Abstract
Small, non-coding microRNAs (miRNAs) have been implicated in many biological processes, including the development of the nervous system. However, the roles of miRNAs in natural behavioral and neuronal plasticity are not well understood. To help address this we characterized the microRNA transcriptome in the adult worker honey bee head and investigated whether changes in microRNA expression levels in the brain are associated with division of labor among honey bees, a well-established model for socially regulated behavior. We determined that several miRNAs were downregulated in bees that specialize on brood care (nurses) relative to foragers. Additional experiments showed that this downregulation is dependent upon social context; it only occurred when nurse bees were in colonies that also contained foragers. Analyses of conservation patterns of brain-expressed miRNAs across Hymenoptera suggest a role for certain miRNAs in the evolution of the Aculeata, which includes all the eusocial hymenopteran species. Our results support the intriguing hypothesis that miRNAs are important regulators of social behavior at both developmental and evolutionary time scales.
Keywords: Division of labor, honey bee, microRNA, phylogenetics, social behavior
Diverse small, non-coding RNAs play a role in controlling gene function (Djuranovic et al . 2011; Fabian et al . 2010; Stefani & Slack 2008). Generally, approximately 22 nucleotides long, microRNAs (miRNAs) are thought to repress protein production either by promoting mRNA degradation or by inhibiting translation (Huntzinger & Izaurralde 2011; Stefani & Slack 2008). miRNAs are pleiotropic, and an individual miRNA can inhibit the translation of many mRNAs (Selbach et al . 2008).
miRNAs affect a wide variety of biological processes including regulation of cell proliferation and apoptosis and tumor suppression (Brennecke et al . 2005; Nolo et al . 2006), and are thought to potentially function as developmental switches (Stefani & Slack 2008). Furthermore, findings indicate that miRNAs also play an important role in regulating gene expression in the nervous system (Ashraf & Kunes 2006; Ashraf et al . 2006; Cao et al . 2006; Perkins et al . 2007), including behavioral responses to drugs (Chandrasekar & Dreyer 2011; Schaefer et al . 2010) and mammalian synaptic plasticity (Schratt 2009). While roles for miRNAs in development and pathological states are well established, how miRNAs influence natural behavioral plasticity has not been investigated thoroughly (Warren et al . 2010).
The honey bee (Apis mellifera) serves as an excellent model to study the role of genes affecting natural behavioral plasticity because of the well characterized, age-related division of labor (DOL) displayed by worker bees. Soon after eclosion, bees assume brood care (nursing) functions in their colony (Ben-Shahar et al . 2000, 2002, 2003, 2004; Ben-Shahar & Robinson 2001; Fahrbach & Robinson 1995; Leoncini et al . 2004; Whitfield et al . 2003, 2006). After about 1 week, bees begin to assume new roles, such as storing and processing food (e.g. turning nectar into honey). Most bees begin foraging for pollen and nectar at around 3 weeks of age (Ben-Shahar 2005; Fahrbach & Robinson 1995; Leoncini et al . 2004; Whitfield et al . 2006). In addition to this basic pattern, honey bee behavioral maturation also is flexible and depends on the needs of the colony, which relate to colony age demography (Huang & Robinson 1996). Honey bee behavioral maturation is associated with changes in expression of many genes (Grozinger et al . 2003; Whitfield et al . 2003, 2006), and proteins (Wolschin & Amdam 2007) in the brain. Taking advantage of this powerful behavioral model, we hypothesized that differences in miRNA expression levels in the brain are associated with behavioral maturation.
Currently, information about miRNA expression in honey bees is limited in scope. Previous work investigated differences in miRNA expression associated with DOL (Behura & Whitfield 2010), relying only on predicted, rather than experimentally characterized, miRNA sequences, especially miRNA precursor sequences, which, given the extensive processing miRNAs undergo during their maturation (Kim 2005), may have different expression patterns from mature miRNAs. Other studies cataloged miRNA expression across different honey bee developmental stages, but did not focus on brain-related expression or DOL-related differences (Chen et al . 2010). Although some recent studies have investigated the tissue specificity of certain miRNAs and characterized their neuroanatomic localization within the bee brain, no associations with behavior have been reported (Hori et al . 2011). Thus, any potential role for miRNAs in regulating honey bee DOL remains unclear.
In this investigation, we used next-generation sequencing and northern blot analyses to perform the first comprehensive analysis of brain expression levels of specific miRNAs in association with DOL. We also performed bioinformatics analyses to study the conservation of these miRNAs across animal species, especially insects. Results of these analyses together provide a portrait of behavior-related miRNA activity at both the developmental and evolutionary time scales.
Materials and methods
Honey bee collections and colony assembly
Bees were obtained from colonies maintained with normal beekeeping practices at the University of Illinois Bee Research Facility, Urbana, IL. Nurses and foragers were collected from typical colonies derived from queens mated naturally (with many males) as previously described (Whitfield et al . 2003). In typical colonies, nurses are roughly one week old or less, and foragers are 3–4-weeks old.
With honey bees it is possible to uncouple behavioral maturation and chronological age. To collect nurses and foragers that were of the same age, we utilized the single-cohort colony (SCC) technique as previously described (Ben-Shahar et al . 2002, 2004; Whitfield et al . 2003). In short, we obtained 1-day-old bees by removing honeycomb frames containing sealed brood from colonies and placing them in a laboratory incubator set to 33°C. Each SCC (N – 3) was made with approximately 1000, 1-day-old bees, an unrelated (naturally) mated queen, an empty frame in which the queen could lay eggs, and a frame containing some honey and pollen that served as the initial food supply for the colony, all placed in a small beehive. One-day-old bees were marked with a paint dot on their thoraces. The absence of older bees in a SCC induces precocious foraging behavior as early as about 7 days of age. We collected young foragers from SCCs at around 1 week of age, along with normal-age nurses of the same age. After these collections, frames containing sealed brood were periodically removed and replaced by empty ones to prevent the emergence of any new bees. As a result, some older workers continued nursing behavior at ages when they would normally begin foraging. We collected these over-age nurses when they were around 3–4-weeks old, along with foragers of the same age.
Small RNA library construction
Small RNAs were gel-purified from 100 µg total RNA isolated from nurse and forager heads, and separate nurse and forager libraries were constructed as previously described (Lu et al . 2007). Deep sequencing of the small RNA libraries was carried out at Illumina Inc. (San Diego, CA, USA).
