Abstract
The ability to specifically engineer the genome of living cells at precise locations using rare-cutting designer endonucleases has broad implications for biotechnology and medicine, particularly for functional genomics, transgenics and gene therapy. However, the potential impact of chromosomal context and epigenetics on designer endonuclease-mediated genome editing is poorly understood. To address this question, we conducted a comprehensive analysis on the efficacy of 37 endonucleases derived from the quintessential I-CreI meganuclease that were specifically designed to cleave 39 different genomic targets. The analysis revealed that the efficiency of targeted mutagenesis at a given chromosomal locus is predictive of that of homologous gene targeting. Consequently, a strong genome-wide correlation was apparent between the efficiency of targeted mutagenesis (≤0.1% to ∼6%) with that of homologous gene targeting (≤0.1% to ∼15%). In contrast, the efficiency of targeted mutagenesis or homologous gene targeting at a given chromosomal locus does not correlate with the activity of individual endonucleases on transiently transfected substrates. Finally, we demonstrate that chromatin accessibility modulates the efficacy of rare-cutting endonucleases, accounting for strong position effects. Thus, chromosomal context and epigenetic mechanisms may play a major role in the efficiency rare-cutting endonuclease-induced genome engineering.
INTRODUCTION
The development of a comprehensive technology platform to efficiently engineer the genome at specific sites in living cells opens new opportunity for functional genomics, generation of transgenic models, gene therapy, stem cell engineering and regenerative medicine. Such targeted genome modification has typically been achieved using different types of designer rare-cutting endonucleases that induce a DNA double-strand break (DSB) at a desired specific location in the genome. Different scaffolds have been used to generate these endonucleases (1,2), and at least four families of such endonucleases have been documented so far: zinc finger nucleases (ZFNs) (3–5), meganucleases (MNs) from the LAGLIDADG family (6,7), chemical nucleases (8,9) and transcription activator-like nucleases (TALENs) (10–12). From a structural or chemical perspective, these designer rare-cutting endonucleases differ in several respects, which have been reviewed elsewhere (1,2). Consequently, one can expect that each of these platforms will have different properties in terms of efficacy, specificity, range of cleavable sequences, spectrum of induced events (homologous gene targeting versus targeted mutagenesis, see below), and therefore, of potential applications.
MNs and ZFNs are currently the most well documented and widely used reagents. The I-SceI MN has been used since the 1990s in genome engineering experiments (13,14). Redesign of the DNA binding interface of the I-CreI MN by protein engineering has substantially increased the number of sequences that can be cleaved (15,16). Similarly, a large number of ZFNs with tailored specificities have been generated and used by several laboratories in a wide range of cell types (3–5,17–21). Both MNs and ZFNs have the ability to deliver targeted DSBs that subsequently undergo either non-homologous end-joining (NHEJ) or homologous recombination (HR) (1). NHEJ, an error prone DSB repair pathway, has been used to inactivate the open reading frames of selected genes by targeted mutagenesis (TM) (22,23). In contrast, HR is used mainly for targeted gene insertion or correction, by homologous gene targeting (HGT). Although cleavage of promiscuous or ‘off-target’ sequences have also been observed, MNs and ZFNs have both been used to induce precise gene modifications without affecting global genome integrity, even in organisms with large genomes such as fish, mammals and plants (1).
Though genome editing based on these designer rare-cutting endonucleases is determined by the recognition of a specific nucleotide target sequence, it is likely that chromosomal context and epigenetic modifications of target loci could also play an important role. Consequently, chromatin accessibility and/or the positioning of nucleosomes in vivo (24,25) at the target locus may impact TM and/or HGT. Alternatively, the location of different target loci in distinct ‘chromosomal territories’ could possibly influence genome editing by modulating access to limiting factors required for DNA repair (26). To address these outstanding questions and assess the potential impact of chromosomal position effects and epigenetics on designer rare-cutting endonucleases, we have conducted a comprehensive analysis of the properties of MNs in human cells. We therefore monitored TM and HGT efficacies in an unprecedented large number of chromosomal loci across the human genome and determined the effect of chromatin accessibility on genome editing by MNs. The current study contributes to a better understanding of the properties of designer rare-cutter endonucleases which has important fundamental biological and translational implications.
MATERIALS AND METHODS
Meganucleases
MNs derived from I-CreI were engineered as described previously (16,27,28). All MNs were used under a single chain format, as described in Grizot et al. (16), except for CAPNS1m and NACAm which were used in 293-H cells under a previously described heterodimeric format (16,28).
293-H cells culture and transfections
Human 293-H cells (Life Technologies, Carlsbad, CA, USA) and hamster CHO-KI cells (ATCC) were cultured at 37°C with 5% CO2 in complete medium DMEM and F12-K, respectively, supplemented with 2 mM l-glutamine, penicillin (100 IU/ml), streptomycin (100 µg/ml), amphotericin B (Fongizone: 0.25 µg/ml, Life Technologies,) and 10% FBS. For extrachromosomal and survival assays, CHO-KI cells were plated at 2500 cells per well in 96-well plate. The next day, cells were transfected with 200 ng of total DNA using Polyfect transfection reagent according to the supplier’s (Qiagen) protocol. In the survival assay, a constant amount of GFP-encoding plasmid (10 ng) was added to various amounts of MN expression vectors. For mutagenesis and gene targeting assays, 293-H cells were plated at a density of 1.2 × 106 cells per 10 cm dish. The next day, cells were transfected with 3 or 5 µg of plasmid DNA using Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer’s protocol.
Extrachromosomal activity assays in CHO-KI and HEK293 cells
Activity in mammalian cells was measured as previously reported by Grizot et al. (29) These assays are used to monitor the activity of the MNs in a non-chromatinized template, an activity that is a function of MN expression and specific activity.
Cell survival assay
The CHO-KI cell line was transfected as described above, with varying amounts of MN expression vector and a constant amount of GFP-encoding plasmid. GFP levels were monitored by flow cytometry (Guava EasyCyte, Guava Technologies) on Days 1 and 6 post-transfection. Cell survival was calculated as a ratio (MN-transfected cells expressing GFP on Day 6/control transfected cells expressing GFP on Day 6) corrected for the transfection efficiency determined on Day 1.
Area under the curve scores
Area under the curve (AUC) score is used to quantify (i) the activity of MN in extrachromosomal assays in CHO-KI and 293-H cells; (ii) cell survival in the toxicity assay; (iii) chromatin resistance to microccocal nuclease digestion at a given locus. Examples are given in Figure 2b and Supplementary Figure S2c. The classical approach based on the use of log-normal model (30) showed a very good fitting quality for both survival curves and activity curves (after normalization with respect to the maximal activity level). All the statistics done in this study are summarized in Supplementary Table S6.
