise physiological function of the P2Y14 receptor has not yet been clearly established, although it appears to be involved in immune function.17 In contrast to other P2Y receptors, the P2Y14 receptor is not activated by uridine 5′-di- or triphosphates or by adenine nucleotides.21 Extracellular release of UDPG has been demonstrated, and UDPG does not activate any other P2Y receptor.17–23 The SAR (structure activity relationship) of synthetic nucleotides for activation of the human P2Y14 receptor has not previously been probed in a systematic fashion. Thus, we synthesized novel analogues of UDPG and characterized their pharmacological activities at the recombinant human P2Y14 receptor. The nucleobase, ribose, and glucose moieties were modified in this study. Molecular modeling was carried out to predict sites of interaction of the ligands with the P2Y14 receptor and to provide recognition hypotheses to be used in the design of additional analogues.
Chart 1.
Structures of three naturally-occurring UDP-sugars (1, 2, and 4a) and related derivatives that activate the P2Y14 receptor.
Results and Discussion
Chemical Synthesis
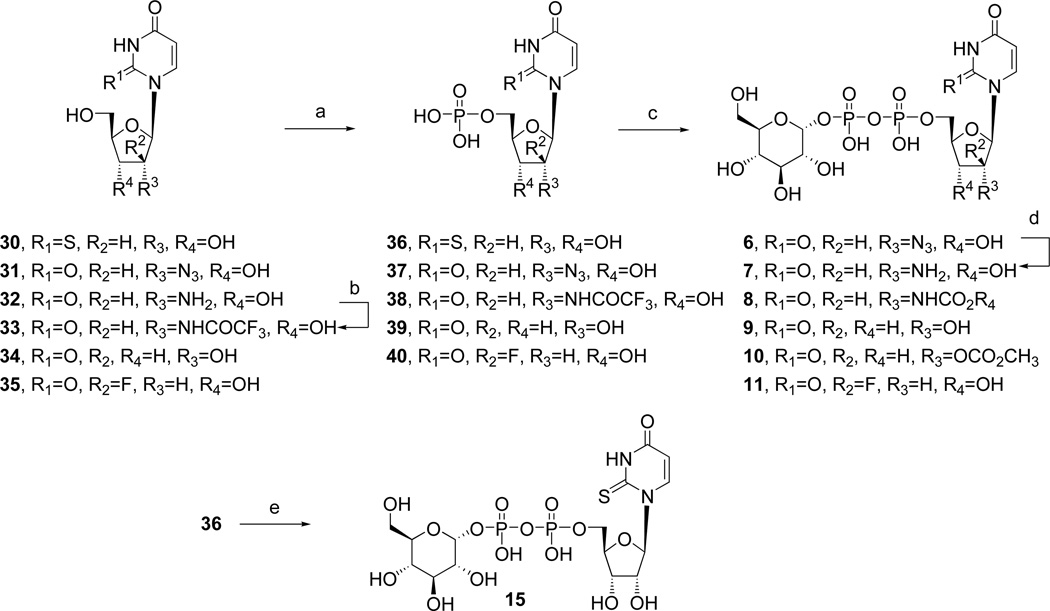
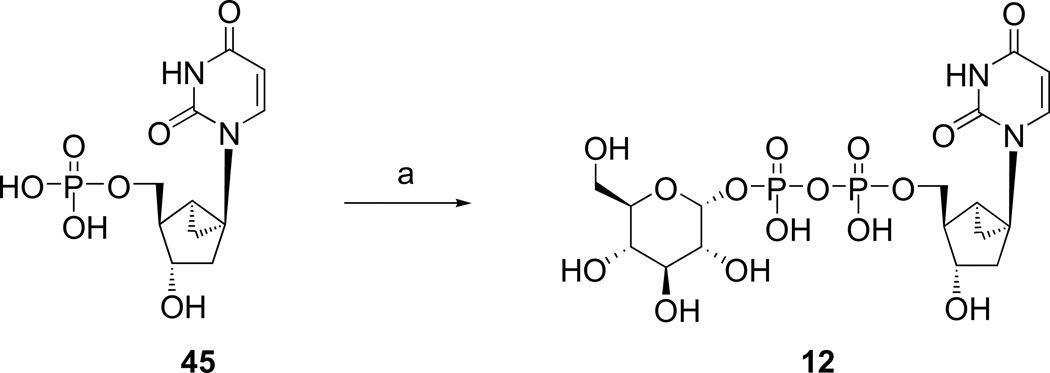
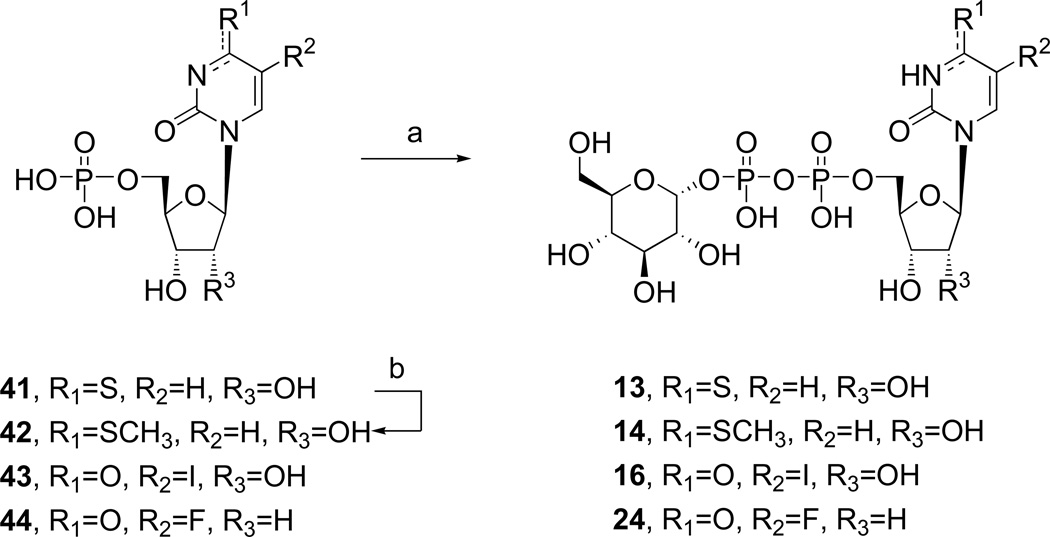
Analogues of UDPG (1) with modifications in the ribose ring, uracil moiety, and sugar moiety as well as dinucleotides (Table 1), were synthesized. The ribose ring was modified by removal of the hydroxyl group at the 2′ position 5 and replacement of the hydroxyl at the 2′ position with various functional groups, 6 – 8, 10, and 11. The hydroxyl group at the 3′ position was also modified in compounds 9 and 10. Replacement of the ribose ring with a 2′-deoxy (S)-methanocarba bicyclic ring24 (12) also was performed. The uracil ring was modified at the 2 and 4 positions, with the synthesis of 4-thiouridine-5′-diphosphoglucose (13) and 2-thiouridine-5′-diphosphoglucose (15),25 and at the 5 position, with iodide (16) and fluoride (24). Sugar-modified compounds (25-27) were synthesized with commercially available sugar monophosphates. Dinucleotides (29) were obtained as byproducts during the synthesis of UDPG analogues and of CDP-glucose 19. All the nucleotide analogues were prepared in their ammonium or triethylammonium salt form according to the methods shown in Schemes 1 – 3 and tested in functional assays of the P2Y14 receptor (Table 1). The nucleotide analogues were characterized using HPLC, nuclear magnetic resonance (1H NMR, 31P NMR), and high-resolution mass spectrometry.
Table 1.
In vitro pharmacological data for UDP-glucose, 1, and its analogues in the stimulation of phospholipase C in COS-7 cells expressing recombinant human P2Y14 receptors
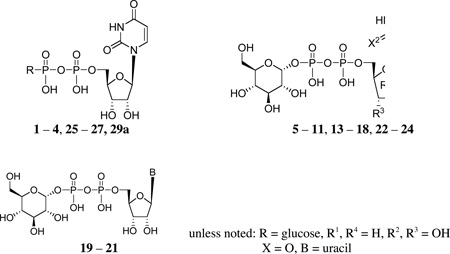 | |||
|---|---|---|---|
| Compound | Modification | R = or other substitution |
EC50 at hP2Y14 receptor, µMa |
| 1 | (= UDP- glucose) |
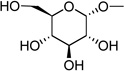 |
0.35 ± 0.12 |
| 2 | (= UDP- galactose) |
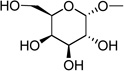 |
0.67 ± 0.09d |
| 3 | (= UDP- glucuronic acid) |
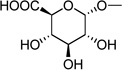 |
0.37 ± 0.07d |
| 4a | (= UDP-N- acetylglucos- amine) |
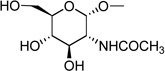 |
4.38 ± 1.05e |
| 4b | (= UDP-N- acetylgalactos- amine) |
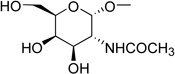 |
0.81 ± 0.09d |
| Ribose-modified | |||
| 5 | 2′-deoxy | R2 = H | NE |
| 6 | 2′-deoxy-2′ -azido |
R2 = N3 | NE |
| 7 | 2′-deoxy-2′- amino |
R2 = NH2 | NE |
| 8 | cyclic-2′- deoxy-2′- aminocarbonyl- 3′-O |
R2, R3 = cyclic- NHCOO |
NE |
| 9 | 3′-deoxy | R3 = H | NE |
| 10 | 2′,3′-dideoxy- 2′-methoxy- carbonyl |
R2 = CH3OCOO, R3 = H |
NE |
| 11 | 2′-fluoro-2′- deoxyara |
R2 = H, R4 = F |
NE |
| 12 | (S)-mc-2′- deoxy |
f | NE |
| Base-modified | |||
| 13b | 4-thio | X1 = S | 0.29 ± 0.16e |
| 14 | 4-methylthio | X1 = SCH3 | >10c |
| 15b | 2-thio | X2 = S | 0.049 ± 0.002e |
| 16 | 5-iodo | R1 = I | NE |
| 17 | 5-azido | R1 = N3 | NE |
| 18 | 5-amino | R1 = NH2 | NE |
| 19 | Base = C | 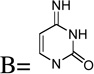 |
NE |
| 20 | Base = A | 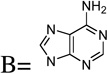 |
NE |
| 21 | Base = G | 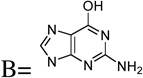 |
NE |
| Base-and ribose modified | |||
| 22 | 2′-deoxy-C | R2 = H, X1 = NH |
NE |
| 23 | 2′-deoxy-T | R1 = CH3, R2 = H |
NE |
| 24 | 2′-deoxy- 5-F-U |
R1 = F, R2 = H |
NE |
| Glucose-modified | |||
| 25 | UDP-inositol | 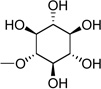 |
1.88 ± 1.1e |
| 26 | UDP-fructose | 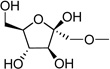 |
0.88 ± 0.21e |
| 27 | UDP-mannose | 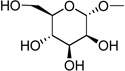 |
0.91 ± 0.15e |
| 28 | 2-thio-UDP- glucuronic acid |
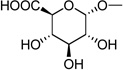 |
0.42 ± 0.19 |
| Dinucleotides | |||
| 29a | Up2U | 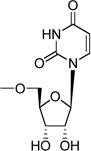 |
NE |
| 29b | Cp2C | 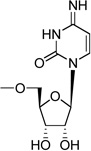 |
NE |
COS-7 cells were transiently transfected with expression vectors for both the human P2Y14 receptor and an engineered Gα-q/i protein that allows coupling of Gi-coupled receptors to activation of phospholipase C.35 Agonist potencies were calculated using a four-parameter logistic equation and the GraphPad software package (GraphPad, San Diego, CA). EC50 values (mean ± standard error) represent the concentration at which 50% of the maximal effect is achieved. N = 3, unless noted.