Initial processing of sequencing data
Each of the two small RNA sequencing libraries was processed independently. Raw sequence reads were first processed to remove reads with no 3′ sequencing adaptor, of low quality, or shorter than 17-nt. The adaptor trimming was performed by an in-house method. If a raw sequence read did not have a substring of the sequencing adaptor longer than 6-nt, it was considered to have no adaptor. The adaptor-trimmed, high-quality sequence reads were referred to as qualified reads. The qualified reads were then mapped perfectly with zero mismatches to the honey bee genome using Bowtie (Langmead et al . 2009), either to 3′- and 5′-UTRs (Consortium 2006) or in intergenic, intronic or exonic regions, using known miRNAs listed in miRBase (Kozomara & Griffiths-Jones 2011) and previously identified miRNAs in the honey bee genome (Chen et al . 2010).
Identification of novel miRNAs
A list of genomic loci that adaptor-trimmed sequence reads mapped to with no mismatches were first obtained. The loci of known miRNAs were removed from the list. The remaining loci were processed to merge neighboring loci if they were adjacent to one another within 30-nt. The folding structures of the (merged) loci were then examined. As the average length of a miRNA precursor in animals is around 80-nt, 100-nt was used as the length of putative pre-miRNAs in our analysis. At each genomic locus to be analyzed, a series of DNA sequence segments covering the sequence reads were extracted for secondary structure analysis. The starting sequence segment extended 220-nt upstream of each merged locus, and subsequent segments were extracted by a sliding window of 100-nt, with an increment of 30-nt, until the window reached 220-nt downstream of the merged locus. Each of these 100-nt segments was folded by the RNAfold program (Hofacker 2003). Segments lacking stems of at least 18-nt or segments lacking sequencing reads that mapped to any of their stems were excluded. Finally, candidate miRNAs were chosen based on the following criteria: (1) occurrence of sequencing reads on the arms of a predicted hairpin structure; (2) number of the peak read on a predicted hairpin is greater than 10; (3) presence of a possible miRNA* sequencing read; and (4) presence of possible 2- or 3-nt 3′ overhangs on the miRNA/miRNA* duplex. The rationale for these criteria is that miRNA precursors are known to be processed by RNase III enzyme, Dicer, yielding a duplex of approximately 22-nt miRNA/miRNA* duplexes with 2 to 3-nt 3′ overhangs (Cullen 2004; Filipowicz et al . 2005; Lund et al . 2004; Zeng & Cullen 2004).
Detection of miRNA expression and normalization of expression levels
Qualified reads were mapped to the genomic loci of annotated miRNAs with perfect matches. The total number of the mapped reads that start within the interval of six nucleotides centered around the annotated starting position of a miRNA sequence was then taken as the raw expression level of the miRNA. The miRNA raw expression levels were then normalized under the assumption that the total amount of small RNAs in a cell was a constant. Let m be the total number of sequencing reads mapped to the genome, n be the constant total number of small RNAs in the cell, and w be the raw expression level of a miRNA. The normalized expression level of the miRNA is then w*n/m.
Phylogenetic analysis
To investigate whether specific mature miRNA sequences were present in genomes of other species we performed BLAST searches with default parameters against reference genomes, using the NCBI database. We considered a sequence ‘conserved’ in a given species if an identical sequence was found, or if a sequence missing by up to two bases at the ends was found. This criterion was based on recent studies, which indicated that in animals the ‘seed’ sequence, which determines miRNA binding specificity is typically found in the middle of the small RNA (Bartel 2009; Brennecke et al . 2005; Brodersen & Voinnet 2009). We considered a sequence ‘similar’ to the gene in question if it was the same length, but had a base substitution in the middle of the sequence. If no such matches were found, we considered a gene ‘not present’.
RNA isolation and expression profiling
Individual brains were dissected on dry ice (Ben-Shahar et al . 2002). Trizol Reagent (Life Technologies, Grand Island, NY, USA) was used to extract total RNA according to the manufacturer’s instructions. For head tissue comparisons, brains and hypopharyngeal glands were dissected from six freshly collected forager heads in chilled phosphate-buffered saline, and then pooled into three groups by tissue (Brain, Gland, rest of head carcass). Northern blot analyses were performed as previously described (Valoczi et al . 2004) except that probes were standard DNA oligos (IDT, Iowa City, IA, USA) labeled with DIG (Roche, Indianapolis, IN, USA).
For mRNA reverse transcription (RT), random hexamers and SuperScript II (Life Technologies, Grand Island, NY, USA) were used according to manufacturer’s instructions. A 0.34 mg of RNA was used per sample (N = 4 per group).
An Applied Biosystems 7500 Real Time PCR System and Applied Biosystems PowerSybr Green PCR Master Mix were used for strand amplification and measurement. Baseline and threshold cycle numbers were determined automatically according to default parameters, unless otherwise noted. To ensure assay specificity, dissociation curves were run for each primer set according to default parameters. Technical triplicates were performed for each sample, and technical replicates were generally discarded if they differed by more than 0.5 cycles from the sample’s average Ct.
For each new set of cDNAs generated, loading controls were tested along with experimental samples to account for technical errors introduced by the RT step. The Ct value for the loading control was subtracted from the Ct value for the sample to obtain each sample’s corrected Ct value (Ben-Shahar et al . 2002). For miRNA expression, U6snRNA was used as a loading control (Li et al . 2009; Marsit et al . 2006; Tazawa et al . 2007). For mRNA expression, the geometric mean of actin and eIFS-8 was used as a loading control (Fischer & Grozinger 2008; Vandesompele et al . 2002). To calculate relative expression levels different behavioral groups, we set the nurses group in each colony as a calibrator. The calibrator was given a value of 1, allowing the other sample to be reported as an n-fold difference relative to the calibrator. Relative values were calculated for the other genes as 2−(CY−CX), where CX is the corrected Ct value for the calibrator and CY is the corrected Ct value for the group being compared to the calibrator (Shpigler et al . 2010).
miRNA target prediction
To identify possible target sequences of ame-miR-2796 in PLC-epsilon transcripts, we used the RNAhybrid algorithm (Rehmsmeier et al . 2004), which provided us with a single predicted site of interaction with a minimum free energy.
Statistical analyses
Statistical analyses were performed using SPSS version 16 (IBM, Armonk, NY, USA). Independent-sample t tests were performed to determine the significance of nurse–forager differences within each colony. In interpreting the results of the t tests, equal variance was assumed for both groups. To test for the overall effects of age and task on the expression levels of each mRNA, two-way analyses of variance were performed. Significance was set at P < 0.05.