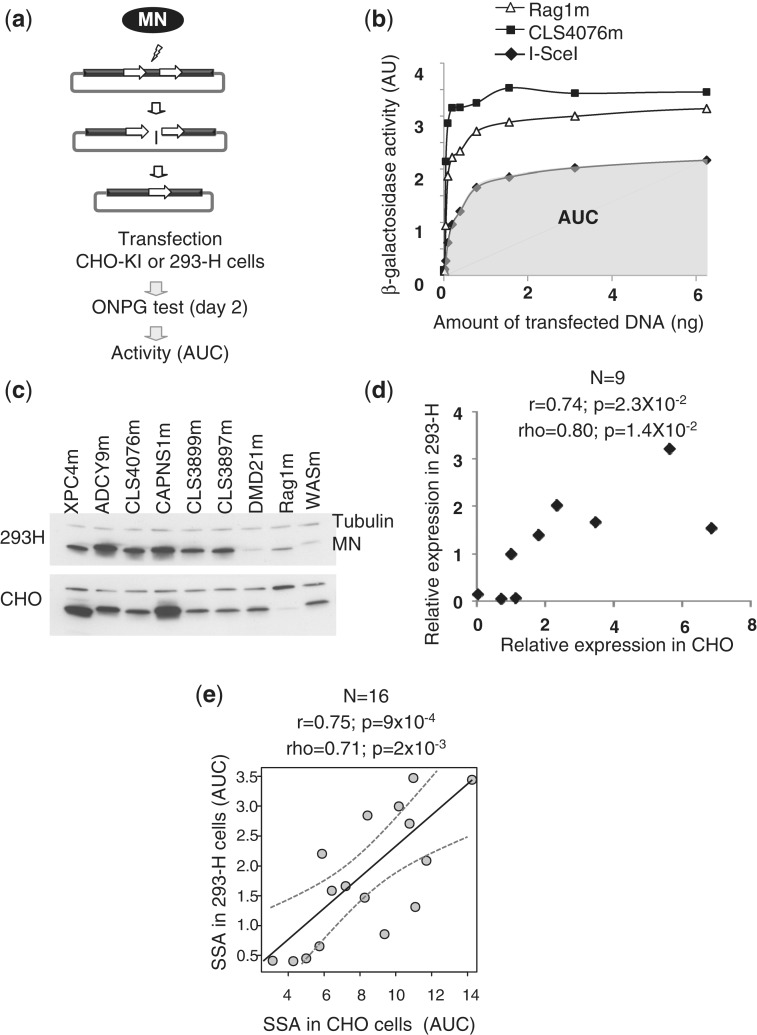
Figure 2.
Characterization of MNs properties using cell-based assays. (a) An extrachromosomal assay for the characterization of MN activity in mammalian cells. Briefly, MN expressing vectors and reporter vector are co-transfected into CHO-KI or 293-H cells. Upon cleavage of the target site, tandem repeat recombination by SSA between two truncated copies of the LacZ gene restores a functional β-galactosidase gene, which can be monitored by standard assays. For each MN, a dose response is performed and AUC is used as a quantitative measure of MN activity. (b) Example of read out of the extrachromosomal assay described in panel. The activity of the CLS4076m, I-SceIm and Rag1m MNs is featured as an example. AUC is used as a quantitative measure of MN activity in this type of extrachromosomal assay. For CLS4076, AUC is the area in grey. (c) Monitoring of MN expression by western blotting. 293-H cells and CHO-K1 were transfected with a plasmid-encoding MN. The MN expression level was monitored by western blot. (d) Quantification of MN expression level. The MN expression level from western blot (c) was quantified and normalized to β-tubulin signal. These ratios were plotted for both cell lines and statistical analysis was performed. r, Pearson coefficient (linear correlation coefficient); ρ, Spearman coefficient (non-linear correlation coefficient); N, sample size; P, probability of finding a given correlation when the underlying variables are not correlated. (e) Comparison between the extra-chromosomal SSA assays in CHO-KI and HEK293 cells. Sixteen MNs were tested in both assays (Table 1). Dotted grey curves represent 95% confidence interval for the regression line. r, Pearson coefficient (linear correlation coefficient); ρ, Spearman coefficient (non-linear correlation coefficient); N, sample size; P, probability of finding a given correlation when the underlying variables are not correlated.
Western blot
293-H and CHO-KI cells were plated at 1.2 × 106 and 2 × 105 cells per 10 cm dish, respectively. These two cell lines were transfected with the same DNA plasmid-encoding MN: 3 µg for 293-H cells and 1 µg for CHO-K1 cells, according to the usual transfection condition. Two days post-transfection, total protein extraction was performed with RIPA (150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 01% SDS, 50 mM Tris pH 8.0) buffer. Western blot was performed on 10 µg of total protein extract. The MN expression was revealed using a specific I-CreI antibody. Beta-tubulin antibody (Cell signaling 2128) was used as loading control. Quantification of signal was performed on low exposed blots using ImageJ software. Results were expressed as a ratio between MN and tubulin signals.
Monitoring of MN-induced TM
293-H cells were transfected with 3 µg of MN expressing vector or empty vector, except for ADCY9-induced mutagenesis where 293-H cells were transfected with 5 µg of MN-encoding plasmid. Two days post-transfection, genomic DNA was extracted and targets studied were amplified with specific primers flanked by specific adaptators needed for HTS sequencing (For: CCATCTCATCCCTGCGTGTCTCCGACTCAG-Tag and Rev: BiotinTEG/ CCTATCCCCTGTGTGCCTTGGCAGTCTCAG) on the 454 sequencing system (454 Life Sciences). An average of 10 000 sequences per sample were analysed. Sequence of the primers used to amplify the targeted locus are indicated on Supplementary Table S3. In clonal experiments (Rag1m and DMD21m), cells were plated after treatment and individual clones were grown. After 21 days, DNA extraction was performed with the ZR-96 genomic DNA kit (Zymo research) according to the supplier’s protocol. The region of the target was amplified by PCR as described above, and PCR product was sequenced.
Monitoring of MN-induced HGT
293-H cells were co-transfected with 3 µg of MN expressing vector, and 2 µg of DNA matrix and seeded at 10 cells per well in 96-well plate, and HGT was monitored by PCR 21 days later. Alternatively, cells were plated and individual colonies were picked 2–3 weeks later in 96-well plates and analysed (Supplementary Table S1). All DNA matrices comprise 1 kb of homologous sequences located on both sides of the MN recognition site. The two homology arms are separated by a heterologous fragment of 29 bp or 1.7 kb. DNA extraction was performed with the ZR-96 genomic DNA kit (Zymo research) according to the supplier’s protocol. The detection of targeted integration is performed by specific PCR amplification using a primer located within the heterologous insert of the DNA repair matrix, and another one located on the genomic sequence outside of the homology. In pool experiments (10 cells/well), HGT frequencies were normalized to plating efficiencies, e.g. 0.33. This method was validated by the good correlation between the frequencies in clonal analysis and the frequencies in pool analysis (Supplementary Table S1). Sequence of the primers used to amplify the homologous regions is available upon request. Sequence of the primers used to diagnostic targeted integrations are indicated on Supplementary Table S3. HGT was also confirmed by Southern Blot for Rag1m, DMD21m, ADCY9m, XPC4m and CLS3690m.
Simultaneous monitoring of MN-induced HGT and TM
HEK293-H cells were co-transfected with plasmids expressing the MN and 2 µg of repair matrix. In each experiment, we amplified the region surrounding the target site by nested PCR using deep sequencing primers, with the first set of primers being located outside of the repair matrix (see Supplementary Table S3 for primer sequence). We measured both HGT and TM by deep sequencing of the amplicons.
CHART-PCR
Cells were resuspended in 1 ml of sucrose buffer supplemented with 0.5% of NP-40. The pellet nuclei was resuspended in ice-cold nuclei buffer supplemented with DTT and PMSF at a concentration of 2 × 104 nuclei/µl. Digestion was performed on 1.5 × 105 nuclei at room temperature for 15 min with 10–500 U of micrococcal nuclease (Biolabs). Restriction cleavage was stopped by the addition of 10 mM EDTA. DNA was extracted in the presence of proteinase K and then quantify with picogreen (Quant-iT™ PicoGreen® dsDNA Assay, Molecular Probes). Q-PCR was performed in duplicate with specific primers described in Supplementary Table S3. The data are expressed as percentage of Q-PCR signal of undigested sample.