13, MRS2670; 15, MRS2690 (n = 4).
Less than or equal to 50% of the maximal effect at 10 µM.
Exhibited a maximal effect somewhat less than 1 in some but not all experiments.
100% of the maximal effect of 1 was achieved.
Compound 12 =
NE - no effect at 100 µM.
Scheme 1.
Synthesis of UDPG analogues substituted on the ribose moiety or the 2 position of the uracil moiety. Reagents and conditions: (a) (i) POCl3, Proton Sponge, PO(OMe)3, 0 °C, (ii) 0.2 M triethylammonium bicarbonate, rt; (b) CF3CO2Et, DIEA, DMF, rt; (c) (i) 1,1′-carbonyldiimidazole, DMF, rt, (ii) Et3N 5% in H2O/MeOH 1/1, rt, (iii) glucose-1-monophosphate tributylammonium salt, DMF, rt; (d) H2, Pd/C, MeOH, rt; (e) (i) morpholine, DCC, t-BuOH, H2O, reflux, (ii) glucose-1-monophosphate tributylammonium salt, DMF, rt.
Scheme 3.
Synthesis of an (S)-methanocarba analogue of 2′-deoxy-UDPG. Reagents and conditions: (a) (i) 1,1′-carbonyldiimidazole, DMF, rt, (ii) Et3N 5% in H2O/MeOH 1/1, rt, (iii) glucose-1-monophosphate tributylammonium salt, DMF, rt.
The nucleoside-5′-diphosphoglucose derivatives were obtained by the following methods. 5′-Monophosphorylation by reaction of the nucleoside with phosphorous oxychloride provided the nucleoside 5′-monophosphate analogues 36 – 40 (Scheme 1).26 4-Methylthiouridine-5′-monophosphate (42) was obtained by the treatment of 41 with 0.25 M NaOH in methanol for 2 h at room temperature followed by reacting with an excess of iodomethane at room temperature (Scheme 2).27 All synthesized and commercially available nucleoside 5′-monophosphate analogues were passed through cation-exchange resin and neutralized with tributylamine. The salt form was changed to a tributylammonium salt to allow the subsequent reaction to proceed in an anhydrous organic solvent. The nucleoside 5′-monophosphate was activated with 1,1′-carbonyldiimidazole in DMF for 6 h at room temperature. After the reaction was quenched and the intermediate containing a 2′-cyclic carbonyl group was hydrolyzed with methanol and triethylamine, the activated intermediate was directly condensed with a glucose 1-monophosphate tributylammonium salt in DMF to afford the corresponding nucleoside-5′-diphosphoglucose derivative (5, 6, 8 – 14, 16, 19, 22, 24).28,29 Several intermediates, including inositol-1-monophosphate, fructose-1-monophosphate and mannose-1-monophosphate, were condensed with uridine-5′-monophosphoimidazolide to afford the compounds 25-27.
Scheme 2.
Synthesis of UDPG analogues substituted on the 4 or 5 position of the uracil moiety. Reagents and conditions: (a) (i) 1,1′-carbonyldiimidazole, DMF, rt, (ii) Et3N 5% in H2O/MeOH 1/1, rt, (iii) glucose-1-monophosphate tributylammonium salt, DMF, rt; (b) (i) 0.25 M NaOH, H2O, MeOH, rt, (ii) iodomethane, DMF, rt.
The monophosphorylation of the 2′-amino-2′-deoxyuridine 32 using the above method provided only the undesired 3′-monophosphate derivative.27 As an alternate approach to synthesize 2′-deoxy-2′-aminouridine-5′-diphosphoglucose (7), the 2′-trifluoroacetylamino derivative 33, formed after protection of the amino group in 32 with a trifluoroacetyl group, was phosphorylated to give 38. However, the protecting group of 3 8 was removed during the reaction to activate the phosphate with 1,1′-carbonyldiimidazole, and this reagent formed a side product containing a cyclic carbonyl group between the 2′-amino and 3′-oxygen. This intermediate was the precursor of compound 8, and it was necessary to prepare compound 7 by another route, i.e. by reduction of the 2′-deoxy-2′-azidouridine-5′-diphosphoglucose (6) with H2 and Pd/C.
1,1′-Carbonyldiimidazole activated not only the 5′-phosphate but also partially activated the 2′-hydroxy group of the intermediate 3′-deoxyuridine-5′-monophosphate (39). The activated 2′-(imidazole-1-carbonyl) group was transformed to a methylcarbonate group when it was exposed to MeOH. After these two transient intermediates were reacted with glucose 1-monophosphate, compounds 9 and 10 were readily separated. When compound 36 was treated with 1,1′-carbonyldiimidazole, a cyclic 3′,5′-monophosphate imidazolide was produced instead of the desired 5′-monophosphate imidazolide. Alternatively, compound 15 could be prepared from nucleoside 5′-phosphoromorpholidate using morpholine and DCC30 followed by the same method of condensing with glucose 1-monophosphate (Scheme 1).
Scheme 3 shows the synthesis of a sterically constrained analogue of 2′-deoxy-UDPG, the (S)-methanocarba analogue 12. This analogue was prepared in an attempt to define the preferred conformation of the ribose moiety in the receptor binding site. The corresponding 2′-hydroxy analogue, which perhaps would be more definitive, was not included in this study.
To test the ability to combine the potency enhancing 2-thio modification with a substituted hexose moiety, 2-thio-UDP-glucuronic acid 28, analogue was prepared. The method used was an adaptation of a one-step selective enzymatic oxidation starting from the corresponding glucose derivative 15, and the product was purified using HPLC.41
Quantification of Pharmacological Activity
Activation of phospholipase C was quantified in COS-7 cells transiently expressing the human P2Y14 receptor and an engineered G protein, Gα-q/i protein (Gαqi5) that allows coupling of Gi-coupled receptors to activation of phospholipase C.35 Thus, inositol lipid hydrolysis33,34 served as a measure of agonist activity at the Gi-coupled P2Y14 receptor.
The high potency of analogues 2 and 3 suggested a flexibility of structural modification at the glucose 4 and 6 positions. The 12-fold lower potency of 4a in comparison to 1 suggested a limited tolerance for steric bulk at the glucose 2 position. Most of the novel UDPG analogues modified on the uracil or ribose moieties were inactive. Thus, the SAR of the P2Y14 receptor is among the most restrictive of all of the P2Y receptors. For example, at the P2Y6 receptor, most modifications of the uracil or ribose moieties reduced, but did not entirely abolish activity. At the P2Y2/P2Y4 receptors, some of the same ribose modifications introduced here (e.g. 2′-amino, 2′-azido) enhanced potency or selectivity.36 Analogues with purine or pyrimidine base substitutions 19 – 21 and the dinucleotide U[5′]p2[5′]U 29a were inactive at the P2Y14 receptor. Also, UMP, UDP, UTP, and U[5′]p4[5′]U failed to activate the P2Y14 receptor (data not shown).
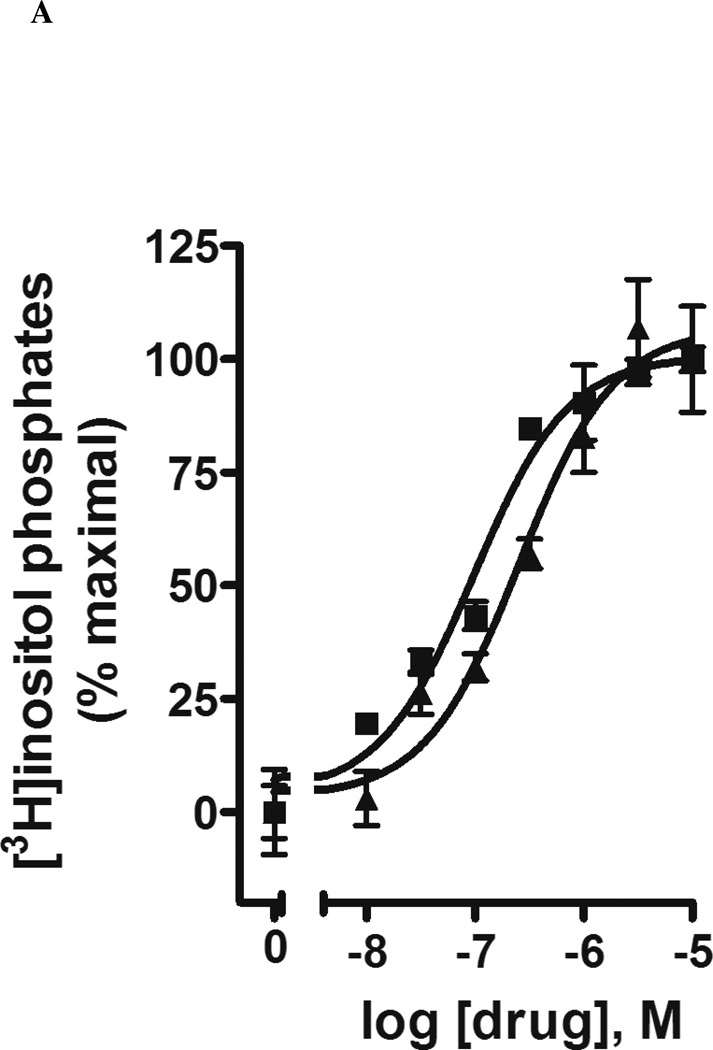
The SAR of the uracil moiety indicated that substitution of this entity was somewhat more permissive than that of the ribose moiety. Among the novel analogues prepared, a 2-thiouracil analogue 15 had a 7-fold higher potency (EC50 49 ± 2 nM) than that of UDPG (Figure 1). A 4-thio analogue (13) was equipotent to UDPG, but its S-methylation (14) was not tolerated for receptor activation. Also, three modifications at the 5 position of the uracil moiety (16 – 18) resulted in inactivity.
Figure 1.
Activation by potent agonists 13 (A) and 15 (B) of phospholipase C in COS-7 cells expressing both the human P2Y14 receptor and an engineered Gα-q/i protein that allows the Gi-coupled receptor to stimulate inositol phosphate hydrolysis by phospholipase C.