Results
The honey bee head miRNA transcriptome
We obtained 7.3 and 6.7 million raw sequencing reads from small RNA libraries from forager and nurse heads, respectively. After filtering out low-quality reads and adapter trimming, there remained 6.7 and 6.1 million qualified reads for foragers and nurses, respectively, for downstream analysis (Fig. S1). We mapped the qualified reads to various parts of the honey bee genome and to known and novel miRNAs (Table S2). Overall, 2.0 and 0.6 million reads were perfectly mapped to known and novel miRNA precursors from the forager and nurse libraries, respectively.
We observed that the majority of the reads mapped to miRNAs are of 22–24nt (Figs S1 and S2), and have an overwhelming U bias at the first nucleotide in the nurse library, which is in accordance with canonical miRNAs (Bartel 2004), while the forager library has a G bias at the first nucleotide (Fig. S2). A close inspection of the results showed that the dominating G at the first position for the forager library was from ame-miR-2796 (about three times more abundant in the forager library than in the nurse library, see ‘Expression of miRNA processing proteins’ section for details).
Seventeen novel miRNAs were identified from the two small RNA sequencing libraries (see ‘Methods’ section). Combining the 17 novel miRNAs with those previously reported for honey bees (Chen et al . 2010; Weaver et al . 2007) resulted in a total of 97 annotated miRNAs that were expressed in the heads of honey bees in nurses, foragers, or both (Tables S3, S4).
We next analyzed the genomic distribution of novel miRNAs. Six out of 17 (35.3%) novel miRNAs reside in intronic regions (Table S5). Interestingly, the precursor of ame-miR#36 can be aligned perfectly to the entire region of the third intron of the Itpr1 gene. Furthermore, the 3′-end of mature ame-miR#36 matches the acceptor site of the exon–intron splice junction of this gene. These findings suggest that ame-miR#36 is potentially a miRtron and could undergo non-canonical processing by the splicing machinery instead of by Drosha (Ruby et al . 2007). The remaining novel miRNAs are intergenic or overlap with introns in an antisense orientation. Notably, the genomic distribution of novel miRNAs is similar to that of mammalian miRNAs (Olena & Patton 2010).
miRNA conservation
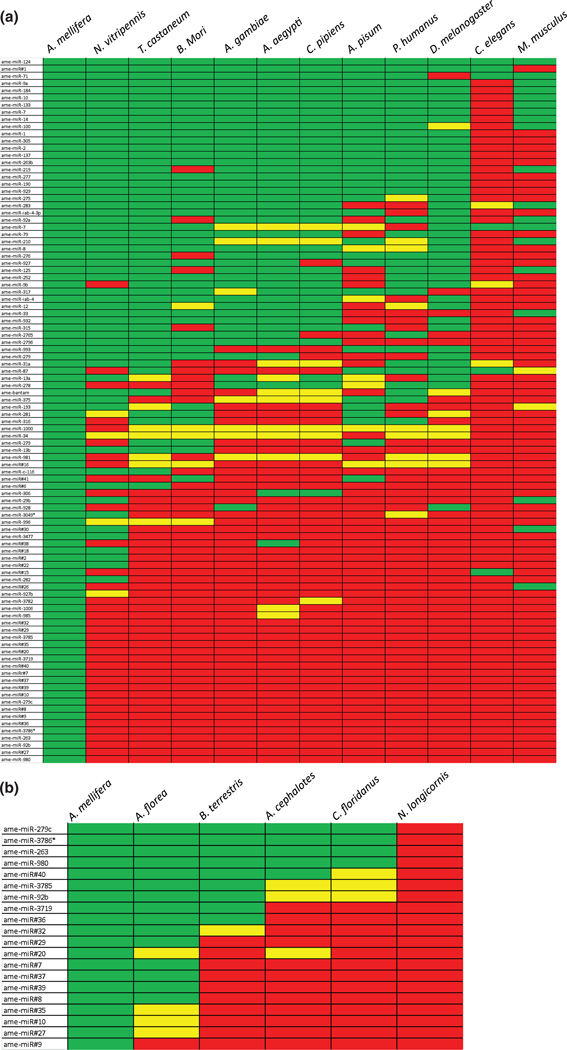
Owing to the general conservation of miRNA functions between taxa, we expected that many of the miRNA genes present in honey bees would be also conserved in other species. To test this hypothesis, we performed a phylogenetic analysis testing the extent to which the miRNAs identified in the honey bee head are conserved in other insects, as well as in two out-groups represented by the round worm Caenorhabditis elegans, and the laboratory mouse, Mus musculus. For miRNAs that had single-base changes mid-sequence in a given species, we considered the gene ‘similar’, but not ‘identical’. Our search criteria were conservative, thus the true degree of conservation is likely to be higher than what we report here. Nonetheless, the results from this analysis showed varying degrees of conservation among miRNAs (Fig. 1a). For instance, one gene, ame-miR-124, is conserved across insects as well as C. elegans and M. musculus. Eight other miRNAs are conserved across insects but are missing in C. elegans and M. musculus. The well-studied let-7 gene (Pasquinelli et al . 2000; Reinhart et al . 2000; Wulczyn et al . 2007), is highly conserved in many species, but is missing in the body louse Pediculus humanus corporis. Similarly, bantam, a well-conserved miRNA in the Drosophila lineage, is not present in several insect species, as well as in the roundworm and the laboratory mouse (Fig. 1a).
Figure 1. Conservation of miRNAs across Insecta.
(a) All identified miRNAs in the honey bee head were used as probes in BLAST searches of various representative insect species, as well as to Caenorhabditis elegans, and Mus musculus, which represented non-insect outgroups. Green highlighting indicates the miRNA is conserved. Yellow highlighting indicates similarity with a single, mid-sequence base changed. Red highlighting indicates a miRNA is not present. (b) Conservation of miRNAs that appeared honey bee-specific in eusocial as well as non-social Hymenoptera. Color coding as in (a). Note that no miRNAs that appeared honey bee-specific in (a) were conserved in Nasonia longicornis, a nonsocial wasp.