RESULTS
Target selection
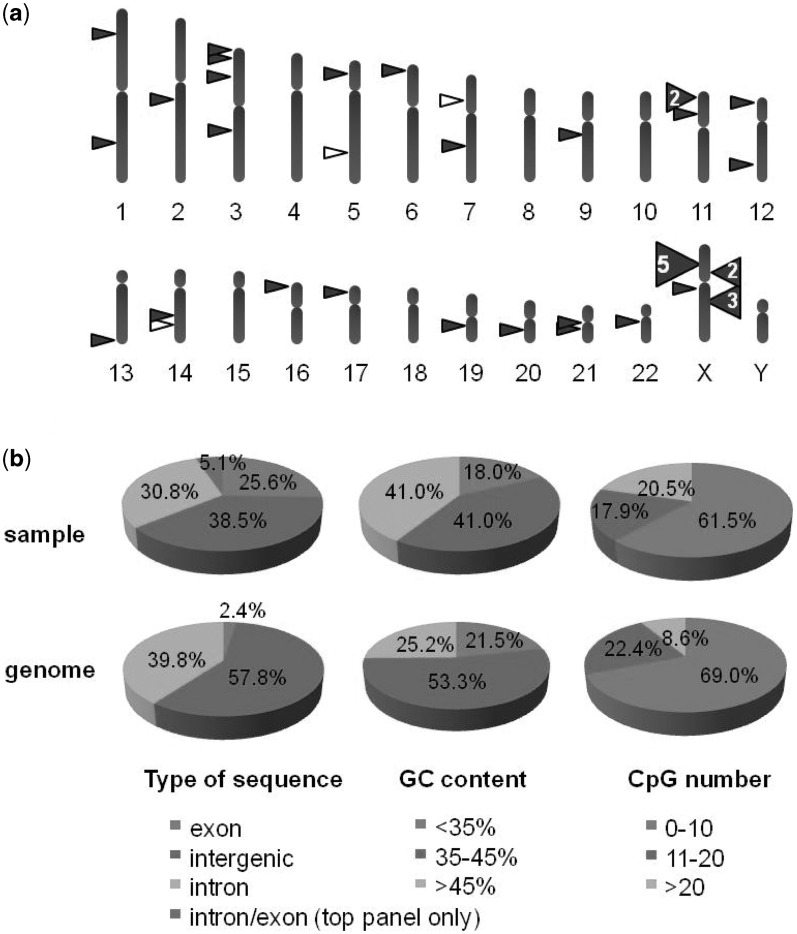
We have previously described a combinatorial method to generate MNs that cleave selected sequences in virtually any gene or locus (16,28,31). Briefly, this approach relies on the use of an archive of locally engineered derivatives of the I-CreI MN. These MNs recognize targets differing from the I-CreI target by a few base pairs only, but can be used as building blocks to generate combinatorial libraries of fully redesigned MNs. MNs recognizing specific chosen targets, unrelated to the sequence recognized by the parental I-CreI scaffold, are identified using a high-throughput yeast cell assay. Typically, a MN cleavage site is placed between two direct repeats in a β-galactosidase (LacZ) reporter plasmid, as extra-chromosomal target (Supplementary Figure S1). Cleavage of the target site by the MN in this cell-based assay induces tandem-repeat recombination, by single-strand annealing (SSA) between two truncated copies of the LacZ gene thereby restoring a functional LacZ gene. This method allowed for the identification of a potential cleavable target every 250 bp, with a success rate of 40% at the protein engineering step (32). Based on this principle, we generated a large collection of 37 different engineered MNs, derived from the parental I-CreI natural protein, that cleaved 39 different sequences, specifically designed for genome editing in coding and non-coding sequences of the human genome (Figure 1 and Table 1). In this study, m refers to the MN and t its respective target sequence. For instance, Rag1 designates the chosen locus, Rag1m the MN, and Rag1t, the 22 bp target sequences cleaved by the Rag1 MN. Our selection was prioritized towards coding sequences, given their high potential interest (i.e. gene inactivation and gene correction) and translational relevance (i.e. gene therapy). Consequently, this prioritization resulted in a higher proportion of targets in GC- and CpG-rich regions (Figure 1b).
Figure 1.
Genome-wide distribution of the targets of the MNs used in this study. (a) Schematic representation of the distribution per chromosome. Thirty-seven MNs cleaving 39 targets were used in this study, including 36 MNs cleaving each single target (represented by filled-in triangles) and 1 MN (CLS3902m), cleaving 3 (represented by open triangles). Target sequences and MN properties are described in Table 1. (b) Distribution per type of genomic sequence, per GC content and per CpG number. Distributions for target locations (top row) are compared with the distribution found for the whole genome (bottom row, on assembly GRCh37/hg19). For the type of genomic sequences, two MN target sites are overlapping with an intron/exon boundary and a special class was made for them. For GC content and CpG number, windows of 1 kb are taken around the recognition site (top row) or successively on the whole genome (bottom row) and the percentage of GC or the number of CpG dinucleotides are computed. Percentages in each class are displayed. The estimation of the percentage of sequences in exon, intron and intergenic regions on the whole genome is based on available annotations. The human genome version we used was GRCh37/hg19.
Table 1.
Meganucleases and targets
| Target characterization |
Extra- chromosomal test (AUC) |
Tox test |
Chromatin accessibility (AUC) |
Targeted mutagenesis in 293 cells (% indels) |
Homologous gene targeting in 293 cells (% KI) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MNa | Geneb | Target sequence | Chromosome | Seq. type | CHOc | 293d | CHO AUCe | act. dosef | Toxg | 293h | CD34h | T cellh | d7 w/ MNi | d2 w/ MNj | d7 w/o MNk | w/ MNl | w/o MNm |
| CLS3906m | TTTTCCTGTCTTACCAGGTTTTGT | 1p34.3 | Intergenic | 6.63 | 5.04 | 6 | 70 | 0.10 | 0.04 | ||||||||
| CLS3903m | TTAAAACTTCTTAAAGGATCCCAG | 1q31.2 | Intergenic | 8.98 | 3.14 | 2.5 | 60 | 0.80 | 0.10 | 2.00 | 0.00 | ||||||
| CLS3898m | CTAGGCTGGATTACAGCGGCTTGA | 2q14.1 | Intergenic | 8.36 | 5.37 | 2.5 | 80 | 0.00 | 0.00 | ||||||||
| CLS4633m | CCR5 | TCGAAATGAGAAGAAGAGGCACAG | 3p21 | Exon | 3.70 | 1.78 | 1.2 | 40 | 0.04 | 0.00 | |||||||
| XPC4m | XPC | TCGAGATGTCACACAGAGGTACGA | 3p25 | Exon | 10.80 | 3.52 | 5.84 | 2.5 | 80 | 188 | 0.40 | 0.36 | 0.00 | 1.05 | 0.00 | ||
| CLS3719m | ATTTCATCACCTAACTAATCTCAC | 3p25.3 | Intergenic | 2.57 | 0.00 | 0.02 | |||||||||||