Based on the potency of compounds 2 – 4 and on the modeling results (see below) indicating as predominance of hydrophilic, H-bonding residues in the region of the hexose moiety, we proposed that the glucose moiety of UDPG should be amenable to extensive structural modification. Consequently, we synthesized UDP-fructose, UDP-mannose, and UDP-inositol and tested these derivatives as agonists of the P2Y14 receptor. Indeed, such variation of the glucose moiety of UDPG in the form of inositol 25, fructose 26, and mannose 27 preserved the agonist potency at the P2Y14 receptor.
UDP-galactose 2, UDP-glucuronic acid 3, and UDP-N-acetylglucosamine 4a also were agonists with potencies similar to that of UDP-glucose 1 at the P2Y14 receptor. The maximal effect observed with 2, 3, and 4a was somewhat variable. Maximal effects identical to that of UDP-glucose were observed in some experiments, whereas the maximal effects observed with these three molecules were somewhat less than that of UDP-glucose in other experiments. 2-Thio-UDP-glucuronic acid 28 acted as an agonist at the P2Y14 receptor and was equipotent to compounds 1 and 3. Thus, the favorable 2-thio modification preserved but did not enhance the potency of a hexose sugar-modified analogue.
Molecular Modeling
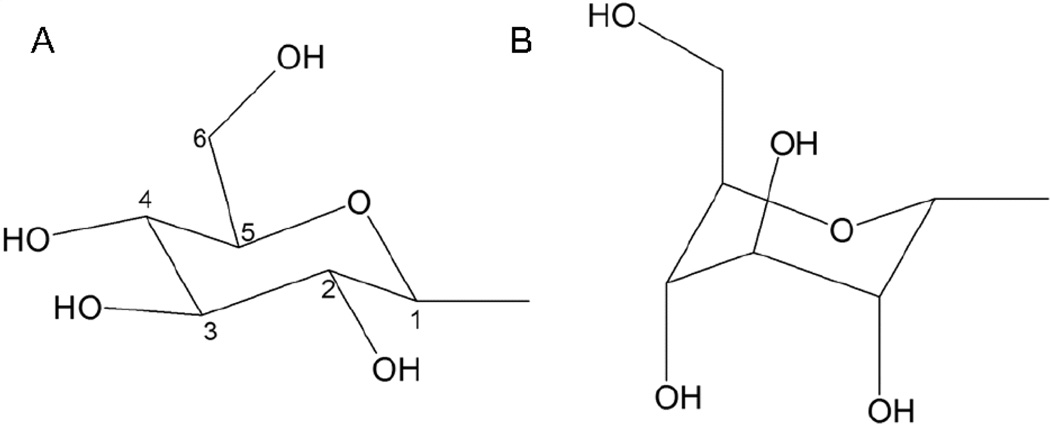
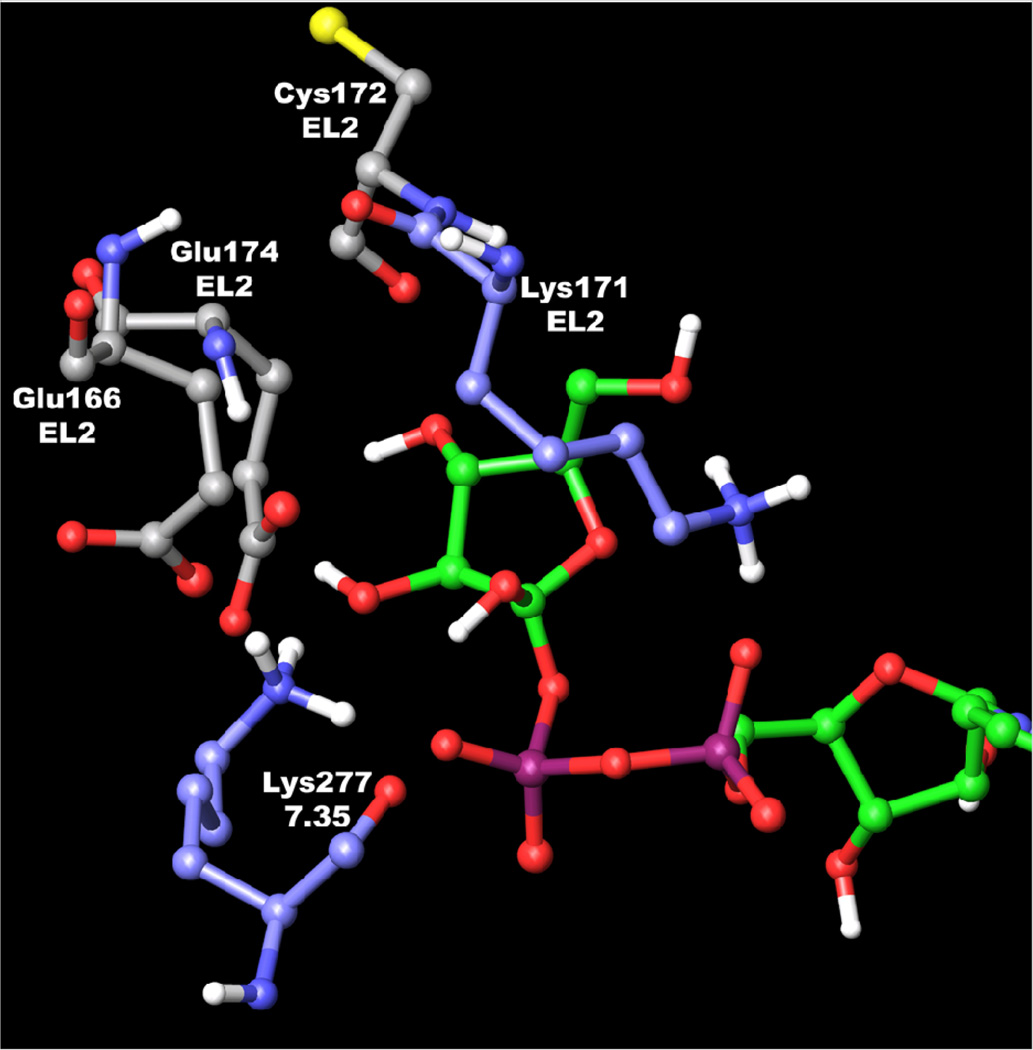
We recently reported a rhodopsin-based homology model of the human P2Y14 receptor inserted into a phospholipid bilayer and refined by molecular dynamics (MD) simulation.31 The model was utilized to study the binding mode of UDPG at the P2Y14 receptor. Ligand docking modes were based on automatic molecular docking protocols combined with the Monte Carlo Multiple Minimum calculations. Despite being based on the ground-state of rhodopsin, the model seems to be applicable to study ligand-receptor interactions for agonists. For example, similar approaches to study the interactions of agonists with P2Y1 and P2Y6 receptors were recently reported24,37 The diphosphate moiety was found to interact with only one cationic residue in the P2Y14 receptor, namely Lys171 of EL2, while in other P2Y receptor subtypes three positively charged residues interact with the phosphate chain. It should be noted that in nature the phosphate groups of UDPG could be also coordinated by a metal cation. The model of the P2Y1 receptor complex with UTP-Mg2+ was recently reported by Major et al.38 The distal hexose moiety appeared to be H-bonded by other conserved cationic residues, namely Arg253 (6.55) and Lys277 (7.35). To facilitate the comparison among receptors, throughout this paper we use the GPCR residue indexing system, as explained in detail elsewhere.32 To refine the binding mode of UDPG, especially the binding mode of the hexose ring, the Monte Carlo Multiple Minimum (MCMM) calculations were continued as described in the Methods section. The results of MCMM calculations suggested that two conformations of the hexose ring are possible at the receptor binding site (Figure 2). However, values of total energy of the entire ligand-receptor complex calculated for both models demonstrated that conformation A is more favorable. This is in a good agreement with the hexose conformation in X-ray data published for complexes of UDPG with various proteins.39 On the other hand, conformation B also seems to be possible due to the interactions between Lys277 (7.35) and hydroxyl groups of the hexose ring. These interactions can lock the hexose ring in its otherwise less favorable conformation.
Figure 2.
Possible conformations of the hexose ring of UDPG.
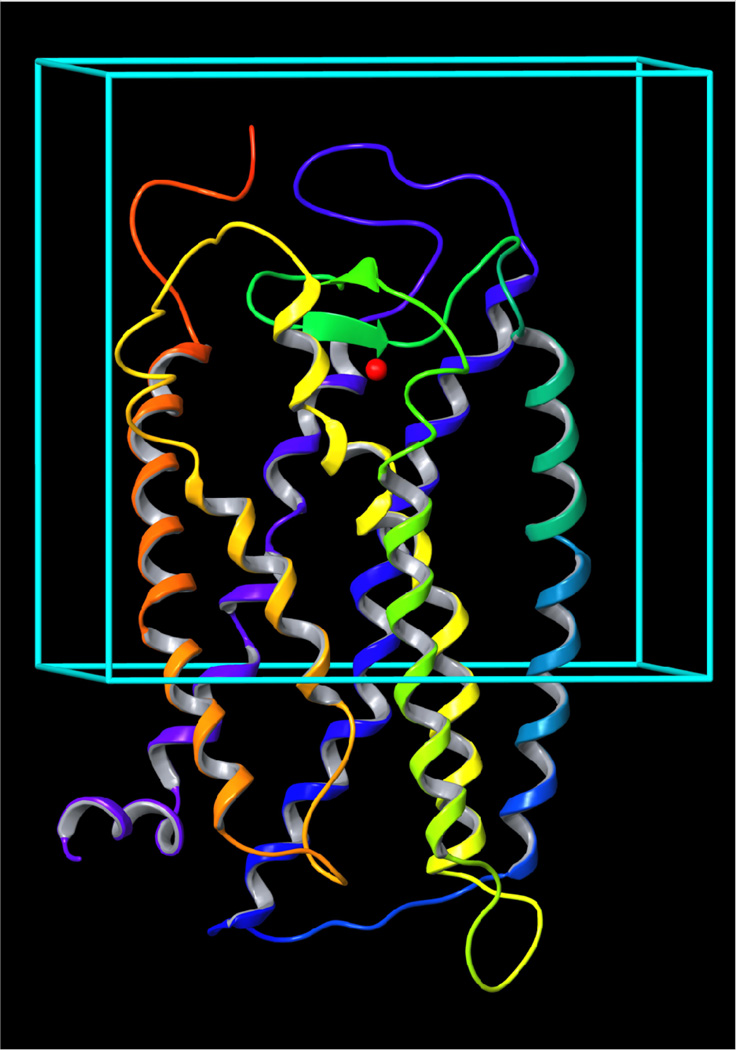
The UDPG binding mode A was proved by independently performed automatic docking of the ligand with the Glide program of MacroModel.40 As mentioned in the Methods section, a box with a side of 46 Å around the centroid of three conserved cationic residues was used for the docking study covering more than half of the entire receptor (Figure 3). The binding mode of UDPG obtained after the Glide-aided molecular docking was very similar to the binding mode of UDPG obtained after MCMM calculations.
Figure 3.
The molecular model of the human P2Y14 receptor obtained after 20 ns of molecular dynamics simulation. A centroid of Lys171, Arg253, and Lys277 (red sphere) was used as a center of the box for molecular docking of UDPG.