Our analysis also identified 25 miRNAs that appear to be Hymenoptera specific, i.e. present only in A. mellifera and the parasitic wasp Nasonia vitripennis, of which 20 were honey bee specific (Fig. 1b). The recent sequencing of the genomes of several additional eusocial insects, all belonging to themonophyletic Aculeata group within the Hymenoptera, allowed us to ask whether honey bee-specific miRNAs might be associated with eusocial traits. To investigate this question, we examined the conservation of 20 miRNAs identified above as honey bee specific in four other eusocial hymenopteran genomes (Apis florea–Asian dwarf bee; Bombus terrestris – bumble bee; Atta cephalotes – leafcutter ant; Camponotus floridanus – carpenter ant), as well as in the genome of the non-social parasitic wasp, Nasonia longicornis, as an additional comparison to a non-social Hymenopteran. A total of 19 out of the 20 miRNAs that initially appeared to be honey bee-specific were also identified in the genomes of other eusocial insects. Furthermore, five miRNAs were conserved in all eusocial hymenoptera we examined and found in no other species. None of these 20 miRNAs were identified in N. longicornis consistent with the fact they were not identified in Nasonia vitripennis, also a non-social wasp (Fig. 1b).
miRNA expression
We first used RNA sequencing analysis to catalog all miRNAs expressed in the honey bee head, which includes neural, muscle, and glandular tissues, in order to obtain a comprehensive view of the honey bee miRNA transcriptome. Our analyses of the expressed miRNA reads from the head libraries suggested that the majority of transcripts were biased toward one of the two behavioral states, nursing or foraging. However, to establish a more direct connection between miRNA differential expression and behavior we focused our detailed analysis specifically on the brain transcriptome. Subsequently, we further investigated the hypothesis that some miRNAs are specifically associated with honey bee division of labor using northern blot analyses. We compared brain miRNA levels from nurses and foragers from SCC colonies composed of only young or old bees, to separate effects of behavioral maturation and age.
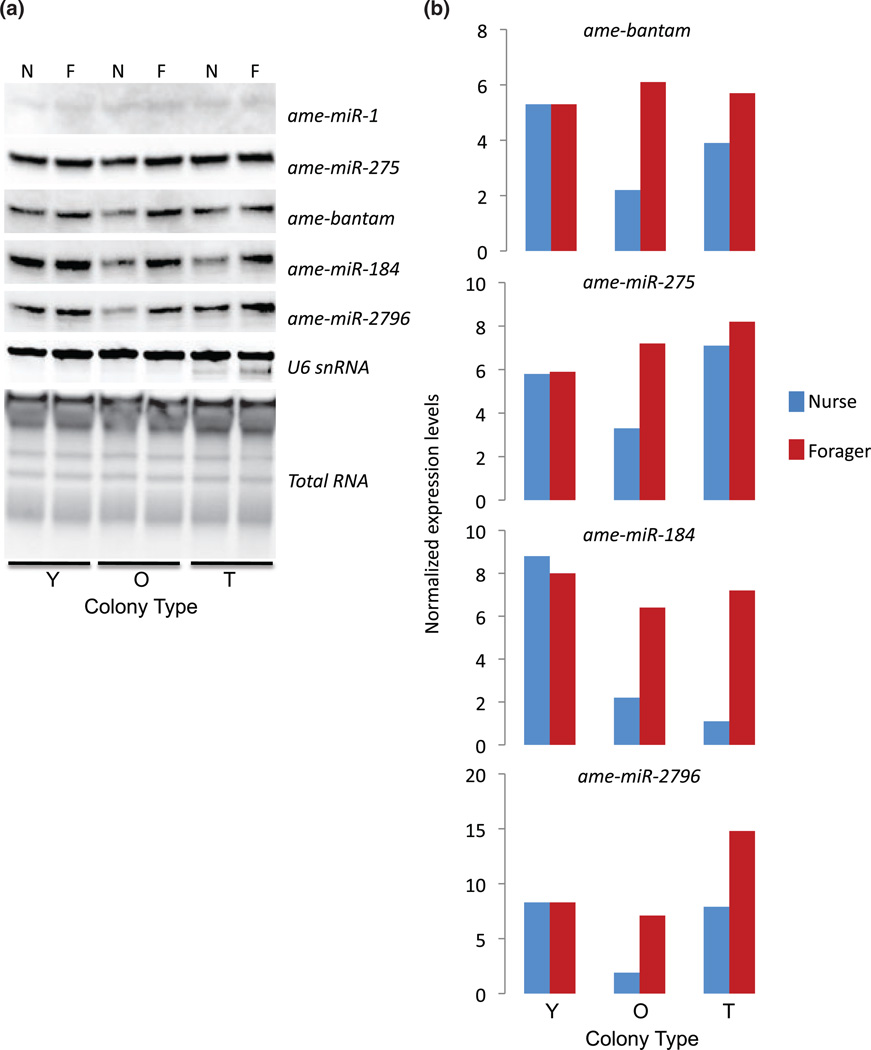
Out of the 97 miRNAs that were identified in the bee head (Table S1), we chose to further analyze five specific miRNA genes. All of these genes were highly expressed in the bee head and four appeared to be strongly regulated according to the RNA sequencing data. ame-miR-184 and ame-miR-2796 appeared upregulated in foragers, and ame-miR-1 and ame-miR-275 appeared upregulated in nurses. Bantam appeared to occur at similar levels in nurses and foragers.
Results from northern blot analyses showed that four of the five miRNAs we tested showed increased expression in forgers relative to nurses in both typical and old SCCs (Fig. 2). These findings are consistent with the RNA sequencing data for ame-miR-2796 and ame-miR-184, but contradict the RNA sequencing data for ame-miR-275 and Bantam. Similar discrepancies between next generation sequencing and northern blot analyses have been reported in other systems as well (Baker 2010; Zhang et al . 2011). In contrast, miRNA expression in young SCCs appeared unchanged. We were not able to detect a quantifiable signal from ame-miR-1 by northern blot analysis, suggesting its expression in the brain is very low. These results suggest the possibility that at least some of the miRNA genes we have examined are regulated in opposite directions in different tissues (brain vs. other head tissues) during the transition from nursing to foraging behavior.
Figure 2. Relative brain expression levels of miRNAs as a function of behavioral maturation.
Bees were collected from either typical (T), or young (Y) or old (O) single-cohort colonies as either nurses (N) or foragers (F). Bees from young single-cohort colonies were 7–10-days old, while bees from old single-cohort colonies were >3-weeks old. (a) Northern blot expression for five miRNAs, along with the control U6snRNA and total RNA expression. (b) Quantified miRNA expression (total area of RNA band) normalized to U6snRNA.