| CLS3191m | Rho | ACTTCCTCACGCTCTACGTCACCG | 3q21-q24 | Exon | 4.82 | 0.40 | 5.37 | 6 | 95 | 0.10 | 0.00 | ||||||
| CLS3894m | ACAATCTGAGGTAAGTAATACTGA | 5p14.3 | Intergenic | 8.09 | 1.45 | 3.87 | 6 | 45 | 0.50 | 0.02 | 0.60 | 0.00 | |||||
| CLS3902m | TCAAACCATTGTACTCCAGCCTGG | 14q24 | Intergenic | 9.99 | 3.03 | 5.36 | 2.5 | 90 | 247 | 0.19 | 0.70 | 0.02 | |||||
| TCAAACCATTGTACTCCAGCCTGG | 5q31 | Intergenic | 337 | 0.11 | 0.39 | 0.02 | |||||||||||
| HECW1 | TCAAACCATTGTACTCCAGCCTGG | 7p13 | Intron | 163 | 0.53 | 1.17 | 0.02 | ||||||||||
| CLS2697m | CCAATACAAGGTACAAAGTCCTGA | 6p25.1 | Intergenic | 9.09 | 4.79 | 2.5 | 80 | 0.70 | 0.00 | ||||||||
| CLS2705m | TTAAAACACTGTACACCATTTTGA | 7q31.2 | Intergenic | 6.41 | 4.49 | 2.5 | 75 | 0.20 | 0.02 | 0.90 | 0.00 | ||||||
| CLS2760m | SMC5L1 | CAACTCCGTCTTACAGAGGTAAAA | 9q21.11 | Intron/Exon | 8.09 | 4.96 | 6 | 60 | 0.10 | 0.00 | 0.00 | 0.00 | |||||
| Rag1m | RAG1 | TTGTTCTCAGGTACCTCAGCCAGC | 11p13 | Intron/Exon | 9.19 | 0.82 | 5.16 | 2.5 | 95 | 106 | 78 | 70 | 1.30 | 1.28 | 0.03 | 8.40 | 0.00 |
| CLS2607m | HBB | TCAGACTTCTCCTCAGGAGTCAGA | 11p15.5 | Exon | 2.04 | 5.80 | 6 | 95 | 0.01 | 0.04 | |||||||
| CLS3769m | HBB | AGGGGATAAGTAACAGGGTACAGT | 11p15.5 | Intron | 7.08 | 4.66 | 2.5 | 45 | 0.06 | 0.00 | |||||||
| CLS4422m | NANOG | ACTGAACGCTGTAAAATAGCTTAA | 12p13.31 | Intron | 11.10 | 5.04 | 2.5 | 50 | 0.10 | 0.03 | |||||||
| CLS3884m | NACA | GGGAACTGCTTCACAGGATCCAGA | 12q23-q24.1 | Exon | 5.88 | 2.5 | 0.02 | 0.06 | |||||||||
| CLS3896m | ATAAAACAAGTCACGTTATTTTGG | 13q34 | Intergenic | 4.24 | 5.76 | 6 | 85 | 0.10 | 0.05 | ||||||||
| CLS4076m | FUT8 | TTGTAATGTCTTACAAGGTTTTAA | 14q23.3 | Promoteur | 10.91 | 1.29 | 3.21 | 2.5 | 40 | 55 | 43 | 41 | 0.70 | 5.09 | 0.06 | 0.37 | 0.00 |
| ADCY9m | ADCY9 | CCCAGATGTCGTACAGCAGCTTGG | 16p13.3 | Exon | 8.24 | 2.87 | 5.97 | 2.5 | 95 | 410 | 201 | 179 | 0.20 | 0.17 | 0.06 | 0.70 | 0.00 |
| CLS2276m | p53.7 | TCTGGCCCCTCCTCAGCATCTTAA | 17p13.1 | Exon | 9.19 | 0.10 | 0.03 | 0.99 | 0.00 | ||||||||
| CAPNS1m | CAPNS1 | CAGGGCCGCGGTGCAGTGTCCGAC | 19q13.12 | 5′UTR | 14.08 | 3.49 | 5.15 | 24.23 | 21 | 52 | 2.40 | 6.10 | 0.01 | 12.00 | 0.26 | ||
| scCAPNS1m | 13.96 | 13.96 | 3.98 | 1.2 | 70 | ND | 6.20 | 0.01 | 15.00 | 0.26 | |||||||
| CLS2752m | CTSZ | ATTACCCCCTTCACAGGAGCCAGC | 20q13 | Intron | 9.99 | 3.89 | 2.5 | 40 | 0.30 | 0.20 | 0.00 | 0.00 | |||||
| CLS3895m | AGAAGCCCAGGTAAAACAGCCTGG | 21q21 | Intergenic | 3.74 | 4.07 | 6 | 50 | 0.10 | 0.10 | ||||||||
| CLS3690m | TTAATACCCCGTACCTAATATTGC | 21q21.1 | Intergenic | 5.71 | 2.21 | 4.94 | 2.5 | 80 | 0.50 | 0.06 | 1.50 | 0.00 | |||||
| CLS3899m | GCAAAACATTGTAAGACATGCCAA | 22q13.33 | Intergenic | 10.59 | 2.73 | 1.74 | 2.5 | 40 | 0.10 | 0.01 | |||||||
| CLS3702m | WAS | TCCAAACCTCTCACAAAAGTGTAT | Xp11.23- p11.22 | Intron | 7.70 | 2.40 | 2.5 | 50 | 0.10 | 0.04 | |||||||
| CLS3759m | WAS | CCGGGCCCTCGTGCAGGAGAAGAT | Xp11.23- p11.22 | Exon | 10.52 | 5.54 | 2.5 | 95 | 286 | 0.50 | 0.48 | 0.00 | 4.50 | 0.40 | |||
| DMD21m | DMD | GAAACCTCAAGTACCAAATGTAAA | Xp21.2 | Intron | 4.10 | 0.35 | 5.62 | 6 | 95 | 61 | 25 | 35 | 1.40 | 1.20 | 0.05 | 8.40 | 0.00 |
| CLS3326m | DMD | AAATCCTGCCTTAAAGTATCTCAT | Xp21.2 | Intron | 6.76 | 5.59 | 2.5 | 100 | 0.10 | 0.07 | 0.09 | 0.00 | |||||
| CLS3402m | DMD | TTTACCTATTTTAAGTCAGATACA | Xp21.2 | Intron | 2.95 | 0.36 | 3.75 | 2.5 | 65 | 0.00 | 0.03 | ||||||
| CLS3631m | DMD | AATGTCTGATGTTCAATGTGTTGA | Xp21.2 | Intron | 7.03 | 1.65 | 3.54 | 6 | 50 | 0.20 | 0.01 | 0.60 | 0.00 | ||||
| CLS4607m | DMD | GAATCCTGTTGTTCATCATCCTAG | Xp21.2 | Intron | 9.19 | 0.90 | 0.08 | 4.00 | 0.00 | ||||||||
| CLS4418m | CCAACATCCTGAACCTCAGCTAC | Xq11 | Pseudogene | 5.61 | 3.61 | 1.2 | 85 | 0.30 | ND | ||||||||
| CLS3368m | IL2RG | CCAGGCTTGAGTGCAGTGGGGCGA | Xq13.1 | Intron | 10.17 | 4.35 | 1.2 | 85 | 0.02 | 0.02 | |||||||
| CLS3640m | IL2RG | ATTGCACTTGGAACAGCAGCTCTG | Xq13.1 | Exon | 6.25 | 1.57 | 5.13 | 2,5 | 80 | 0.40 | 0.00 | ||||||
| CLS3676m | IL2RG | CCTAAACTTGGTGCCCCATCTCTC | Xq13.1 | Intron | 11.53 | 2.09 | 3.98 | 6 | 50 | 0.20 | 0.04 | 1.08 | 0.00 | ||||
| I-CreI | NA | TAGGGATAACAGGGTAAT | NA | NA | 12.39 | 2.20 | 1 | 95 | NA | NA | NA | NA | NA | NA | NA | NA | |
| I-SceI | NA | TCAAAACGTCGTGAGACAGTTTGG | NA | NA | 5.56 | 0.61 | 5.98 | 6 | 95 | NA | NA | NA | NA | NA | NA | NA | NA |
bWhen in a gene, or in the promoter.