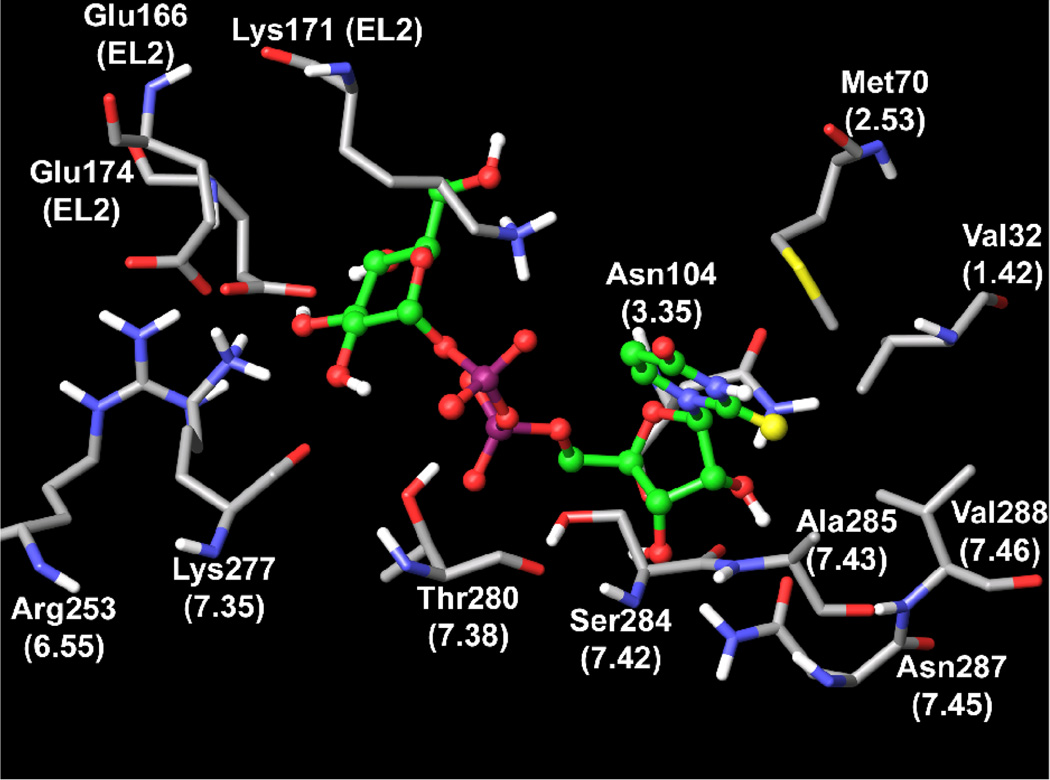
It was observed in the docking model obtained for UDPG (Figure 4) that, in the case of conformation A, Lys171 (EL2) can form an electrostatic interaction with the β-phosphate group and an additional hydrogen bond with the hydroxyl group at the 5 position of the hexose ring. Also this hydroxyl group of UDPG can interact with the backbone oxygen atom of a conserved Cys172 (EL2). The β-phosphate group formed a H-bond with the hydroxyl group of Thr280 (7.38), while Ser284 (7.42) appeared in proximity to the α-phosphate group of UDPG. K277 (7.35) interacted with hydroxyl groups at the hexose 2 and 3 positions, while Arg253 (6.55) was found near the hydroxyl group at position 3. The glutamate residue Glu174 (EL2) can interact with hydroxyl groups at the 3 and 4 positions of the hexose ring. The second glutamate residue Glu166 (EL2) appeared near hydroxyl groups at the 2 and 3 positions and in proximity to Lys277 (7.35). Similarly, hexose binding regions in protein crystallographic structures typically contain charged amino acid side chains, which H-bond to the hydroxyl groups.39
Figure 4.
Molecular model of the P2Y14-compound 15 complex. A rhodopsin-based molecular model of the human P2Y14 receptor11,31 was refined using 20 ns molecular dynamics (MD) simulation in the phospholipid bilayer. The complex obtained after the automatic docking of 15 was used as a starting point for a Monte Carlo Multiple Minimum (MCMM) conformational search analysis of the ligand inside the putative binding site of the P2Y14 receptor.
The hydroxyl groups of the ribose moiety of UDPG were found to be H-bonded with two asparagine residues. In particular, it was observed that the 2′-hydroxyl group can interact with the sidechain amino group of Asn104 (3.35) and the sidechain C=O group of Asn287 (7.45). The 3′-hydrogen group of UDPG seemed to be H-bonded with the sidechain amino group of Asn287 (7.45). These results are in a good agreement with the experimental values of EC50 measured for the ribose-modified UDPG derivatives suggesting the importance of both 2′- and 3′-hydroxyl groups.
Similar to our previous model,31 the ribose ring oxygen atom was not involved in hydrogen bonding with the receptor. This observation makes it possible to propose that replacement of this oxygen atom of UDPG by a carbon atom will not decrease the potency of a ligand. Several examples of such modifications of the ribose ring were already published for other subtypes of P2Y receptors.
As shown in Table 1, replacement of the oxygen atom at the uracil 2 position of UDPG by a sulfur atom increases agonist potency. This finding is in good agreement with the receptor docking models obtained for UDPG and 2-thio-UDPG 15 (Figure 4), in which there is a mainly hydrophobic pocket surrounding the 2 position of the uracil ring. The oxygen atom at the 2 position of UDPG was not involved in H-bonding with the receptor, implying that this oxygen is not critical for ligand recognition. The more hydrophobic sulfur atom of larger radius at the 2 position of 15 would be expected to favorably occupy more free space inside this pocket and result in improved potency of the ligand. The sulfur atom at the 2 position of the uracil ring occupied a space between several residues, namely Val32 (1.42), Met70 (2.53), Ala285 (7.43), and Val288 (7.46).
The binding modes of several UDP derivatives with various sugar moieties were examined with molecular docking studies. In particular, binding modes of UDP-mannose 27 and UDP-inositol 25 were studied. In general, the position and conformation of these two ligands were found to be very similar to UDPG. In the case of 27, the 2-OH group was located closer to Glu166 than to Lys277 (7.35), which formed a H-bond with the 3-OH group. Also, hydroxyl groups at the 3 and 4 positions of 27 formed H-bonds with Glu174 (EL2). The hydroxyl group at the 5 position can form a H-bond with Lys171 (EL2), while the oxygen atom of the mannose ring of 27 was not involved in H-bonding with the receptor.
The molecular model of the P2Y14 receptor with UDP-inositol 25 obtained after MCMM calculations indicated that similar to UDPG and 27, the ligand 25 can interact with Lys277 (7.35) and Glu174 (EL2) due to H-bonding between these residues and the hydroxyl groups at the 3 and 4 positions of the inositol moiety. In addition, the 3-OH group of the inositol moiety can interact with Arg253 (6.55). In contrast, Glu166 (EL2) appeared far from the hydroxyl groups of 25 and did not form H-bonds with the ligand. The hydroxyl group at the 6 position of the inositol ring was found near Lys171 (EL2). We propose that introduction of negatively charged groups at 2, 3, and 6 positions of the inositol ring could provide more favorable interactions with cationic residues.
Among UDP derivatives is a five-membered sugar ring derivative, UDP-fructose 26. The docking complex of 26 in the P2Y14 receptor indicated that the ligand can form several H-bonds with the P2Y14 receptor (Figure 5). In particular, similar to UDPG, the oxygen atom of the hydroxyl group at the 5 position of 26 can be H-bonded to Lys171 (EL2). In addition, Lys171 can interact with the ring oxygen atom of the fructose moiety. The H-bonds between the ligand and Glu166 (EL2) were not observed in the model obtained after MCMM calculations. However, the hydroxyl group at the 4 position was H-bonded to Glu174 (EL2) and could interact with the backbone oxygen atom of Cys172 (EL2). The 3-hydroxyl group of the fructose moiety was also found to be H-bonded with Glu174 (EL2) and with Lys277.
Figure 5.
The fructose moiety of 26 docked within the binding site of the human P2Y14 receptor.
The results of molecular docking performed for ligands containing various other sugar moieties 25 – 27 indicated that all these structures display similar binding modes at the P2Y14 receptor. In general, the sugar moieties of these ligands were involved in similar interactions with the receptor as UDPG. For this reason, compounds 25 – 27 were proposed as potential agonists of the P2Y14 receptor and subsequently synthesized and tested biologically (see above).
Conclusions
We have identified various modifications that deselect for this receptor subtype in relation to other uracil nucleotide-responsive receptors and one modification (i.e. 2-thio) that provides higher potency at the P2Y14 receptor. The 4-thio analogue of UDPG was equipotent to the parent nucleotide. Most other modifications of the uracil or ribose moieties led to inactivity. Thus, the P2Y14 receptor is among the least permissive P2Y receptors with respect to SAR. The molecular modeling studies allowed us to propose that the glucose moiety can be replaced by various six-membered or five-membered sugar rings. This prediction was tested experimentally and it was shown that variation of the glucose moiety of UDPG in the form of inositol, fructose, and mannose preserved the potency. In the models all these sugar moieties were able to interact with various hydrophilic and H-bonding side chains located in the vicinity. Therefore, the hexose moiety provides a fertile region for chemical modification of P2Y14 receptor agonists.
Experimental Section
Chemical Synthesis
2-Thiouridine (30) was purchased from Berry & Associates, Inc. (Dexter, MI). 2′-Azido-2′-deoxyuridine (31) and 2′-amino-2′-deoxyuridine (32) were purchased from CMS Chemicals Ltd. (Oxfordshire, UK). 3′-Deoxyuridine (34) was purchased from T.R.C., Inc. (North York, Ontario, Canada). 2′-Ara-fluoro-2′-deoxyuridine (35) was purchased from R.I. Chemical, Inc. (Orange, CA). 5-Azidouridine diphosphoglucose (17) and 5-aminouridine diphosphoglucose (18) were purchased from A.L.T., Inc. (Lexington, KY). Compounds 1 – 4, 20, 21, 23, and all reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO). Compounds 5, 19, 22, 29a, and 29b were synthesized as reported.28
1H NMR spectra were obtained with a Varian Gemini 300 spectrometer using D2O as a solvent. The chemical shifts are expressed as relative ppm from HOD (4.78 ppm). 31P NMR spectra were recorded at room temperature by use of Varian XL 300 spectrometer (121.42 MHz); orthophosphoric acid (85%) was used as an external standard. Purity of compounds was checked using a Hewlett–Packard 1100 HPLC equipped with a Luna 5 µ RP-C18(2) analytical column (250 × 4.6 mm; Phenomenex, Torrance, CA). System A: linear gradient solvent system: 5 mM TBAP (tetrabutylammonium dihydrogenphosphate)-CH3CN from 80:20 to 40:60 in 20 min, then isocratic for 2 min; the flow rate was 1 mL/min. System B: linear gradient solvent system: 10 mM TEAA (triethylammonium acetate)-CH3CN from 100:0 to 85:15 in 20 min, then isocratic for 2 min; the flow rate was 1 mL/min. Purity of compounds 6 – 16 and 24 – 27 in system B was checked using a Zorbax Eclipse 5 µm XDB-C18 analytical column (250 × 4.6 mm; Agilent Technologies Inc, Palo Alto, CA). Peaks were detected by UV absorption with a diode array detector. All derivatives tested for biological activity showed >98% purity in the HPLC systems.