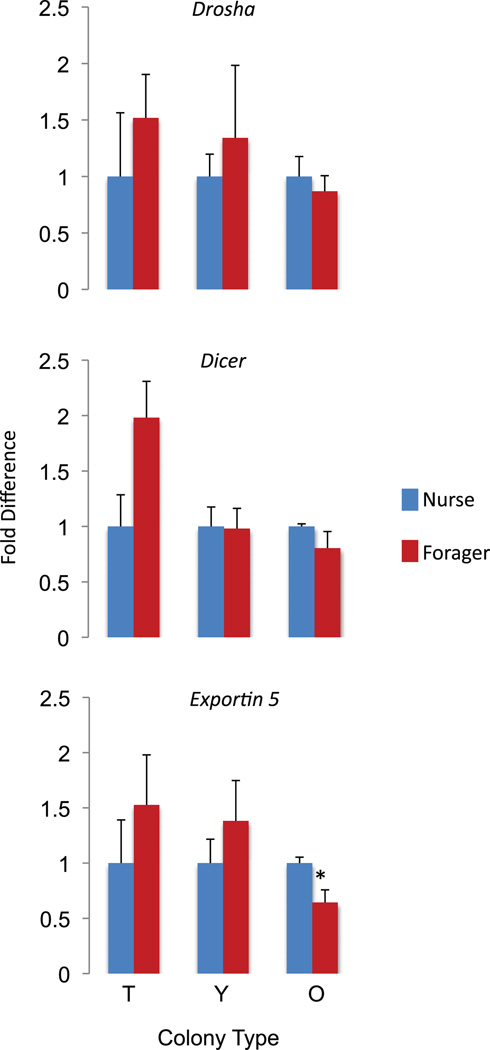
Expression of miRNA processing proteins
The global changes we observed in miRNA levels between nurses and foragers could have arisen from differences in rates of transcription, processing of the pri-miRNA or pre-miRNA, or stability of the mature miRNAs. To explore this possibility, we examined the expression of the miRNA processing proteins, Dicer, Drosha and Exportin (Esquela-Kerscher & Slack 2006). We used qRT-PCR to measure the relative differences between nurse and forager brain expression in typical, young, and old colonies for each of these miRNAs. Overall, expression of the miRNA processing proteins was not associated with behavior, with the exception of Exportin 5, which was significantly elevated in old nurses relative to old foragers (Fig. 3), but not in the other nurse–forager comparisons (Independent samples Student’s two-tailed t tests were used for testing for differences in expression levels between nurses and foragers at each colony type. Drosha: typical nurse vs. typical forager, P = 0.44, t(6) = 0.83; young nurse vs. young forager, P = 0.52, t(6) = 0.68; old nurse vs. old forager, P = 0.55, t(6) = −0.64; Dicer: typical nurse vs. typical forager: P = 0.06, t(6) = 2.33; young nurse vs. young forager, P = 0.94, t(6) = −0.08; old nurse vs. old forager, P = 0.25, t(6) = −1.26; Exportin 5: typical nurse vs. forager, P = 0.35, t(6) = 1.00; young nurse vs. young forager, P = 0.33, t(6) = 1.06; old nurse vs. old forager, P = 0.04, t(6) = −2.57). It is thus unlikely that the abovementioned differences in miRNA expression are due to differences in processing rates.
Figure 3. Relative brain mRNA expression levels of miRNA processing proteins as a function of behavioral maturation.
Bees were the same as those depicted in Fig. 4 (independent samples t test for each colony). df = 6 for typical, young and old colonies). N = 4 per group. * indicates P < 0.05.
ame-miR-2796 and its host gene, PLC-epsilon
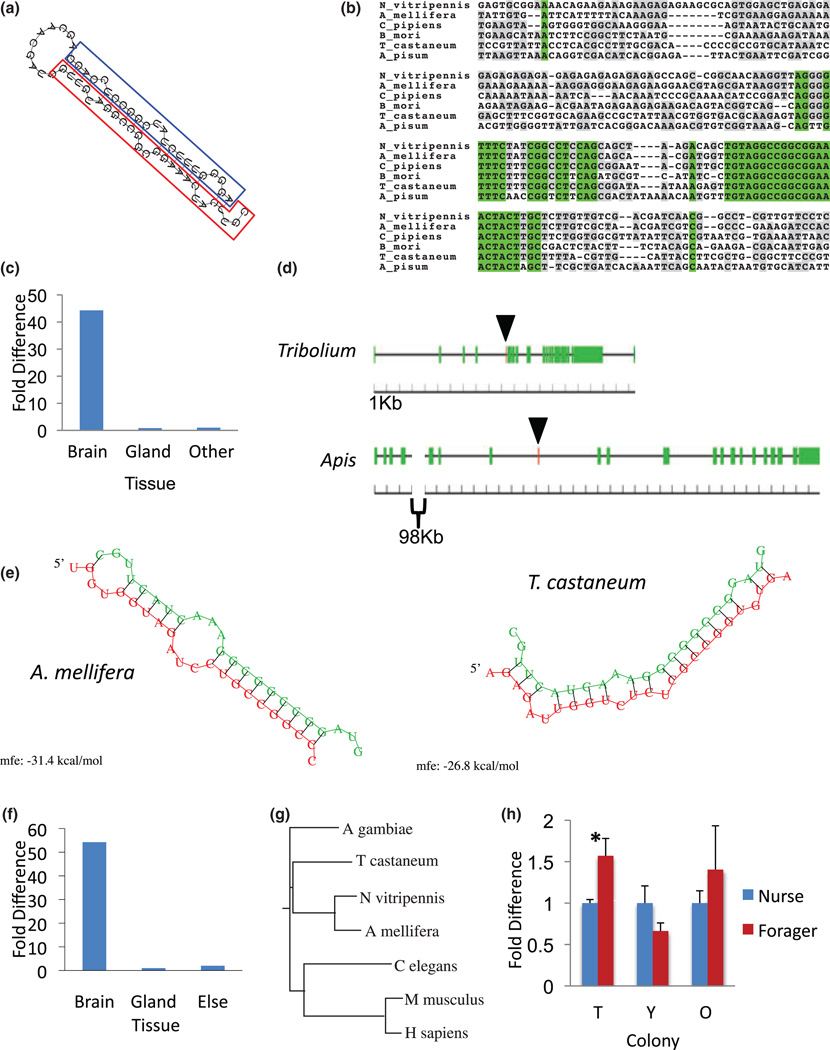
The sequencing-based profiling experiment showed ame-miR-2796 (Fig. 4a) as the most abundant miRNA expressed in the honey bee head. Remarkably, out of 528 370 total normalized nurse transcripts, 60.3% of these represented ame-miR-2796. Similarly, out of 413 879 total forager transcripts, 60.7% of these represented ame-miR-2796. Although sequencing data showed that ame-miR-2796 is found abundantly in the honey bee head, that analysis did not provide any specific evidence on where in the head the miRNA is expressed. In order to study the potential functional site of this most abundant miRNA, we used qRT-PCR to compare the relative expression of ame-miR-2796 in bee brain, head glands, and all other head tissue. These data showed that ame-miR-2796 is highly enriched in the brain relative to gland and all other head tissue (Fig. 4c).
