SUMMARY
MicroRNAs (miRNAs) comprise a large family of small RNA molecules that post-transcriptionally regulate gene expression in many biological pathways1. Most miRNAs are derived from long primary transcripts that undergo processing by Drosha to produce ~65 nucleotide (nt) precursors that are then cleaved by Dicer, resulting in the mature 22 nt forms2,3. Serving as guides in Argonaute protein complexes, mature miRNAs use imperfect base-pairing to recognize sequences in mRNA transcripts, leading to translational repression and destabilization of the target mRNAs4,5. Here we show that the miRNA complex also targets and regulates non-coding RNAs (ncRNAs) that serve as substrates for the miRNA processing pathway. We found that the C. elegans Argonaute, ALG-1, binds to a specific site at the 3′ end of let-7 miRNA primary transcripts and promotes downstream processing events. This interaction is mediated by mature let-7 miRNA via a conserved complementary site in its own primary transcript, thus creating a positive feedback loop. We further show that ALG-1 associates with let-7 primary transcripts in nuclear fractions. Argonaute also binds let-7 primary transcripts in human cells, demonstrating that the miRNA pathway targets non-coding RNAs in addition to protein-coding mRNAs across species. Moreover, our studies in C. elegans reveal a novel role for Argonaute in promoting biogenesis of a targeted transcript, expanding the functions of the miRNA pathway in gene regulation. This discovery of auto-regulation of let-7 biogenesis sets a new paradigm for controlling miRNA expression.
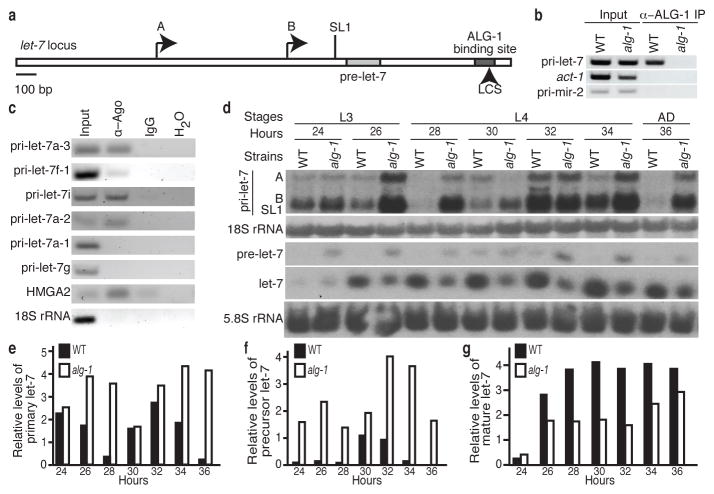
Recent studies from our lab provided a global map of interactions between Argonaute and endogenous mRNAs in C. elegans at the fourth larval (L4) stage of development6. Using cross-linking immunopurification with high-throughput sequencing (CLIP-seq), over 3,000 mRNA transcripts were found to have sequences bound by the C. elegans Argonaute Like Gene 1 (ALG-1). Surprisingly, the non-coding let-7 primary transcripts (pri-let-7), which are processed into the mature let-7 miRNA, were among the RNAs targeted by Argonaute, as indicated by the presence of an ALG-1 binding site towards the 3′ end of the transcripts (Fig. 1a and Supplementary Fig. 1). To verify that Argonaute associates with pri-let-7, we performed RNA Immunopurification (RIP) assays, using an anti-ALG-1 antibody, in L4 stage lysates from wild-type (WT) worms and alg-1(gk214) mutants, which lack the anti-ALG-1 epitope and exhibit slightly delayed development (Supplementary Fig. 2). A robust signal for pri-let-7 sequences was observed in WT but not alg-1(gk214) extracts (Fig. 1b and Supplementary Fig. 3a). Two unspliced (A and B) and one SL1 trans-spliced primary let-7 isoforms are produced by the let-7 gene in C. elegans7, and they all appear to associate with Argonaute (Supplementary Fig. 3a). Control actin mRNA or pri-mir-2 transcripts were not detected in ALG-1 immunopurifications, demonstrating the specificity of Argonaute association with primary let-7 transcripts (Fig. 1b and Supplementary Fig. 3a). Human Argonaute (Ago) proteins also associate with a subset of let-7 primary transcripts, suggesting that this interaction is conserved in higher organisms (Fig. 1c and Supplementary Fig. 3b).