High-resolution mass measurements were performed on Micromass/Waters LCT Premier Electrospray Time of Flight (TOF) mass spectometer coupled with a Waters HPLC system. Purification of the nucleotide analogues for biological testing was carried out on (diethylamino)ethyl (DEAE)-A25 Sephadex columns as described below. Compounds 6, 7, 9, 10, 12, 16 and 25 – 27 were additionally purified by HPLC using system C. System C: gradient solvent system: 10 mM TEAA-CH3CN from 100:0 to 90:10 in 30 min, then isocratic for 2 min; the flow rate was 2 mL/min with a Luna 5 µ RP-C18(2) semi-preparative column (250 × 10.0 mm; Phenomenex, Torrance, CA).
General Procedure for the Preparation of Nucleoside 5′-monophosphates. Procedure A
A solution of the corresponding nucleoside (0.04–0.19 mmol) and Proton Sponge (1.5 equiv) in trimethyl phosphate (2 mL) was stirred for 10 min at 0 °C. Then phosphorous oxychloride (2 equiv) was added drop wise, and the reaction mixture was stirred for 2 hours at 0 °C. 0.2 M triethylammonium bicarbonate solution (2 mL) was added to the reaction mixture, and the clear solution was stirred at room temperature for 1 h. The latter was lyophilized overnight. The residue was purified by ion-exchange column chromatography using a Sephadex-DEAE A-25 resin with a linear gradient (0.01–0.5 M) of 0.5 M ammonium bicarbonate as the mobile phase. The portions of desired compounds were collected, frozen and lyophilized to give the corresponding nucleoside 5′-monophosphates as the ammonium salts.
General Procedure for the Preparation of Nucleoside 5′-diphosphoglucose Procedure B
An ammonium salt or sodium salt of the nucleoside 5′-monophosphate (0.01–0.02 mmol) was treated with ion-exchange resin (DOWEX® 50WX2-200 (H)) followed by neutralization with tributylamine. The mixture was evaporated and lyophilized to afford the nucleoside 5′-monophosphate tributylamine salt. To a solution of the dried corresponding nucleoside 5′-monophosphates tributylamine salt in DMF (1.5–2.0 mL) was added 1,1′-carbonyldiimidazole (3 equiv). The reaction mixture was stirred at room temperature for 6 h. Then 5% triethylamine solution in 1/1 water/methanol (2 mL) was added and stirring was continued at room temperature for additional 2 h. After removal of the solvent, the residue was dried in high vacuum and dissolved in DMF (1 mL). Glucose-1-monophosphate tributylammonium salt (1.5 equiv, inositol-1-monophsphate for compound 25, fructose-1-monophosphate for compound 26, and mannose-1-monophosphate for compound 27 were used.) was added to a solution of the nucleotide imidazolide in DMF. The reaction mixture was stirred at room temperature for 2 days. After removal of the solvent, the residue was purified by ion-exchange column chromatography using a Sephadex-DEAE A-25 resin with a linear gradient (0.01–0.5 M) of 0.5 M ammonium bicarbonate as the mobile phase. The portion of desired compounds were collected, frozen and lyophilized to give the corresponding nucleoside 5′-diphosphoglucoses as the ammonium salts. Some of the products were additionally purified by HPLC using system C.
((2R,3S,4R,5R)-3,4-dihydroxy-5-(4-oxo-2-thioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl)methyl phosphate, Ammonium salt (36). Procedure A
Compound 36 (8 mg, 35%) was obtained from 30 (15 mg, 0.06 mmol). 1H NMR (D2O) δ 8.21 (d, J=8.1 Hz, 1H), 6.64 (d, J=2.7 Hz, 1H), 6.22 (d, J=8.1 Hz, 1H), 4.23 (dd, J=4.5, 3.0 Hz, 1H), 4.31 (br d, J=3.0 Hz, 1H), 4.26 (dd, J=12.3, 3.9 Hz, 1H), 4.11 (dd, J=12.0, 5.0 Hz ,1H); 31P NMR (D2O) δ 0.81 (s); HRMS m/z found 339.0056 (M – H+)−. C9H12N2O8P2S requires 339.0052.
((2R,3S,4R,5R)-4-azido-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3-hydroxytetrahydrofuran-2-yl)methyl phosphate, Ammonium salt (37). Procedure A
Compound 37 (7 mg, 41%) was obtained from 31 (12 mg, 0.045 mmol). 1H NMR (D2O) δ 7.91 (d, J=8.1 Hz, 1H), 5.98 (d, J=5.1 Hz, 1H), 5.91 (d, J=8.1 Hz, 1H), 4.52 (t, J=5.4 Hz, 1H) 4.32 (t, J=5.4 Hz, 1H), 4.23 (m, 1H), 4.18 (ddd, J=12.0, 4.5, 2.7 Hz, 1H), 4.08 (ddd, J=12.0, 5.1, 2.7 Hz, 1H); 31P NMR (D2O) δ −3.52 (s); HRMS m/z found 348.0366 (M – H+)−. C9H11N5O8P requires 348.0345.
((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3-hydroxy-4-(2,2,2-trifluoroacetamido)tetrahydrofuran-2-yl)methyl phosphate, Ammonium salt (38). Procedure A
Compound 38 (3.2 mg, 45%) was obtained from 33 (5.8 mg, 0.017 mmol). 1H NMR (D2O) δ 8.09 (dd, J=8.4, 1.8 Hz, 1H), 6.05 (d, J=8.1 Hz, 1H), 6.00 (dd, J=7.8, 1.5 Hz, 1H), 4.41 (m, 1H), 4.31 (m, 1H), 3.96 (m, 2H), 3.80 (m, 1H); 31P NMR (D2O) δ 3.85 (s); HRMS m/z found 418.0251 (M – H+)−. C11H12N3O9F3P1 requires 418.0263.
((2S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-hydroxytetrahydrofuran-2-yl)methyl phosphate, Ammonium salt (39). Procedure A
Compound 39 (9.2 mg, 41%) was obtained from 34 (15 mg, 0.066 mmol). 1H NMR (D2O) δ 8.09 (d, J=8.1 Hz, 1H), 5.88 (d, J=7.8 Hz, 1H), 5.79 (d, J=1.2 Hz, 1H), 4.59 (m, 1H), 4.48 (m, 1H), 4.09 (ddd, J=11.7, 4.5, 2.7 Hz, 1H), 3.86 (ddd, J=11.7, 4.5, 4.5 Hz, 1H), 2.14 (ddd, J=14.1, 9.6, 5.7 Hz, 1H), 2.00 (ddd, J=13.8, 6.3, 2.7 Hz, 1H); 31P NMR (D2O) δ 2.35 (s); HRMS m/z found 307.0309 (M – H+)−. C9H12N2O8P requires 307.0331.
((2R,3R,4S,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-fluoro-3-hydroxytetrahydrofuran-2-yl)methyl phosphate, Ammonium salt (40). Procedure A
Compound 40 (9.5 mg, 33%) was obtained from 35 (20 mg, 0.08 mmol). 1H NMR (D2O) δ 7.90 (dd, J=8.1, 1.2 Hz, 1H), 6.31 (dd, J=15.9, 4.2 Hz, 1H), 5.89 (dd, J=8.1, 1.2 Hz, 1H), 5.21 (d, J=51.5, 3.0 Hz, 1H), 4.52 (d, J=19.2, 4.2 Hz, 1H), 4.18 (m, 3H); 31P NMR (D2O) δ 4.75 (s); HRMS m/z found 325.0240 (M – H+)−. C9H11N2O8FP requires 325.0237.
((2R,3S,4R,5R)-3,4-dihydroxy-5-(4-(methylthio)-2-oxopyrimidin-1(2H)-yl)tetrahydrofuran-2-yl)methyl phosphate, Ammonium salt (42)
To a suspension of 4-thiouridine 5′-monophosphate, 41 (sodium salt, 10 mg, 0.03 mmol) in MeOH/H2O (1.0 mL/0.5 mL) was added 0.25 M NaOH (0.5 mL). The reaction mixture was stirred at room temperature for 2 h and then the solvent was removed under the high vacuum. The resulting residue was dissolved in dry DMF (2.0 mL) and iodomethane (0.09 mL, 1.5 mmol) was added. The reaction mixture was stirred at room temperature for 1 h, and an additional 0.09 mL of iodomethane was added. After 2 h, the solvent was removed under the reduced pressure. The crude compound was purified by the same method in procedure A to give 42 (7 mg, 62%). 1H NMR (D2O) δ 8.40 (d, J=6.9 Hz, 1H), 6.78 (d, J=7.5 Hz, 1H), 5.97 (d, J=2.4 Hz, 1H), 4.34-4.37 (m, 2H), 4.30 (m, 1H), 4.10-4.17 (m, 1H), 3.96-4.02 (m, 1H); 31P NMR (D2O) δ 2.09 (s); HRMS m/z found 353.0238 (M – H+)−. C10H14N2O8SP requires 325.0209.