Figure 4. The ame-miR-2796 and PLC-epsilon loci.
(a) Structure of ame-miR-2796 pre-miRNA with hairpin loop, mature sequence (red box), and miRNA* (blue box). (b) Alignment of the genomic region around ame-miR-2796 in five insect species. (c) Relative expression of ame-miR-2796 in honey bee brain, gland, and all other head tissues left in the carcass (Other). (d) The genomic architecture of the PLC-epsilon gene in Apis and Tribolium. Green boxes are exons and the red box (marked by arrow) shows the location of ame-miR-2796. (e) Predicted targeting sites for ame-miR-2796 in the PLC-epsilon transcripts of Apis and Tribolium. No targeting sites were identified in the PLC-epsilon 3′ UTR. Minimum free energy of hybridization is shown below each duplex prediction. (f) Relative expression of PLC-epsilon in honey bee brain, gland, and all other head tissue (Other). (g) Protein tree of PLC-epsilon. (h) Relative expression of PLC-epsilon in typical, young, and old honeybee colonies. (Independent samples t test. df = 6 for typical, young, and old colonies). N = 4 per group. * indicates P < 0.05.
The extreme abundance of ame-miR-2796 in the honey bee brain suggested that this miRNA might be essential and thus conserved in other insect species. Surprisingly, we found that ame-miR-2796 is conserved in six insect species in addition to honey bees, including several dipterans, but is entirely missing in the Drosophila lineage (Fig. 1a; data not shown). These data suggest that this miRNA has been lost in some insect lineages. Among species in which ame-miR-2796 is conserved, the mature sequence is identical, despite the fact that the genomic regions flanking the mature sequence show significant variability (Fig. 4b). However, the conservative nature of our search criteria potentially limited our ability to detect all species in which this miRNA is found. For instance, both the pea aphid Acyrthosiphon pisum and P. humanus corporis contain genomic sequences that resemble the first 20 out of 23 nucleotides of ame-miR-2796, and hence were considered not conserved according to our criteria, but may represent a highly related miRNA gene with a conserved seed sequence (Selbach et al . 2008).
ame-miR-2796 is located within an intron of a locus encoding the sole representative of a conserved Phospholipase C (PLC)-epsilon gene in honey bees as well as in a number of the other insects, such as Tribolium castaneum (Fig. 4d). PLC-epsilon has been implicated in neuronal development and differentiation in mammals (Wing et al . 2003), and has been reported to be transcriptionally regulated in association with division of labor in honey bees (Tsuchimoto et al . 2004). There is evidence that intronic miRNAs can share functional connections with their host coding genes (Barik 2008; Liu & Olson 2010; Musiyenko et al . 2008). Hence, we hypothesized that both ame-miR-2796 and its host gene might be evolutionarily linked.
To test this hypothesis we first characterized the spatial distribution of PLC-epsilon within the bee head. Similar to the nested ame-miR-2796 gene, PLC-epsilon expression was highly enriched in the brain relative to gland and all other head tissue (Fig. 4f). These data suggest a functional association between the host gene PLC-epsilon and the nested ame-miR-2796 gene. This suggestion is supported by computational predictions that indicate ame-miR-2796 can effectively bind to coding regions of PLC-epsilon in honey bees as well as other species (Fig. 4e), and also by the apparent loss of both loci in all currently sequenced genomes of the Drosophila lineage.
In contrast to its spatial distribution, the temporal expression patterns of PLC-epsilon in honey bee brains were not consistently associated with behavior across the three colony types (Fig. 4h) (Independent Student’s t tests; typical nurse vs. typical forager, P = 0.01, t(6) = 3.42; young nurse vs. young forager, P = 0.13, t(6) = −1.76; old nurse vs. old forager, P = 0.37, t(6) = 0.98). These results suggest that transcription of the nested miRNA gene might be independent of the transcriptional regulation of the host mRNA or that other post-transcriptional processes decoupled the transcriptional relationship between the host and nested genes.
Discussion
Our idea that miRNAs might play a role in regulating social behaviors is based upon a series of recent studies implicating miRNAs in complex nervous system functions. Specifically, miRNA transcriptional changes have been shown to play a role in neurodevelopment (Cheng & Obrietan 2007; Makeyev et al . 2007), learning (Olde Loohuis et al . 2012), complex cognitive and behavioral pathologies – such as psychiatric disease (Edbauer et al . 2010; Perkins et al . 2007; Qurashi et al . 2007), and circadian clock regulation (Alvarez-Saavedra et al . 2011; Cheng et al . 2007; Kadener et al . 2009; Yang et al . 2008).
We found that several miRNAs were upregulated in foragers relative to nurses, in both the typical cases (young nurses vs. old foragers) and when comparing old nurses and foragers, but not when comparing young nurses and foragers. To the extent that these findings do not agree with the RNA sequencing results, it is likely that the RNA sequencing data is biased due to genes expressed in high levels in the hypopharyngeal glands or muscle tissue relative to the brain. The discrepancy with the young bee comparison might relate to the following two possibilities. First, some aspects of behavioral maturation might differ when it is experimentally accelerated compared to when it occurs more naturally; this has been observed for aspects of learning behavior (Ben-Shahar & Robinson 2001; Ben-Shahar et al . 2000). Precocious foragers have much less foraging experience than older foragers; they are typically collected on their first day of foraging. Perhaps the expression of these miRNAs is experience-dependent; (Lutz et al . 2012) showed that the expression of many genes in the mushroom bodies of the bee brain relate to the number of days of foraging that occurred prior to sampling. Alternatively, these data may suggest that the absolute expression levels of specific miRNAs are not causally associated with the transition to foraging, but rather represent a more complex relationship involving both behavioral state and colony conditions. One possibility is that the presence of older foragers in the colony suppresses the transcription of specific miRNAs, and as old foragers are absent in a young SCC, this would lead to no differences between nurses and foragers. Together our results suggest that changes in brain miRNA expression are associated with behavioral maturation, but additional studies are required to better understand the relationship.