cMeasured as shown in Figure 2A and B, in CHO-KI cells.
dMeasured as shown in Figure 2A and B, in 293-H cells.
fMeasured in nanograms, amount of transfected DNA corresponding to MN maximal activity.
gMeasured in percentage of survived cells at the active dose.
hMeasured as shown in Figure 5, see also ‘Materials and Methods’ section.
iMeasured in percentage of chromatids, as shown in Figure 3A, 7 days after transfection with MN expressing vector in 293-H cells.
jMeasured in percentage of chromatids, as shown in Figure 3A, 2 days after transfection with MN expressing vector in 293-H cells.
kMeasured in percentage of chromatids, as shown in Figure 3A, 7 days after mock transfection in 293-H cells.
lMeasured in percentage of cells, as shown in Figure 3B, 21 days after transfection with MN expressing vector vector in 293-H cells. HGT frequencies were normalized to plating efficiencies, e.g. 0.33.
mMeasured in percentage of cells, as shown in Figure 3B, 21 days after mock transfection in 293-H cells HGT frequencies were normalized to plating efficiencies, e.g. 0.33.
Characterization of MNs activity based on extrachromosomal cellular assays
A quantitative cell-based assay in CHO-KI and 293-H cells was used to characterize the MN activity of this MN collection was used (Figure 2a and Table 1). This extrachromosomal assay (29,31) is similar to the cellular assay in yeast and allows for a quantitative assessment of MN activity based on LacZ reporter gene expression (Figure 2b). Importantly, the activity measured in this extrachromosomal in vivo cellular assay depends not only on the intrinsic specific activity of the MN, but also on the MN expression level in the target cell. Whereas specific activity can vary widely even between closely related I-CreI derivatives (28,33), MN expression levels can also be strongly affected by the few amino acid substitutions that differ from one engineered MN to another (Figure 2c). Nevertheless, there is a strong correlation in relative MN expression levels in different cell types, at least based on 293-H and CHO-KI cells (Figure 2d).
Most importantly, when comparing MN activity in CHO-KI and 293-H cells, we observed very consistent results (Figure 2e), with linear (Pearson’s r) and non-linear (Spearman’s ρ) correlation coefficients ranging between 0.71 and 0.75, and P ≤ 0.002. We also compared MN activity measured in our CHO-KI cell assay with that obtained with aforementioned yeast assay, that also measures MN-induced tandem repeat recombination on a plasmid-borne reporter system (10,16,21,27). Here again, we observed a very good consistency (r = 0.8; and ρ = 0.79 with P < 2.2 × 10−16 in both cases; Supplementary Figure S1b), suggesting that our extrachromosomal assay in CHO cells is predictive for a wide range of different cell types. This MN activity assay in CHO cells therefore serves as an appropriate and representative assay for MN activity.
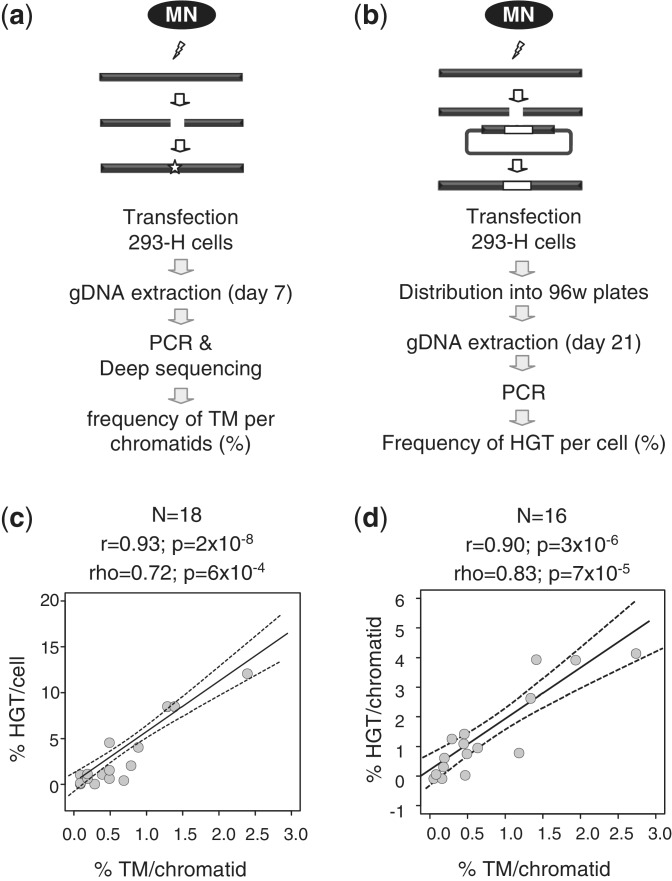
MN-induced TM and HGT efficiencies are strongly correlated
MNs were then tested for their efficacy, defined in this study as their ability to induce targeted gene modification at their cognate chromosomal locus in human cells. Results are summarized in Table 1. TM was monitored for all MNs by pyrosequencing of the targeted locus 7 days after transfection of 3 µg of MN-expressing vector (Figure 3a). The locus was amplified by PCR, and an average of 10 000 molecules was sequenced per amplicon. The MN-induced rate varied between background level (up to 0.06% due to PCR and sequencing errors) and 2.4% of the analyzed chromatids (Table 1). HGT was monitored for 18 MNs by PCR in pools of 10 cells grown in 96-well plates, 21 days after transfection, as described previously (16) (Figure 3b). HGT was also monitored in clonal experiments for 4 MNs, with the results confirming the results from pools in all cases (Supplementary Table S1). For the 18 MNs that were characterized for both TM and HGT, a strong linear correlation (r = 0.93 with P = 2 × 10−8) was observed between the frequency of TM ‘per chromatid’ and the frequency of HGT ‘per cell’ throughout the target genome (Figure 3c). Most importantly, this comprehensive analysis revealed that the efficiency of TM at a given chromosomal locus is predictive of that of HGT and that the HGT/TM ratio is relatively conserved throughout the genome.
Figure 3.
Characterization of MNs efficacy on their chromosomal cognate target. (a) A chromosomal assay for the characterization of MN-induced TM. MN expressing vectors are transfected into 293-H cells; the targeted locus is amplified by PCR and 10 000 molecules (on average) are sequenced to detect indels. (b) A chromosomal test for the detection of MN-induced HGT. MN-expressing vectors are co-transfected into 293-H cells together with a repair matrix; cells are grown in 96-well plates, and targeted alleles are detected by PCR. (c) Comparison between the TM and HGT assays in 293-H cells. Eighteen MNs were characterized in both assays (Table 1). Dotted grey curves represent 95% confidence interval for the regression line. Linear regression fits the formula y = 5.46x − 0.66. r, Pearson coefficient (linear correlation coefficient); ρ, Spearman coefficient (non-linear correlation coefficient); N, sample size; P: probability of finding a given correlation when the underlying variables are not correlated. (d) Comparison between the TM and HGT frequencies measured simultaneously by deep sequencing. Sixteen independent experiments were conducted, using different MNs (Rag1m, ADCY9m, CLS3676m, CLS3759m, CLS3894m, CLS4076m and CLS4607m), promoters (SV40 or CMV) and vector concentrations, all described in Supplementary Table S2. Dotted grey curves represent 95% confidence interval for the regression line. Linear regression fits the formula y = 1.71x + 0.20. r, ρ, P and N have the same meaning as on the previous panel.