Figure 1. Argonaute binds and regulates pri-let-7.
a, The let-7 gene: precursor sequence, two transcriptional start sites (A, B), splice site for SL1 trans-splicing and the ALG-1 binding site, which includes a let-7 complementary site (LCS), are indicated. b–c, Detection of the indicated transcripts by RIP of WT and alg-1(gk214) or HeLa cell extracts. d, Northern analysis of RNA from WT and alg-1(gk214). e–g, Levels of pri-let-7 relative to 18S rRNA and pre- or mature let-7 relative to 5.8S rRNA in WT or alg-1(gk214) quantified from d.
Argonaute-mediated post-transcriptional regulation can result in mRNA degradation4,5, and many established miRNA targets in C. elegans are up-regulated in alg-1(gk214) mutant worms6. To test if alg-1 also regulates the levels of primary let-7 transcripts, we performed Northern blotting on RNA extracted from WT and alg-1(gk214) worms at every two hours of development from mid L3, when mature let-7 starts to accumulate, until early adulthood. The three primary let-7 transcripts were detected at similar levels in WT and alg-1(gk214) worms at 24 hours of development (Fig. 1d). However, by four hours later the total let-7 primary transcript levels in WT had decreased to almost undetectable levels whereas the levels in alg-1(gk214) remained high (Fig. 1d–e). While the expression of let-7 primary transcripts in both strains still followed the previously described oscillation behaviour8, the cycling was muted in the alg-1(gk214) worms. Overall the pattern is consistent in independent biological replicates though the rapid cycling of let-7 can shift the expression peaks of individual time courses (Supplementary Fig. 3d–e). In contrast to let-7, the levels of other primary miRNA transcripts exhibited modest, if any, changes in alg-1(gk214) versus WT at the L4 stage, and none of these primary miRNAs were detected in ALG-1 RIP assays (Supplementary Fig. 3c and data not shown). At all time points, precursor let-7 (pre-let-7) accumulated to higher levels in alg-1(gk214) compared to WT, whereas mature let-7 was more abundant in WT (Fig. 1d,f–g and Supplementary Fig. 3d,f–g). An accumulation of precursor and diminished level of mature was observed for all miRNAs tested (mir-58, mir-90, lin-4), regardless of changes in primary transcript levels, in alg-1(gk241) compared to WT worms (Supplementary Fig. 3d and data not shown). These observations are consistent with previous reports of a general role for Argonaute in precursor processing and mature miRNA stabilization9–12.
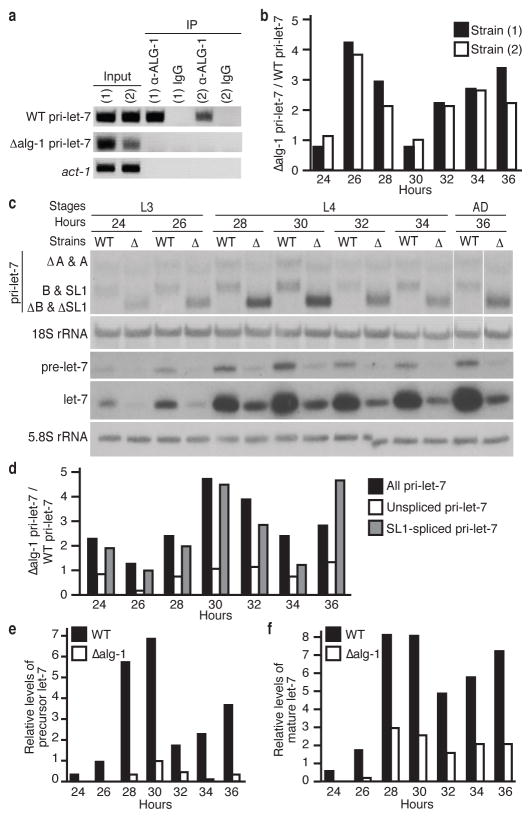
The Argonaute-bound region in let-7 primary transcripts is about 500 nucleotides downstream of the mature let-7 sequence and covers about 100 nucleotides, as mapped by ALG-1 CLIP-seq6 (Fig. 1a). To test if this site is required for interaction of ALG-1 with let-7 primary transcripts, transgenic strains with a single copy insertion of the let-7 locus lacking the ALG-1 binding site were created. These animals express the endogenous primary let-7 transcripts from the X Chromosome (WT pri-let-7) and primary let-7 transcripts with the ALG-1 binding site deleted (Δalg-1 pri-let-7) from Chromosome II. Both the WT and Δalg-1 pri-let-7 transcripts were expressed in the transgenic animals, but only the WT pri-let-7 with the ALG-1 binding site intact immunopurified with Argonaute (Fig. 2a and Supplementary Fig. 4a).