Analysis of mRNAs that encode miRNA processing proteins in nurse and forager bees suggest that it is unlikely that the observed changes in miRNA expression with behavioral maturation are due to variation in levels of processing proteins. Instead, we hypothesize that miRNA expression is most likely regulated at the level of transcription, or by cytoplasmic inhibitory mechanisms, such as the Lin-28 pathway in which a miRNA is uridylated, which inhibits Dicer processing and leads to degradation (Heo et al . 2008).
Our study also identified ame-miR-2796 as a remarkable, abundantly expressed miRNA in the bee brain. PLC-epsilon, the host protein-coding gene, and its nested ame-miR-2796 gene were either jointly conserved or missing in most insect species examined. Importantly, both PLC-epsilon and ame-miR-2796 are missing from all species in the Drosophila group, despite the fact that both genes are found in other dipterans. This finding is puzzling as one would expect such a highly abundant miRNA and thus apparently important signaling molecule to be widely conserved. In addition to this shared evolutionary fate, ame-miR-2796 is predicted to bind to sequences in the coding region of PLC-epsilon in Apis and Tribolium. Such binding to a coding region is atypical, as most miRNAs are thought to recognize complementary sequences in the 3′ UTRs of their target mRNAs (Huntzinger & Izaurralde 2011). This finding suggests that ame-miR-2796 might regulate its host gene by affecting mRNA stability or splicing rather than via canonical translational repression, and may explain the evolutionary linkage between the two loci.
As miRNAs are thought to act as pleiotropic factors affecting a wide range of genes, we expected many miRNAs to show a wide degree of conservation. As expected, a phylogenetic analysis of the expressed miRNAs showed that several are conserved across insects, and one in worms and mammals as well. In contrast, most miRNA genes are missing in at least one insect, worm, or mammal species, and 20 are restricted to Aculeata. Surprisingly, let-7, and to a greater extent bantam, was missing in one or more insect species examined. These miRNAs have been shown to be critical for essential cellular processes, often across distant taxa. For instance, let-7 plays a critical developmental role in the worm and the mouse (Peter 2009), and bantam controls cell proliferation in Drosophila (Brennecke et al . 2003). Like the loss of ame-miR-2796 in the Drosophila group, the absence of these apparently essential miRNAs – along with many less studied miRNAs – in a number of species suggests that gain or loss of miRNA genes may have served as an important agent of evolutionary divergence.
Eusociality, the most extreme form of altruism, has arisen multiple times in the monophyletic aculeate hymenoptera, separately in ant and bee lineages (Andersson 1984; Nowak et al . 2010; Wilson & Holldobler 2005). Our data suggest that certain miRNA loci are specific to the aculeate hymenoptera, but present only in the eusocial taxa, or at least absent in the non-eusocial wasps N. longicornis and N. vitripennis. If this pattern holds, it would suggest the tantalizing possibility that some of them have been involved in the multiple evolutions of eusociality in the Hymenoptera. Such a finding would be consistent with recent work showing differences in miRNA expression between different eusocial ant species (Bonasio et al . 2010).
The results of this study provide the first evidence that the striking differences in neural and behavioral plasticity associated with DOL in honey bees are correlated with changes in miRNA expression in the brain. We showed that the expression of some miRNA genes in the honey bee brain is closely associated with behavior independent of age, and that across species, patterns of miRNA conservation are consistent with the idea that some miRNAs may have served a role in evolutionary divergence, including the multiple evolutions of eusociality in the Hymenoptera. Building on previous work implicating miRNAs in neuronal development and animal learning, these results are consistent with the possibility that miRNAs are involved in regulating social behavior.
Supplementary Material
Acknowledgments
We thank John Timmons and Karen Pruiett for assistance with beekeeping. The Ben-Shahar laboratory was supported by NIH grant R03DC010244, and an award from the Klingenstein Fund to Y.B. J.K.G. was supported in part by a Summer Undergraduate Research Fellowship (SURF) from the HHMI. S.R.T. was supported by NIH/NIGMS CBI Training Grant 5T32 GM 08550-15, and NSF grants MCB#0548569 and MCB#0946326 to P.J.G. Research in the Zhang laboratory was supported by NSF grant DBI-0743797 and NIH grants 5RC1AR058681 and R01GM086512. Research in the Robinson laboratory was supported by 1R01DK082605-01A1. S.A.A. was supported by an NSF Graduate Research Fellowship.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1: Length and nucleotide distributions of all qualified reads in nurse and forager samples. (a) Distribution of all qualified reads. (b, c) First nucleotide distributions.
Figure S2: Length and nucleotide distributions of all mappable reads in nurse and forager samples. (a) Distribution of all qualified reads. (b, c) First nucleotide distributions.
Table S1: All miRNAs identified in the honey bee head by RNA-seq. miR#(X) represents novel miRNA genes, which have not yet received a formal miRBase designation. Gene name, sequence, and conservation score are shown. Conservation score was calculated by adding one point for each species in which a gene is conserved, 0.5 points for each species in which a similar gene was found, and zero points for species in which the gene is not present. Genes are ordered from most- to lest-conserved genes as in Fig. 2.
Table S2: Distributions of the sequencing reads from small RNA libraries of forager and nurse. Shown in the table are the total number of raw sequencing reads, the number of qualified reads that can perfectly map to the corresponding honeybee genome, known and novel miRNA sequences, coding sequencing (CDS), 3′- and 5′-UTRs, intergenic regions, and intron and exon sequences. No mismatches were allowed for the mapping. The second number for each condition (column) is the percentage of reads relative to the total qualified reads.
Table S3: Previously annotated genes: raw and normalized (norm) transcript numbers for each miRNA in the nurse and forager library, along with total number of qualified and unqualified reads for each library.
Table S4: Novel miRNA genes: raw and normalized transcript numbers for each gene in the nurse and forager library, along with total number of qualified and unqualified reads for each library.