In Figure 3c, we compared HGT events per cell and TM events ‘per chromatid’ in 293-H cells. However, these immortalized cells typically do not have a normal diploid genome. Therefore, we refined our analysis and conducted additional studies to simultaneous monitor HGT and TM ‘per chromatid’ by deep sequencing. Events were monitored at Day 2, to alleviate toxicity effect over time. Sixteen measurements were made with seven different MNs (Supplementary Table S2). Again, a very strong linear correlation (r = −0.90 with P = 3 × 10−6) was observed between the frequency of HGT ‘per chromatid’ and frequency of TM ‘per chromatid’ (Figure 3d, Supplementary Table S2), and in this case, a global HGT/TM ratio of 1.7 could be inferred from the slope of the regression line.
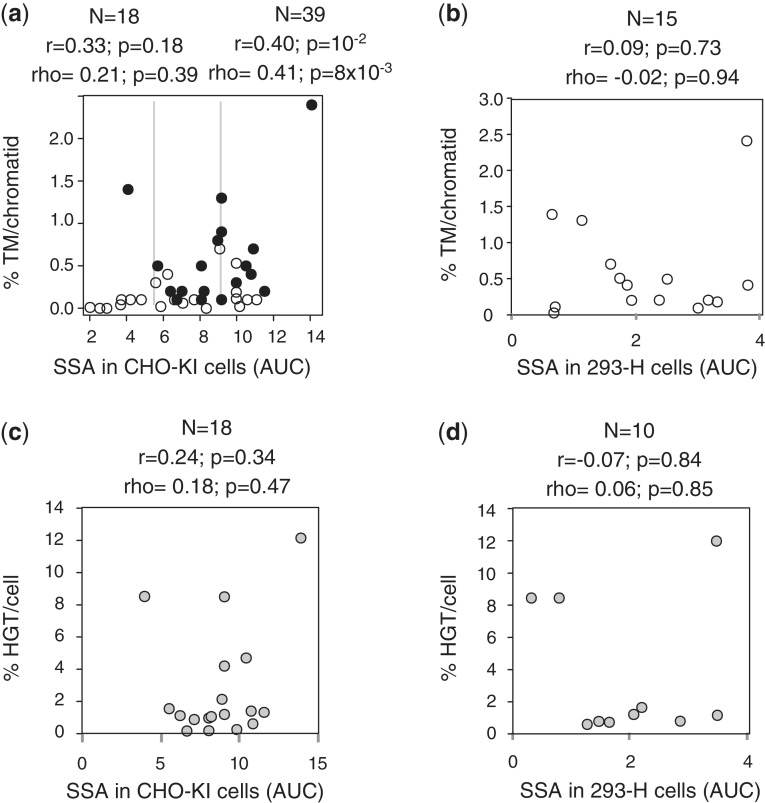
MN efficacies are poorly correlated with their activities based on extrachromosomal substrates
The differences in MN efficacy on chromosomal target loci may reflect different parameters, particularly the activity of a given MN, as measured using extrachromosomal substrates, and/or MN-associated cellular toxicities. However, other parameters may also contribute to these differences in MN efficacy, such as bona fide target site-dependent chromosomal position effects. Consequently, MN efficacy (i.e. TM, HGT) may result from a combination of ‘intrinsic’ factors (i.e. MN activity, MN toxicity) and ‘extrinsic’ factors (i.e. MN target locus).
To assess the contribution of each of these distinct parameters on the overall MN efficacy, we performed a comprehensive quantitative analysis. We first compared MN efficacy based on TM and HGT assays and MN activity based on the extra-chromosomal assay, as measured in CHO-KI and 293-H cells (Figure 4a). Overall, MN efficacies based on the frequency of TM per chromatid correlated poorly with their respective intrinsic activities on extrachromosomal substrates (Figure 4a and b), consistent with the low correlation coefficients and P-values. A somewhat higher correlation was observed only when comparing MN efficacies (based on the frequency of TM per chromatid) with activity in CHO-KI cells (r = 0.40 with P = 10−2, ρ = 0.41 with P = 8 × 10−3), and was statistically significant only when a large sample (n = 39) was characterized (compare with n = 18 on Figure 4a), though the correlation coefficient remained low (r = 0.40). Notably, some of the highest frequencies of indels (as a measure of the frequency of TM per chromatid) were achieved with DMD21m, which has relatively moderate activities in the extrachromosomal assay (Figure 4a and Table 1). Similarly, HGT frequencies were poorly correlated with MN activity in CHO-KI and 293-H cells (Figure 4c and d). These modest correlations contrast with the high consistency observed between the extrachromosomal tests (Figure 2e and Supplementary Figure S1b) and between the two types of efficacy tests (Figure 3c and d).
Figure 4.
The efficacy of MNs on their chromosomal target is poorly correlated with the activity of the nucleases in extrachromosomal assays. Throughout the figure, r, Pearson coefficient (linear correlation coefficient); ρ, Spearman coefficient (non-linear correlation coefficient); N, sample size; P, probability of finding a given correlation when the underlying variables are not correlated. (a) Comparison between the extrachromosomal SSA assay in CHO-KI and the TM assay in HEK293 cells. Thirty-seven MNs cleaving 39 targets were tested in both assays (Table 1). Correlation was also made with a smaller sample of 18 MNs (black circles), which are the same as the 18 MNs characterized in the HGT assay (Figure 3b), and displayed on Figure 3c. Vertical grey lines represent the activity levels of I-SceIm (left line) and Rag1m (right line). (b) Correlation between the extrachromosomal SSA assays in 293-H cells and the TM assay in 293-H cells. (c) Correlation between the extrachromosomal SSA assays in CHO cells and the HGT assay in 293-H cells. (d) Correlation between the extrachromosomal SSA assays in 293-H cells and the HGT assay in 293-H cells.
Hence, the intrinsic activity of a given MN on extrachromosomal substrates is not predictive for its efficacy on chromosomal targets. Therefore, we decided to test the impact of other factors.
MN efficacies are not correlated with their cellular toxicity
It has been shown before that in a population of cells treated with a rare cutting endonuclease, the proportion of cells modified by the nuclease could vary over time, probably due to nuclease toxicity (34). We actually observed a similar phenomenon, whereas treatment with the non-toxic Rag1m molecule results in a stable rate of ∼1% of TM, most of the events generated by CLS4076m at Day 2 (∼5% of TM) have disappeared by Day 13 (Supplementary Figure S2a). Thus, cell death resulting from MN toxicity could impact with apparent efficacy of MN, especially when measured after day several days.
We assessed the toxicity of all MNs featured in this study, using a cell survival assay that was previously validated and relies on decreased green fluorescent protein (GFP) expression as a read-out for cellular toxicity (16,20,35; Supplementary Figure S2b and S2c). Toxicity was MN dose-dependent and typically ranged between that of I-SceI (the most specific reported rare-cutting endonuclease) and I-CreI, with the exception of two MNs (CLS3899 and CLS4633, see Table 1). Importantly, toxicity was found not to be related to the level of activity (r = −0. 15 with P = 0.52; and ρ = −0.14 with P = 0.43).