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((2"-deoxy-2"-azido)uridin-5"-yl) ester, Triethylammonium salt (6). Procedure B
Compound 6 (2.5 mg, 63%) was obtained from 37 (ammonium salt, 2 mg, 0.005 mmol). 1H NMR (D2O) δ 7.94 (d, J=8.1 Hz, 1H), 6.01 (d, J=5.4 Hz, 1H), 5.96 (d, J=7.8 Hz, 1H), 5.58 (dd, J=7.2, 3.6 Hz, 1H), 4.80 (m, 1H), 4.36 (t, J=5.4 Hz, 1H), 4.19-4.25 (m, 3H), 3.82-3.89 (m, 2H), 3.72-3.78 (m, 2H), 3.51 (dd, J=9.9, 3.0 Hz, 1H), 3.44 (t, J=9.6 Hz, 1H); 31P NMR (D2O) δ −10.89 (d, J=20.8 Hz), −12.47 (d, J=20.8 Hz); HRMS m/z found 590.0531 (M – H+)−. C15H22N5O16P2 requires 590.0537; HPLC (System A) 13.7 min (98%), (System B) 7.2 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((2"-deoxy-2"-amino)uridin-5"-yl) ester, Triethylammonium salt (7)
H2 at atmospheric pressure was applied to a mixture of 6 (1.5 mg, 0.002 mmol) and Pd/C 10% (0.3 mg) in MeOH (0.5 mL) for 2 h at room temperature. The reaction mixture was filtered and the filtrate was evaporated under the reduced pressure. The residue was purified by HPLC using system C to afford 7 (1.0 mg, 69%). 1H NMR (D2O) δ 7.96 (d, J=7.8 Hz, 1H), 6.02 (d, J=7.8 Hz, 1H), 5.98 (d, J=8.1 Hz, 1H), 5.62 (dd, J=8.1, 3.6 Hz, 1H), 4.38 (m, 1H), 4.33 (m, 1H), 4.20 (m, 2H), 3.77-3.91 (m, 4H), 3.46-3.64 (m, 3H); 31P NMR (D2O) δ −10.83 (d, J=20.8 Hz), −12.49 (d, J=20.8 Hz); HRMS m/z found 564.0600 (M – H+)−. C15H24N3O16P2 requires 564.0632; HPLC (System A) 5.7 min (98%), (System B) 5.4 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((2"-deoxy-2"-aminocarbonyl-3"-O)uridin-5"-yl) ester, Ammonium salts (8). Procedure B
Compound 8 (2.0 mg, 48%) was obtained from 38 (ammonium salt, 3.0 mg, 0.007 mmol). 1H NMR (D2O) δ 7.84 (d, J=8.4 Hz, 1H), 5.91 (m, 2H), 5.59 (dd, J=7.5, 3.6 Hz, 1H), 5.40 (dd, J=8.4, 3.3 Hz, 1H), 4.67-4.75 (m, 2H), 4.30 (m, 2H), 3.75-3.92 (m, 4H), 3.43-3.55 (m, 2H); 31P NMR (D2O) δ −11.25 (br s), −12.52 (br s); HRMS m/z found 590.0449 (M – H+)−. C16H22N3O17P2 requires 590.0424; HPLC (System A) 11.7 min (98%), (System B) 5.5 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((3"-deoxy)uridin-5"-yl) ester, Triethylammonium salt (9) and Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((2",3"-dideoxy-2"-acetyloxy)uridin-5"-yl) ester, Triethylammonium salt (10). Procedure B
Compounds 9 (1.3 mg, 7%) and 10 (3.4 mg, 16%) were obtained from 39 (ammonium salts, each: 9 mg, 0.026 mmol). Compound 9 1H NMR (D2O) δ 8.02 (d, J=8.1 Hz, 1H), 5.95 (d, J=8.1 Hz, 1H), 5.85 (d, J=2.1 Hz, 1H), 5.62 (dd, J=7.2, 3.3 Hz, 1H), 4.71 (m, 1H), 4.54 (m, 1H), 4.34 (m, 1H), 4.15 (m, 1H), 3.77-3.93 (m, 4H), 3.44-3.57 (m, 2H), 2.07-2.24 (m, 2H); 31P NMR (D2O) δ −10.66 (d, J=21.5 Hz), −12.48 (d, J=20.8 Hz); HRMS m/z found 549.0543 (M – H+)−. C15H23N2O16P2 requires 549.0523; HPLC (System A) 12.9 min (98%), (System B) 8.5 min (98%). Compound 10 1H NMR (D2O) δ 7.96 (d, J=8.1 Hz, 1H), 6.02 (d, J=1.5 Hz, 1H), 5.96 (d, J=7.8 Hz, 1H), 5.61 (dd, J=7.2, 3.6 Hz, 1H), 5.37 (m, 1H), 4.65 (m, 1H), 4.35 (ddd, J=11.7, 5.1, 2.4 Hz, 1H), 4.14 (ddd, J=11.7, 6.3, 4.2 Hz, 1H), 3.92 (m, 1H), 3.84 (s, 3H), 3.76-3.86 (m, 3H), 3.54 (dt, J=9.9, 3.3 Hz, 1H), 3.47 (t, J=9.3 Hz, 1H), 2.44 (m, 1H), 2.31 (ddd, J=14.1, 6.0, 2.1 Hz, 1H); 31P NMR (D2O) δ −10.72 (d, J=20.8 Hz), −12.47 (d, J=20.8 Hz); HRMS m/z found 607.0568 (M – H+)−. C17H25N2O18P2 requires 607.0578; HPLC (System A) 13.1 min (98%), (System B) 8.0 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((2'-fluoro-2"-deoxyara)uridin-5"-yl)ester, Ammonium salts (11). Procedure B
Compound 11 (ammonium salt, 2.4 mg, 24%) was obtained from 40 (ammonium salt, 6 mg, 0.017 mmol). 1H NMR (D2O) δ 7.92 (dd, J=8.1, 1.2 Hz, 1H), 6.33 (dd, J=15.9, 3.9 Hz, 1H), 5.94 (d, J=8.4 Hz, 1H), 5.61 (dd, J=7.2, 3.6 Hz, 1H), 5.23 (dd, J=51.3, 4.2 Hz, 1H), 4.57 (dd, J=18.9, 3.3 Hz, 1H), 4.25 (m, 3H), 3.76-3.92 (m, 4H), 3.44-3.56 (m, 2H); 31P NMR (D2O) δ −10.73 (d, J=20.8 Hz), −12.45 (d, J=20.8 Hz); HRMS m/z found 567.0427 (M – H+)−. C15H22N2O16FP2 requires 567.0429; HPLC (System A) 11.7 min (98%), (System B) 8.4 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-(((S)-methanocarba-2"-deoxy)uridin-5"-yl)ester, Triethylammonium salt (12). Procedure B
Compound 12 (1.4 mg, 59%) was obtained from 4524 (ammonium salt, 1.1mg, 0.003 mmol). 1H NMR (D2O) δ 7.81 (d, J=8.1 Hz, 1H), 5.80 (d, J=7.8 Hz, 1H), 5.59 (dd, J=6.9, 3.3 Hz, 1H), 4.37 (d, J=6.6 Hz, 1H), 4.09 (m, 2H), 3.74-3.87 (m, 4H), 3.42-3.53 (m, 2H), 2.40 (m, 1H), 2.19 (m, 2H), 1.86 (m, 1H), 1.63 (m, 1H), 1.37 (m, 1H); 31P NMR (D2O) δ −10.48 (d, J=20.8 Hz), −12.53 (d, J=20.8 Hz); HRMS m/z found 559.0735 (M – H+)−. C17H25N2O15P2 requires 559.0730; HPLC (System A) 11.6 min (98%), (System B) 7.9 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((4'-thio)uridin-5"-yl)ester, Ammonium salts (13). Procedure B
Compound 13 (1.9 mg, 18 %) was obtained from 41 (12 mg, 0.017 mmol). 1H NMR (D2O) δ 7.86 (d, J=7.8 Hz, 1H), 6.68 (d, J=7.5 Hz, 1H), 5.96 (d, J=3.6 Hz, 1H), 5.62 (dd, J=7.5, 3.6 Hz, 1H), 4.23-4.39 (m, 5H), 3.83-3.90 (m, 2H), 3.76-3.84 (m, 2H), 3.54 (dt, J=9.0, 3.0 Hz, 1H), 3.48 (dd, J=9.9, 9.3 Hz, 1H); 31P NMR (D2O) δ −10.33 (d, J=20.8 Hz), −12.46 (d, J=20.8 Hz); HRMS m/z found 581.0249 (M – H+)−. C15H23N2O16P2S requires 581.0224; HPLC (System A) 13.7 min (98%), (System B) 6.6 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((4'-methylthio)uridin-5"-yl)ester, Ammonium salts (14). Procedure B
Compound 14 (5.1 mg, 40 %) was obtained from 42 (ammonium salt, 6.2 mg, 0.016 mmol). 1H NMR (D2O) δ 8.20 (d, J=7.5 Hz, 1H), 6.76 (d, J=7.2 Hz, 1H), 5.94 (d, J=2.4 Hz, 1H), 5.59 (dd, J=6.9, 3.0 Hz, 1H), 4.18-4.35 (m, 5H), 3.73-3.90 (m, 4H), 3.20-3.53 (m, 2H), 2.53 (s, 3H); 31P NMR (D2O) δ −10.80 (d, J=20.8 Hz), −12.46 (d, J=20.8 Hz); HRMS m/z found 595.0415 (M – H+)−. C16H25N2O16P2S requires 595.0400; HPLC (System A) 14.2 min (98%), (System B) 8.0 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((2'-thio)uridin-5"-yl)ester, Ammonium salts (15)
To a suspension of 36 (tributylammonium salt, 8 mg, 0.01 mmol) in t-BuOH/H2O (0.5 mL/0.5 mL) were added morpholine (0.006 mL, 0.06 mmol) and DCC (14 mg, 0.06 mmol). The reaction mixture was heated to reflux for 4 h and cooled to room temperature. After removal of solvent, the reaction was diluted with H2O and washed with diethyl ether twice. The aqueous layer was concentrated and passed through H2O and ion-exchange resin (DOWEX® 50WX2-200 (H)) followed by treatment with tributylamine. After removal of all solvents, the residue was dried in high vacuum and dissolved in DMF (1 mL). Glucose-1-monophosphate tributylammonium salt was added to a solution of the nucleotide morpholidate in DMF. The reaction mixture was stirred for 2 days followed by the same purification procedure B to give 15 (5 mg, 77%). 1H NMR (D2O) δ 8.15 (d, J=8.4 Hz, 1H), 6.68 (d, J=3.0 Hz, 1H), 6.26 (d, J=8.1 Hz, 1H), 5.61 (dd, J=6.9, 3.3 Hz, 1H), 4.20-4.45 (m, 5H), 3.70-3.95 (m, 4H), 3.54 (dt, J=9.0, 2.7 Hz, 1H), 3.47 (t, J=9.3 Hz, 1H); 31P NMR (D2O) δ −10.23 (d, J=20.8 Hz), −11.83 (d, J=20.8 Hz); HRMS m/z found 581.0248 (M – H+)−. C15H23N2O16P2S requires 581.0244; HPLC (System A) 12.6 min (98%), (System B) 5.0 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((5'-iodo)uridin-5"-yl)ester, Triethylammonium salt (16). Procedure B
Compound 16 (1.4 mg, 11%) was obtained from 43 (sodium salt, 7 mg, 0.014 mmol) 1H NMR (D2O) δ 8.27 (s, 1H), 5.95 (d, J=4.8 Hz, 1H), 5.63 (dd, J=6.9, 4.5 Hz, 1H), 4.23-4.42 (m, 5H), 3.77-4.01 (m, 4H), 3.54 (dt, J=9.0, 3.0 Hz, 1H), 3.47 (t, J=9.6 Hz, 1H); 31P NMR (D2O) δ −10.40 (d, J=20.8 Hz), −11.87 (d, J=20.8 Hz); HRMS m/z found 690.9444 (M – H+)−. C15H22N2O17IP2 requires 690.9439; HPLC (System A) 12.8 min (98%), (System B) 6.9 min (98%).
Diphosphoric acid 1-α-D-glucopyranosyl ester 2-((2"-deoxy-5'-flouro)uridin-5"-yl)ester, Ammonium salts (24). Procedure B
Compound 24 (1.8 mg, 21%) was obtained from 44 (tributylammonium salt, 10 mg, 0.014 mmol).1H NMR (D2O) δ 8.04 (d, J=5.7 Hz, 1H), 6.30 (t, J=6.9 Hz, 1H), 5.59 (dd, J=7.5, 3.2 Hz, 1H), 4.18 (m, 2H), 3.74-3.91 (m, 4H), 3.42-3.53 (m, 2H), 3.10-3.21 (m, 2H), 2.34-2.38 (m, 2H); 31P NMR (D2O) δ −10.89 (d, J=20.8 Hz), −12.51 (d, J=20.8 Hz); HRMS m/z found 386.9995 (M – H+)−. C9H13N2O11P2 requires 386.9995; HPLC (System A) 14.5 min (98%), (System B) 4.8 min (98%).