Table S5: List of 18 novel miRNA genomic loci and function annotation.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. The evolution of eusociality. Annu Rev Ecol Syst. 1984;15:165–189. [Google Scholar]
- Ashraf SI, Kunes S. A trace of silence: memory and microRNA at the synapse. Curr Opin Neurobiol. 2006;16:535–539. doi: 10.1016/j.conb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Baker M. MicroRNA profiling: separating signal from noise. Nat Methods. 2010;7:687–692. doi: 10.1038/nmeth0910-687. [DOI] [PubMed] [Google Scholar]
- Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK, Whitfield CW. Correlated expression patterns of microRNA genes with age-dependent behavioural changes in honeybee. Insect Mol Biol. 2010;19:431–439. doi: 10.1111/j.1365-2583.2010.01010.x. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y. The foraging gene, behavioral plasticity, and honeybee division of labor. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2005;191:987–994. doi: 10.1007/s00359-005-0025-1. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y, Robinson GE. Satiation differentially affects performance in a learning assay by nurse and forager honey bees. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2001;187:891–899. doi: 10.1007/s00359-001-0260-z. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Influence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y, Leung HT, Pak WL, Sokolowski MB, Robinson GE. cGMP-dependent changes in phototaxis: a possible role for the foraging gene in honey bee division of labor. J Exp Biol. 2003;206:2507–2515. doi: 10.1242/jeb.00442. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y, Dudek NL, Robinson GE. Phenotypic deconstruction reveals involvement of manganese transporter malvolio in honey bee division of labor. J Exp Biol. 2004;207:3281–3288. doi: 10.1242/jeb.01151. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y, Thompson CK, Hartz SM, Smith BH, Robinson GE. Differences in performance on a reversal learning test and division of labor in honey bee colonies. Anim Cogn. 2000;3:119–125. [Google Scholar]
- Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C, Zhang P, Huang Z, Berger SL, Reinberg D, Wang J, Liebig J. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science. 2010;329:1068–1071. doi: 10.1126/science.1192428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Chandrasekar V, Dreyer JL. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology. 2011;36:1149–1164. doi: 10.1038/npp.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yu X, Cai Y, Zheng H, Yu D, Liu G, Zhou Q, Hu S, Hu F. Next-generation small RNA sequencing for microRNAs profiling in the honey bee Apis mellifera. Insect Mol Biol. 2010;19:799–805. doi: 10.1111/j.1365-2583.2010.01039.x. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Obrietan K. Revealing a role of microRNAs in the regulation of the biological clock. Cell Cycle. 2007;6:3034–3035. doi: 10.4161/cc.6.24.5106. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium HGS. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Fahrbach SE, Robinson GE. Behavioral development in the honey bee: toward the study of learning under natural conditions. Learn Mem. 1995;2:199–224. doi: 10.1101/lm.2.5.199. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Fischer P, Grozinger CM. Pheromonal regulation of starvation resistance in honey bee workers (Apis mellifera) Naturwissenschaften. 2008;95:723–729. doi: 10.1007/s00114-008-0378-8. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Sharabash NM, Whitfield CW, Robinson GE. Pheromone-mediated gene expression in the honey bee brain. Proc Natl Acad Sci USA. 2003;100(Suppl. 2):14519–14525. doi: 10.1073/pnas.2335884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32 doi: 10.1016/j.molcel.2008.09.014. 276284. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Kaneko K, Saito T, Takeuchi H, Kubo T. Expression of two microRNAs, <i>ame</i>-mir-276 and-1000, in the adult honeybee (Apis mellifera) brain. Apidologie. 2011;42:89–102. [Google Scholar]
- Huang ZY, Robinson GE. Regulation of honey bee division of labor by colony age demography. Behav Ecol Sociobiol. 1996;39:147–158. [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoncini I, Le Conte Y, Costagliola G, Plettner E, Toth AL, Wang M, Huang Z, Becard JM, Crauser D, Slessor KN, Robinson GE. Regulation of behavioral maturation by a primer pheromone produced by adult worker honey bees. Proc Natl Acad Sci USA. 2004;101:17559–17564. doi: 10.1073/pnas.0407652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, Li Y, Zhang CY. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50:1756–1765. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Lutz CC, Rodriguez-Zas SL, Fahrbach SE, Robinson GE. Transcriptional response to foraging experience in the honey bee mushroom bodies. Dev Neurobiol. 2012;72:153–166. doi: 10.1002/dneu.20929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med (Berl) 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Nowak MA, Tarnita CE, Wilson EO. The evolution of eusociality. Nature. 2010;466:1057–1062. doi: 10.1038/nature09205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olena AF, Patton JG. Genomic organization of microRNAs. J Cell Physiol. 2010;222:540–545. doi: 10.1002/jcp.21993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qurashi A, Chang S, Peng J. Role of microRNA pathway in mental retardation. Sci World J. 2007;7:146–154. doi: 10.1100/tsw.2007.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Veno MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O’Carroll D, Greengard P. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med. 2010;207:1843–1851. doi: 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shpigler H, Patch HM, Cohen M, Fan Y, Grozinger CM, Bloch G. The transcription factor Kruppel homolog 1 is linked to hormone mediated social organization in bees. BMC Evol Biol. 2010;10:120. doi: 10.1186/1471-2148-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto M, Aoki M, Takada M, Kanou Y, Sasagawa H, Kitagawa Y, Kadowaki T. The changes of gene expression in honeybee (Apis mellifera) brains associated with ages. Zoolog Sci. 2004;21:23–28. doi: 10.2108/0289-0003(2004)21[23:TCOGEI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DB, Anzola JM, Evans JD, Reid JG, Reese JT, Childs KL, Zdobnov EM, Samanta MP, Miller J, Elsik CG. Computational and transcriptional evidence for microRNAs in the honey bee genome. Genome Biol. 2007;8:R97. doi: 10.1186/gb-2007-8-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Ben-Shahar Y, Brillet C, Leoncini I, Crauser D, Leconte Y, Rodriguez-Zas S, Robinson GE. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EO, Holldobler B. Eusociality: origin and consequences. Proc Natl Acad Sci USA. 2005;102:13367–13371. doi: 10.1073/pnas.0505858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing MR, Bourdon DM, Harden TK. PLC-epsilon: a shared effector protein in Ras-, Rho-, and G alpha beta gamma-mediated signaling. Mol Interv. 2003;3:273–280. doi: 10.1124/mi.3.5.273. [DOI] [PubMed] [Google Scholar]
- Wolschin F, Amdam GV. Plasticity and robustness of protein patterns during reversible development in the honey bee (Apis mellifera) Anal Bioanal Chem. 2007;389:1095–1100. doi: 10.1007/s00216-007-1523-5. [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol. 2011;75:93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.