However, no correlation could be established between toxicity and efficacy. The estimated values of linear (Pearson’s r) and non-linear (Spearman, ρ) correlation coefficients between TM and toxicity are 0.04 and 0.03, respectively, with corresponding P-values of 0.81 and 0.86, respectively. Therefore, we looked for more complex correlations factors. Supplementary Figure S2d features an attempt of correlation between toxicity, on the one hand, and the ratio of the efficacy of TM in 293-H cells versus the activity in the extrachromosomal assay in CHO-KI cell (TM/SSA ratio), on the other. Correlation coefficients are very low, with very high P-values (r = 0.16 with P = 0.42; and ρ = 0.03 with P = 0.86), indicating a lack of correlation, in spite of the relatively large sample size (n = 39). Fitting of TM as a linear function of toxicity and SSA (TM = a*SSA + b*TOX + c), lead to similar results with a = 2.38 (P = 9 × 10−3), b = −0.24 (P = 0.55) and c = 7.9385 (P = 2 × 10−4). This outcome indicates that when measuring the contribution of activity in CHO-KI cells (SSA) and toxicity (TOX) to efficacy, the contribution of activity in CHO-KI cell is significant (P = 9 × 10−3), consistent with the results shown in Figure 4a for the same sample, but the contribution of toxicity is not (P = 0.55), consistent with the results of Supplementary Figure S2d.
While toxicity has no direct correlation with the efficacy of TM, it seems that it might play an important role in the dynamic of TM over time. There is a significant correlation between toxicity and the ratio of TM in 293-H at Day 7 versus Day 2 (r = 0.86 P = 6 × 10−3; ρ = 0.9, P = 5 × 10−3), with more toxic MNs having a bigger drop in the efficacy of TM from Day 2 to Day 7.
In conclusion, whereas toxicity can impact the apparent efficacy of MNs, it cannot account for the global discrepancies observed between the on extrachromosomal and chromosomal targets, and MN efficacy is not solely the consequence of its intrinsic properties (activity and toxicity). Therefore, we decided to test the impact of extrinsic factors, such as chromatin-based position effect.
MN efficacy is strongly correlated with chromosomal position effects
Chromatin-based position effects could impact on MN efficacy by modulating access of the MN, the repair matrix and/or cellular repair factors to the target DNA. Eukaryotic genomes are packaged in chromatin, and nuclease sensitivity remains one of the main methods used to monitor chromatin accessibility and/or the positioning of nucleosomes in vivo (24,25). Nuclease accessibility is known to vary largely across the genome and at least in the promoter region, to correlates with a transcriptionally active state (24,36). In order to assess the possible role of chromatin position effect on genome editing using MN, we first compared (i) the efficacy of different MNs (isoschizomers) at the same locus (ii) the efficacy of the same MN at different loci.
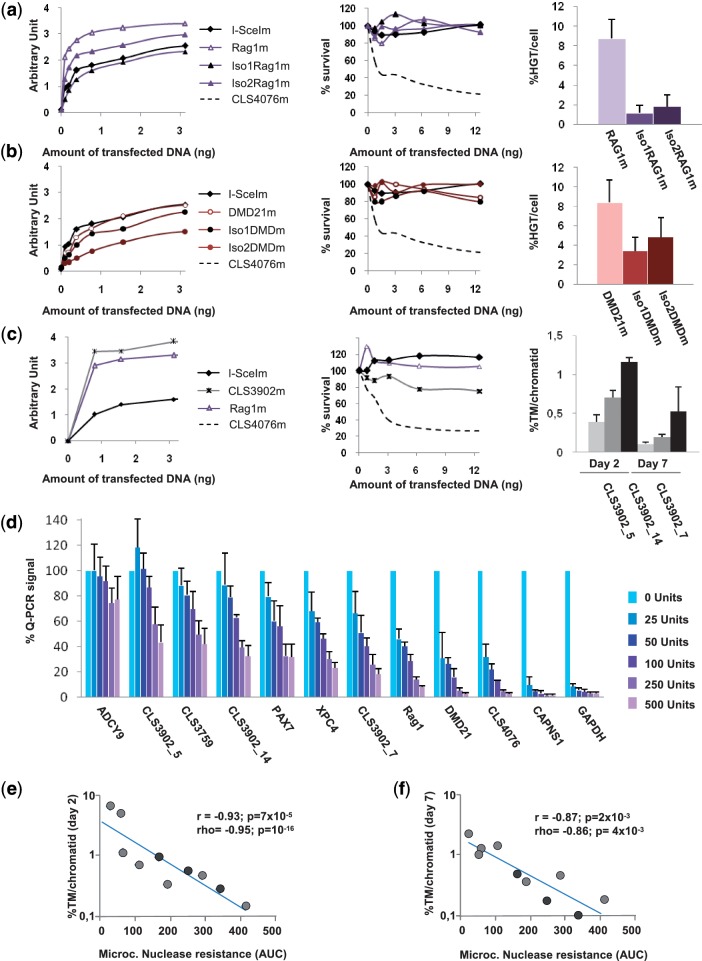
The Rag1m MN cleaves a sequence from the human Rag1 gene (16). DMD21m cleaves a sequence from the 38th intron of the gene (Table 1). Several isoschizomers were obtained when we engineered these proteins, and we compared Rag1m and DMD21m with two additional isoschizomers. At each individual locus, efficacy was found to correlate with activity in CHO-KI cells (Figs. 5a and 5b), with the most active cognate MN giving the highest rates of HGT. However, this correlation was not found when comparing the results achieved at the two loci. For example, the iso2DMDm MN is significantly less active than the iso2RAG1m MN, but has a similar efficacy. Thus, efficacy is correlated with activity in CHO-KI cells only when assessed at the ‘same target locus’. We also characterized CLS3902m, a MN whose cognate target is found four times in the genome. CLS3902m has the same activity as Rag1m, and a slightly higher toxicity (Figure 5c). Examination of TM at three target loci (the fourth one could not be amplified by PCR) gave clearly different frequencies of events, ranging from 0.39% to 1.17% at Day 2 and from 0.13 to 0.75 at Day 7 (Figure 5c), demonstrating locus dependence.
Figure 5.
Impact of position effect on MNs efficacy. (a) The Rag1m MNs as well as Iso1Rag1m and Iso2Rag1m, two isoschizomers were tested for activity in our extrachromosomal assay in CHO-KI cells, for toxicity, and for their ability to induce targeted insertion at the endogenous Rag1 locus. Activity, efficacy and toxicity assays were conducted as described in Figures 2 and 3. (b) Similar studies were conducted with the DMD21m MN and Iso1DMDm and Iso2DMDm cleaving the DMD21t target. (c) The CLS3902m MN cleaves a sequence found in four copies in the human. CLS3902m was tested for activity, toxicity and efficacy of targeted mutagenesis at three of its cognate targets, CLS3902t_5, CLS3902t_7 and CLS3902t_14. In A, B and C, each value is the average of two independent experiments but for Rag1m (n = 7) and DMD21 (n = 3). (d) CHART-PCR assay for micrococcal nuclease accessibility of the Rag1t, DMD21t, CLS3902t_5, CLS3902t_7, CLS3902t_14, CLS4076t, CAPNS1t, ADCY9t, CLS3759t and XPC4t loci. The GAPDH and PAX7 loci are used as controls for accessible and non accessible chromatin, respectively (37). Nuclei were isolated and treated with various amounts of micrococcal nuclease, and for each locus, the amount of non digested DNA was monitored by Q-PCR as described in materials and methods. Data are normalized to the value obtained in absence of nuclease, and each point is the average of four measurements from two independent experiments. (e) Comparison between the micrococcal nuclease accessibility assay and the TM assay at Day 2. The values of panel d were used to construct a curve, and a nuclease resistance index (AUC) was calculated as the AUC, based on the same principle as activity and cell survival index calculations (Figure 2b and h). Efficacy of TM at Day 2 was plotted against this resistance index, using a semi-logarithmic scale. Here, r represents the linear correlation coefficient between log(TM) and AUC. r, Pearson coefficient (linear correlation coefficient); ρ, Spearman coefficient (non-linear correlation coefficient); N, sample size; P, probability of finding a given correlation when the underlying variables are not correlated. (f) Comparison between the micrococcal nuclease accessibility assay and the TM assay at Day 7. Here, r represents the linear correlation coefficient between log(TM) and AUC.