UDP-inositol, Triethylammonium salt (25). Procedure B
Compound 25 (3.2 mg, 30%) was obtained from uridine-5′-monophosphate (tributylammonium salt, 10 mg, 0.014 mmol).1H NMR (D2O) δ 7.97 (d, J=7.8 Hz, 1H), 5.99 (m, 2H), 4.72 (m, 1H), 4.40 (m, 2H), 4.31 (m, 1H), 4.26 (m, 2H), 3.74 (t, J=9.9 Hz, 2H), 3.56 (m, 2H), 3.30 (t, J=9.3 Hz, 1H); 31P NMR (D2O) δ −9.95 (d, J=21.4 Hz), −10.56 (d, J=21.4 Hz); HRMS m/z found 565.0463 (M – H+)−. C15H23N2O17P2 requires 565.0472; HPLC (System A) 12.3 min (98%), (System B) 9.2 min (98%).
UDP-fructose, Triethylammonium salt (26). Procedure
Compound 26 (4.0 mg, 37%) was obtained from uridine-5′-monophosphate (tributylammonium salt, 10 mg, 0.014 mmol). 1H NMR (D2O) δ 7.98 (d, J=8.1 Hz, 1H), 6.00 (m, 2H), 4.40 (m, 2H), 4.31 (m, 1H), 4.26 (m, 2H), 4.04-4.11 (m, 2H), 3.94-4.01 (m, 2H), 3.91 (m, 1H), 3.85 (m, 1H), 3.72 (m, 1H); 31P NMR (D2O) δ −10.82 (d, J=4.2 Hz), −10.96 (s); HRMS m/z found 565.0468 (M – H+)−. C15H23N2O17P2 requires 565.0472; HPLC (System A) 12.4 min (98%), (System B) 8.7 min (98%).
UDP-mannose, Triethylammonium salt (27). Procedure B
Compound 27 (2.8 mg, 26%) was obtained from uridine-5′-monophosphate (tributylammonium salt, 10 mg, 0.014 mmol). 1H NMR (D2O) δ 7.99 (d, J=8.4 Hz, 1H), 6.00 (m, 2H), 5.53 (dd, J=7.2, 2.3 Hz, 1H), 4.40 (m, 2H), 4.31 (m, 1H), 4.25 (m, 2H), 4.07 (m, 1H), 3.67-3.97 (m, 5H); 31P NMR (D2O) δ −11.17 (d, J=20.8 Hz), −13.46 (d, J=20.9 Hz); HRMS m/z found 565.0471 (M – H+)−. C15H23N2O17P2 requires 565.0472; HPLC (System A) 12.5 min (98%), (System B) 7.8 min (98%).
Diphosphoric acid 1-α-D-glucopyranosuronic acid 2-((2'-thio)uridin-5"-yl)ester, Triethylammonium salts (28)
Compound 15 (ammonium salt, 0.7 mg, 0.001 mmol) was dissolved in 0.7 mL of stock solution (stock solution: Tris buffer (pH 7.4, 10 mL), NAD (17 mg, 0.026 mmol), sodium pyruvate (177 mg, 1.6 mmol), and lactate dehydrogenase (67 U, EC 1.1.1.27, from rabbit muscle, Sigma). The pH of this solution was adjusted to 8.7 with 0.05 NaOH). Uridine-5′-diphosphoglucose dehydrogenase (0.3 U, Sigma, from bovine liver) was added to the mixture.41 The reaction was monitored by HPLC (System A). After 20 h at room temperature, an additional 0.1 U of UDPG dehydrogenase was added. After 5 h, the reaction mixture was lyophilized and purified by HPLC (gradient solvent system: 10 mM TEAA-CH3CN from 100:0 to 97:3 in 40 min, then isocratic for 5 min; the flow rate was 2 mL/min with a Luna 5 µ RP-C18(2) semi-preparative column (250 × 10.0 mm; Phenomenex, Torrance, CA)). The portion of desired compound was collected, frozen and lyophilized to give 28 (0.5 mg, 0.56 µmol, 56%, starting material 15 (0.3 mg, 0.38 µmol, 38%) was recovered). 1H NMR (D2O) δ 8.17 (d, J=8.1 Hz, 1H), 6.72 (d, J=3.3 Hz, 1H), 6.24 (d, J=7.5 Hz, 1H), 5.63 (dd, J=7.2, 3.0 Hz, 1H), 4.43(m, 1H), 4.33 (m, 2H), 4.24 (m, 1H), 4.10-4.16 (m, 2H), 3.78 (t, J=9.6 Hz, 1H), 3.57 (m, 1H), 3.51 (t, J=9.6 Hz, 1H); 31P NMR (D2O) δ −10.81 (br s), −12.45 (br s); HRMS m/z found 595.0032 (M – H+)−. C15H21N2O17P2S requires 595.0036; HPLC (System A) 17.1 min (98%), (System B) 12.4 min (98%).
Assay of P2Y14 receptor-stimulated PLC activity
COS-7 cells were transiently transfected with the human P2Y14 receptor and Gαqi5.35 Twenty-four hours after transfection, the inositol lipid pool of the cells was radiolabeled by incubation in 200 µL of serum-free inositol-free Dulbecco's modified Eagle's medium, containing 0.4 µCi of myo-[3H]inositol. No changes of medium were made subsequent to the addition of [3H]inositol. Forty-eight hours after transfection, cells were challenged with 50 µL of the five-fold concentrated solution of receptor agonists in 200 mM N-(2-hydroxyethyl)-piperazine-N′-2-ethanesulfonic acid, pH 7.3, containing 50 mM LiCl for 20 min at 37 °C. Incubations were terminated by aspiration of the drug-containing medium and addition of 450 µL of ice-cold 50 mM formic acid. After 15 min at 4 °C, samples were neutralized with 150 µL of 150 mM NH4OH. [3H]Inositol phosphates were isolated by ion exchange chromatography on Dowex AG 1-X8 columns as previously described.34
Data Analysis
Agonist potencies (EC50 values) were obtained from concentration-response curves by non-linear regression analysis using the GraphPad software package Prism (GraphPad, San Diego, CA). All experiments were performed in triplicate assays and repeated at least three times. The results are presented as mean ± SEM from multiple experiments or in the case of concentration effect curves from a single experiment carried out with triplicate assays that were representative of results from multiple experiments.
Molecular Modeling
Conformational Search
The recently reported complex of the P2Y14 receptor with UDPG was utilized as a starting point for further calculations31. The conformational analysis of the UDP-sugars in the putative binding site of the P2Y14 receptor was performed with MCMM implemented in MacroModel 9.0 software.40
Initially, an MCMM search was performed only on the glucose ring of UDPG. The atoms located within 8 Å from the ring were used as a shell. The bond between carbon atoms at positions 2 and 3 of the glucose ring was “opened” during the conformational search. The following parameters were used: MMFFs force field, water was used as an implicit solvent, a maximum of 1000 iterations of the Polak-Ribier Conjugate Gradient minimization method was used with a convergence threshold of 0.05 kJ·mol−1·Å−1, the number of conformational search steps = 1000, the energy window for saving structures = 1000 kJ·mol−1. An MCMM search then was performed on the entire ligand and all residues located within 6 Å from UDPG using a shell of residues located within 2 Å. Two ligand-receptor complexes with different conformation of the hexose ring were subjected to MCMM calculations. The one hundred steps of conformational search and an energy window for saving structures of 100 kJ·mol−1 were used. This protocol was applied to MCMM calculations performed for all other studied ligands.
Molecular Docking
To prove the binding mode obtained for UDPG at the P2Y14 receptor an additional independent docking study was performed with the Glide program of MacroModel package40. Our initial model of the P2Y14 receptor obtained after 20 ns of MD simulation was utilized for automatic docking of UDPG. The receptor grid generation was performed for the box with a side of 46Å with a center in the centroid of Lys171, Arg253, and Lys277. Extra precision (XP) of docking was used. The flexibility of a ligand was allowed. The conformations of the sidechains of residues located within 6 Å from UDPG were refined by 100 steps of MCMM calculations with an energy window for saving structures of 100 kJ·mol−1. The model obtained after this refinement was used for a Glide-aided automatic docking of all other ligands.
Supplementary Material
Acknowledgments
Mass spectral measurements were carried out by Dr. John Lloyd and NMR by Wesley White (NIDDK). We thank by Dr. Stefano Costanzi (NIDDK) for helpful discussion and to Savitri Maddileti for technical assistance. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases. This work was supported by National Institutes of Health grants GM38213 and HL34322 to T.K. Harden. This work was partly supported by the Korea Research Foundation Grant (E00076) to Hyojin Ko.
Abbreviations
- DCC
dicyclohexylcarbodiimide
- DMF
dimethylformamide
- MCMM
Monte Carlo Multiple Minimum
- PLC
phospholipase C
- SAR
structure activity relationship
- UDPG
UDP-[1]glucose
Footnotes
Supporting Information Available: Graphical views of the P2Y14-compound 15 complex (but not including those of the phospholipid bilayer and aqueous medium) are included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fredholm BB, Abbracchio MP, Burnstock G, Dubyak GR, Harden TK, Jacobson KA, Schwabe U, Williams M. Towards a revised nomenclature for P1 and P2 receptors. Trends Pharmacol. Sci. 1997;18:79–82. doi: 10.1016/s0165-6147(96)01038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.North RA, Barnard EA. Nucleotide receptors. Curr. Opin. Neurobiol. 1997;7:346–357. doi: 10.1016/s0959-4388(97)80062-1. [DOI] [PubMed] [Google Scholar]
- 3.Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, Fumagalli M, King BF, Gachet C, Jacobson KA, Weisman GA. International Union of Pharmacology. Update of the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol. Rev. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobson KA, Costanzi S, Ohno M, Joshi BV, Besada P, Xu B, Tchilibon S. Molecular recognition at purine and pyrimidine nucleotide (P2) receptors. Curr. Top. Med. Chem. 2004;4:805–819. doi: 10.2174/1568026043450961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Kügelgen I. Pharmacological profiles of cloned mammalian P2Y-receptor subtypes. Pharmacology Therapeutics. 2006;110:415–432. doi: 10.1016/j.pharmthera.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Lustig KD, Shiau AK, Brake AJ, Julius D. Expression cloning of an ATP receptor from mouse neuroblasoma cells. Proc. Natl. Acad. Sci. USA. 1993;90:5113–5117. doi: 10.1073/pnas.90.11.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Communi D, Pirotton S, Parmentier M, Boeynaems JM. Cloning and functional expression of a human uridine nucleotide receptor. J. Biol. Chem. 1995;270:30849–30852. doi: 10.1074/jbc.270.52.30849. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen T, Erb L, Weisman GA, Marchese A, Heng HHQ. Cloning, expression, and chromosomal localization of the human uridine nucleotide receptor gene. J. Biol. Chem. 1995;270:30845–30848. doi: 10.1074/jbc.270.52.30845. [DOI] [PubMed] [Google Scholar]
- 9.Chang K, Hanaoka K, Kumada M, Takuwa Y. Molecular cloning and functional analysis of a novel P2 nucleotide receptor. J. Biol. Chem. 1995;270:26152–26158. doi: 10.1074/jbc.270.44.26152. [DOI] [PubMed] [Google Scholar]
- 10.Communi D, Suarez Gonzalez N, Detheux M, Brezillon S, Lannoy V, Parmentier M, Boeynaems JM. Identification of a novel human ADP receptor coupled to Gi. J. Biol. Chem. 2001;276:41479–41485. doi: 10.1074/jbc.M105912200. [DOI] [PubMed] [Google Scholar]
- 11.Costanzi S, Mamedova L, Gao Z-G, Jacobson KA. Architecture of P2Y nucleotide receptors: structural comparison based on sequence analysis, mutagenesis, and homology modeling. J. Med. Chem. 2004;47:5393–5404. doi: 10.1021/jm049914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaver SR. P2Y receptors: biological advances and therapeutic opportunities. Curr. Opin. Drug Disc. 2001;4:665–670. [PubMed] [Google Scholar]
- 13.Yerxa BR, Sabater JR, Davis CW, Stutts MJ, Lang-Furr M, Picher M, Jones AC, Cowlen M, Dougherty R, Boyer J, Abraham WM, Boucher RC. Pharmacology of INS37217 [P(1)-(uridine 5')-P(4)- (2'-deoxycytidine 5')tetraphosphate, tetrasodium salt], a next-generation P2Y2 receptor agonist for the treatment of cystic fibrosis. J. Pharmacol. Exp. Ther. 2002;302:871–880. doi: 10.1124/jpet.102.035485. [DOI] [PubMed] [Google Scholar]
- 14.Burnstock G. Purinergic P2 receptors as targets for novel analgesics. Pharmacology Therapeutics. 2006;110:433–454. doi: 10.1016/j.pharmthera.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson KA, Jarvis MF, Williams M. Perspective: Purine and pyrimidine (P2) receptors as drug targets. J. Med. Chem. 2002;45:4057–4093. doi: 10.1021/jm020046y. [DOI] [PubMed] [Google Scholar]
- 16.Nicholas RA, Lazarowski ER, Watt WC, Li Q, Harden TK. Uridine nucleotide selectivity of three phospholipase C-activating P2 receptors: identification of a UDP-selective, a UTP-selective, and an ATP- and UTP-specific receptor. Mol. Pharmacol. 1996;50:224–229. [PubMed] [Google Scholar]
- 17.Scrivens M, Dickenson JM. Functional expression of the P2Y14 receptor in human neutrophils. Eur. J. Pharmacol. 2006;543:166–173. doi: 10.1016/j.ejphar.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 18.Abbracchio MP, Boeynaems J-M, Barnard EA, Boyer JL, Kennedy C, Miras-Portugal MT, King BF, Gachet C, Jacobson KA, Weisman GA, Burnstock G. Characterization of the UDP-glucose receptor (re-named here the P2Y14 receptor) adds diversity to the P2Y receptor family. Trends Pharmacol. Sci. 2003;24:52–55. doi: 10.1016/S0165-6147(02)00038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller T, Bayer H, Myrtek D, Ferrari D, Sorichter S, Ziegenhagen MW, Zissel G, Virchow JC, Jr, Luttmann W, Norgauer J, Di Virgilio F, Idzko M. The P2Y14 receptor of airway epithelial cells coupling to intracellular Ca2+ and IL-8 secretion. Am. J. Respir. Cell. Mol. Biol. 2005;33:601–609. doi: 10.1165/rcmb.2005-0181OC. [DOI] [PubMed] [Google Scholar]
- 20.Ault AD, Broach JR. Creation of GPCR-based chemical sensors by directed evolution in yeast. Protein Eng. Des. Sel. 2006;19:1–8. doi: 10.1093/protein/gzi069. [DOI] [PubMed] [Google Scholar]
- 21.Chambers JK, Macdonald LE, Sarau HM, Ames RS, Freeman K, Foley JJ, Zhu Y, McLaughlin MM, Murdock P, McMillan L, Trill J, Swift A, Aiyar N, Taylor P, Vawter L, Naheed S, Szekeres P, Hervieu G, Scott C, Watson JM, Murphy AJ, Duzic E, Klein C, Bergsma DJ, Wilson S, Livi GP. A G protein-coupled receptor for UDP-glucose. J. Biol. Chem. 2000;275:10767–10771. doi: 10.1074/jbc.275.15.10767. [DOI] [PubMed] [Google Scholar]
- 22.Skelton L, Cooper M, Murphy M, Platt A. Human immature monocyte-derived dendritic cells express the G protein-coupled receptor GPR105 (KIAA0001, P2Y14 ) and increase intracellular calcium in response to its agonist, uridine diphosphoglucose. J. Immunol. 2003;171:1941–1949. doi: 10.4049/jimmunol.171.4.1941. [DOI] [PubMed] [Google Scholar]
- 23.Lee BC, Cheng T, Adams GB, Attar EC, Miura N, Lee SB, Saito Y, Olszak I, Dombkowski D, Olson DP, Hancock J, Choi PS, Haber DA, Luster AD, Scadden DT. P2Y-like receptor, GPR105 (P2Y14 ), identifies and mediates chemotaxis of bone-marrow hematopoietic stem cells. Genes Dev. 2003;17:1592–1604. doi: 10.1101/gad.1071503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costanzi S, Joshi BV, Maddileti S, Mamedova L, Gonzalez-Moa MJ, Marquez VE, Harden TK, Jacobson KA. Human P2Y6 receptor: Molecular modeling leads to the rational design of a novel agonist based on a unique conformational preference. J. Med. Chem. 2005;48:8108–8111. doi: 10.1021/jm050911p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kochetkov NK, Budowsky EI, Shibaev VN, Yeliseeva GI, Grachev MA, Demushkin VP. Some synthetic analogues of uridine diphosphate glucose. Tetrahedron. 1963;19:1207–1218. doi: 10.1016/s0040-4020(01)98582-5. [DOI] [PubMed] [Google Scholar]
- 26.Fischer B, Chulkin A, Boyer JL, Harden KT, Gendron FP, Beaudoin AR, Chapal J, Hillaire-Buys D, Petit P. 2-Thioether 5'-O-(1-thiotriphosphate)adenosine derivatives as new insulin secretagogues acting through P2Y-receptors. J. Med. Chem. 1999;42:3636–3646. doi: 10.1021/jm990158y. [DOI] [PubMed] [Google Scholar]
- 27.Besada P, Shin DH, Costanzi S, Ko H, Mathé C, Gagneron J, Gosselin G, Maddileti S, Harden TK, Jacobson KA. Structure-activity relationships of uridine 5′-diphosphate analogues at the human P2Y6 receptor. J. Med. Chem. 2006;49:5532–5543. doi: 10.1021/jm060485n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endo T, Kajihara Y, Kodama H, Hashimoto H. Novel aspects of interaction between UDP-gal and GlcNAc β-l,4-galactosyltransferase: Transferability and remarkable inhibitory activity of UDP-(mono-O-methylated gal), UDP-fuc and UDP-man. Bioorg. Med. Chem. 1996;4:1939–1948. doi: 10.1016/s0968-0896(96)00176-9. [DOI] [PubMed] [Google Scholar]
- 29.Kim HS, Ravi RG, Marquez VE, Maddileti S, Wihlborg AK, Erlinge D, Malmsjö M, Boyer JL, Harden TK, Jacobson KA. Methanocarba modification of uracil and adenine nucleotides: High potency of Northern ring conformation at P2Y1, P2Y2, P2Y4 and P2Y11, but not P2Y6 receptors. J. Med. Chem. 2002;45:208–218. doi: 10.1021/jm010369e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen LB, Boswell KH, Khawaja TA, Meyer RB, Sidwell RW, Witkowski JT, Christenesen LF, Robins RK. Synthesis and antiviral activity of some phosphate of the broad-spectrum antiviral nucleoside, 1-α-D-ribofuranosyl-1,2,4- triazole-3-carboxamide (ribavirin) J. Med. Chem. 1978;21:742–746. doi: 10.1021/jm00206a005. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov AA, Fricks I, Harden TK, Jacobson KA. Molecular dynamics simulation of the P2Y14 receptor. Ligand docking and identification of a putative binding site of the distal hexose moiety. Bioorg. Med. Chem. Lett. 2007;17:761–766. doi: 10.1016/j.bmcl.2006.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballesteros J, Weinstein H. Integrated methods for the construction of threedimensional models of structure-function relations in G protein-coupled receptors. Methods. Neurosci. 1995;25:366–428. [Google Scholar]
- 33.Harden TK, Hawkins PT, Stephens L, Boyer JL, Downes P. Phosphoinositide hydrolysis by guanosine 5′-[gamma-thio]triphosphate-activated phospholipase C of turkey erythrocyte membranes. Biochem. J. 1988;252:583–593. doi: 10.1042/bj2520583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer JL, Downes CP, Harden TK. Kinetics of activation of phospholipase C by P2Y purinergic receptor agonists and guanine nucleotides. J. Biol. Chem. 1989;264:884–890. [PubMed] [Google Scholar]
- 35.Lazarowski ER, Shea DA, Boucher RC, Harden TK. Release of cellular UDPglucose as a potential extracellular signaling molecule. Mol. Pharmacol. 2003;63:1190–1197. doi: 10.1124/mol.63.5.1190. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson KA, Costanzi S, Ivanov AA, Tchilibon S, Besada P, Gao ZG, Maddileti S, Harden TK. Structure activity and molecular modeling analyses of ribose- and base-modified uridine 5′-triphosphate analogues at the human P2Y2 and P2Y4 receptors. Biochem. Pharmacol. 2006;71:540–549. doi: 10.1016/j.bcp.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Major DT, Fischer B. Molecular recognition in Purinergic receptors. 1. A comprehensive computational study of the h-P2Y1-receptor. J. Med. Chem. 2004;47:4391–4404. doi: 10.1021/jm049772m. [DOI] [PubMed] [Google Scholar]
- 38.Major DT, Nahum V, Wang Y, Reiser G, Fischer B. Molecular recognition in Purinergic receptors. 2. Diastereoselectivity of the h-P2Y1-receptor. J. Med. Chem. 2004;47:4405–4416. doi: 10.1021/jm049771u. [DOI] [PubMed] [Google Scholar]
- 39.Ramakrishnan B, Boeggeman E, Qasba PK. Mutation of arginine 228 to lysine enhances the glucosyltransferase activity of bovine β-1,4-galactosyltransferase I. Biochemistry. 2005;44:3202–3210. doi: 10.1021/bi0479454. [DOI] [PubMed] [Google Scholar]
- 40.Mohamadi FN, Richards GJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. MacroModel - an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 1990;11:440–467. [Google Scholar]
- 41.Toone EJ, Simon ES, Whitesides GM. Enzymatic synthesis of uridine 5’-diphosphoglucuronic acid on a gram scale. J. Org. Chem. 1991;56:5603–5606. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.