This type of position effect is likely the consequence of chromatin-based differences of target accessibility. We used CHART-PCR (36,37) to test micrococcal nuclease accessibility at 10 loci targeted by eight engineered MNs (Figure 5d), and a nuclease resistance index (AUC) was calculated for each locus, as indicated in figure legend and ‘Materials and Method’ section. CLS4076t and CAPNS1t, two targets for which we observed very high rates of TM, were indeed located in very accessible regions. Interestingly, CAPNS1t is in a non-methylated CpG island in the 5′UTR of the CAPNS1 gene, whereas CLS4076t is close to a CpG island, about 4 kb upstream of the region coding for the FUT8 protein. More generally, a very strong ‘negative’ correlation was observed between nuclease resistance and percentage of TM per chromatid at Day 2 (Figure 5e) and at Day 7 (Figure 5f), fitting with strong linear correlation between log (TM) and accessibility index (r = −0.93 with P = 7 × 10−5and r = −0.87 with P = 2 × 10−3, respectively). Thus, target accessibility is a major determinant of efficacy of rare-cutting designer endonucleases.
DISCUSSION
We describe here the first genome-wide study of a rare-cutting designer endonuclease platform. We showed that a wide range of efficacies of TM and HGT (between ≤0.1% and 15%) can be achieved in human cells using MNs. This provides an effective benchmark for genome editing. In addition, these comprehensive studies allow us to accurately address several long-standing questions. In particular, since both TM and HGT depend on cellular DNA repair mechanisms, it is tempting to hypothesize that HGT and TM would be correlated at the genome level. Moreover, the potential impact of chromosomal context and epigenetics on designer endonuclease-mediated genome editing remains largely unexplored. Our current study is the first of its kind that specifically addresses the role of chromosomal context and epigenetics in genome editing.
Our comprehensive analysis revealed a robust genome-wide correlation between MN-induced HGT and MN-induced TM (r = 0.93; P = 2 × 10−8). Thus, the efficiency of repair of a designer endonuclease-induced DSB by either NHEJ or homologous recombination essentially depends on extrinsic factors at the cellular level (although one cannot exclude subtle locus-specific or MN-specific differences). One of the direct consequences is that the TM rate, which is relatively easy to monitor, is predictive of the HGT rate, which monitoring requires the construction of a repair matrix and a more sophisticated protocol. Therefore, TM can be used as a surrogate assay to investigate the ability of a given locus to be edited by MN-induced HGT. We could actually infer a global relatively constant HGT/TM ratio throughout the genome close to 1.7 in 293-H cells. This ratio is likely to be cell-type dependent, particularly since HGT is thought to be largely dependent on the rate of cell division (38). It would be interesting to determine whether, the average HGT/TM ratio is similar or substantially different with alternative platforms, such as ZFNs or TALENs in the same cell type.
The availability our comprehensive platform of MN allowed us to assess the potential impact of chromosomal context and epigenetics on rare-cutting endonuclease-mediated genome editing. In particular, it has long been assumed that the efficacy of these nucleases could be subject to position effects. We showed here that ‘extrinsic’ factors (i.e. chromosome locus) have a strong impact that can outweighs that of ‘intrinsic’ factors (i.e. MN activity, expression, toxicity) on the net MN efficacy. Most importantly, molecular analysis shows a very robust correlation between chromatin accessibility and MN ‘efficacy’, which appears as a key predictive factor in genome engineering experiments. These results should not hide the fact that at a given individual locus, the most active isoschizomers will have the highest efficacies, as shown on Figure 5a and b. However, these results underscore the potential importance of chromosomal context and epigenetic factors in genome editing using designer rare-cutting endonucleases. Our results also argue that relatively simple techniques such as CHART-PCR, or the simple data mining of existing databases could be used to better predict the loci allowing for the best efficiencies. The promoters or 5′UTR regions of housekeeping genes could be ideal targets for applications requiring high efficacies, as illustrated by the high efficacies observed with the CAPNS1t and CLS4076t targets. Consequently, our results set the stage for a more rational design of genome engineering strategies using designer endonucleases and contribute to a better understanding of their properties at the genome-wide level. It is likely that chromosomal position effects and epigenetics, particularly target site accessibility, will have broad implications for other genome engineering platforms such as those based on ZFNs and TALENs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figures 1–2.
FUNDING
Cellectis; an FWO fellowship (Belgium; to M.L.); Free University of Brussels and University of Leuven; research Project Funding [FWO N° G.0632.07, in part]; Association Nationale de la recherche et de la Technologie, contrat [Cifre 535/2008 to A.D.]. Funding for open access charge: Cellectis.
Conflict of interest statement. All authors but M.L., O.D., M.C., T.V., G.D., L.P., and T.K. are employees of Celectis. F.P. has shares and stock options of Cellectis.
Supplementary Material
ACKOWLEDGEMENTS
We thank E. Samara and Dr E. Belay for technical assistance.
REFERENCES
- 1.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Paques F. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pingoud A, Silva GH. Precision genome surgery. Nat. Biotechnol. 2007;25:743–744. doi: 10.1038/nbt0707-743. [DOI] [PubMed] [Google Scholar]
- 3.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 5.Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo Riso P, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 6.Pâques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 7.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNeer NA, Chin JY, Schleifman EB, Fields RJ, Glazer PM, Saltzman WM. Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol. Ther. 2010;19:172–180. doi: 10.1038/mt.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenschmidt K, Lanio T, Simoncsits A, Jeltsch A, Pingoud V, Wende W, Pingoud A. Developing a programmed restriction endonuclease for highly specific DNA cleavage. Nucleic Acids Res. 2005;33:7039–7047. doi: 10.1093/nar/gki1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 12.Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Prieto J, Redondo P, Blanco FJ, Bravo J, Montoya G, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 19.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 20.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 25.Carey MF, Peterson CL, Smale ST. In vivo DNase I, MNase, and restriction enzyme footprinting via ligation-mediated polymerase chain reaction (LM-PCR) Cold Spring Harb. Protoc. 2009;2009 doi: 10.1101/pdb.prot5277. doi:10.1101/pdb.prot5277. [DOI] [PubMed] [Google Scholar]
- 26.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell. Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 29.Grizot S, Duclert A, Thomas S, Duchateau P, Paques F. Context dependence between subdomains in the DNA binding interface of the I-CreI homing endonuclease. Nucleic Acids Res. 2011;39:6124–6136. doi: 10.1093/nar/gkr186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen AK, Srivastava MS. Regression Analysis: Theory, Methods and Applications. New-York: Springer; 1990. [Google Scholar]
- 31.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 32.Galetto R, Duchateau P, Paques F. Targeted approaches for gene therapy and the emergence of engineered meganucleases. Expert Opin. Biol. Ther. 2009;9:1289–1303. doi: 10.1517/14712590903213669. [DOI] [PubMed] [Google Scholar]
- 33.Redondo P, Prieto J, Munoz IG, Alibes A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 34.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 35.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 36.Rao S, Procko E, Shannon MFJ. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 37.Cruickshank M, Fenwick E, Abraham LJ, Ulgiati D. Quantitative differences in chromatin accessibility across regulatory regions can be directly compared in distinct cell-types. Biochem. Biophys. Res. Commun. 2008;367:349–355. doi: 10.1016/j.bbrc.2007.12.121. [DOI] [PubMed] [Google Scholar]
- 38.Delacote F, Lopez BS. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle. 2008;7:33–38. doi: 10.4161/cc.7.1.5149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.