Figure 2. The ALG-1 binding site in pri-let-7 regulates expression of let-7.
a, Detection of the indicated transcripts by RIP from two independent transgenic strains. b, Ratio of the levels of pri-let-7 inΔalg-1 versus WT determined by qRT-PCR and normalised to 18S rRNA. c, Northern analysis of RNA from WT orΔalg-1 pri-let-7 transgenes in let-7(mn112). d, Ratio of the levels of let-7 transcripts expressed fromΔalg-1 versus WT pri-let-7 transgenes in let-7(mn112) determined by qRT-PCR and normalized to 18S rRNA. e–f, Levels of pre- or mature let-7 relative to 5.8S rRNA expressed from WT or Δalg-1 pri-let-7 transgenes in let-7(mn112) quantified from c.
Compared to the control strain, worms harbouring the Δalg-1 pri-let-7 transgene expressed overall higher levels of primary let-7 (Fig. 2b and Supplementary Fig. 4b). In these assays, primary transcripts expressed from the endogenous let-7 locus are detected in addition to those of transgenic origin. To specifically analyse transgenic let-7 expression, we crossed the transgenic strains into the let-7(mn112) background. The let-7(mn112) mutation removes 190 base pairs just upstream and including the first nucleotide of the let-7 precursor, resulting in undetectable precursor or mature miRNA production13. Northern blotting showed higher levels of primary let-7 transcripts lacking the ALG-1 binding site compared to WT for most time points in the let-7(mn112) background (Fig. 2c and Supplementary Fig. 4c). Using primers that detect the unspliced versus SL1-spliced primary transcripts derived from the transgenes in qRT-PCR assays, we found that the SL1-spliced isoform was responsible for the increase detected in the Δalg-1 pri-let-7 transgenic worms (Fig. 2d and Supplementary Fig. 4d). Surprisingly, although the Δalg-1 pri-let-7 levels were elevated, the precursor and mature let-7 levels in those animals were 2- to 5-fold lower than the levels in their WT counterparts (Fig. 2c,e–f and Supplementary Fig. 4c,e–f). This deficiency reduced the rescue activity of the Δalg-1 pri-let-7 transgene in the let-7(mn112) background (Supplementary Fig. 4g).
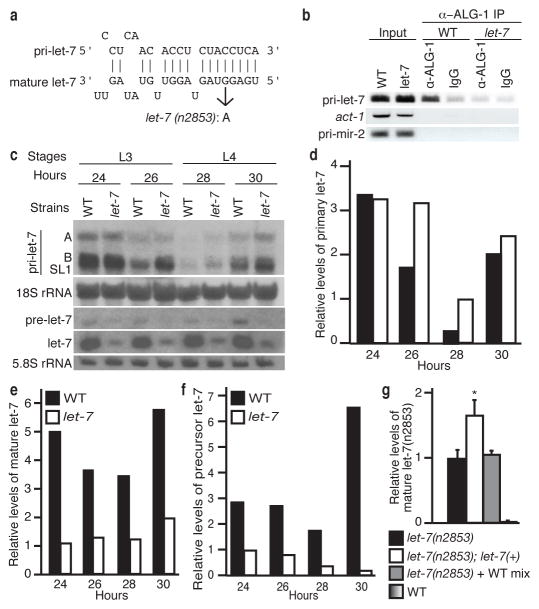
We found a potential let-7 complementary site (LCS) within the ALG-1 binding region of pri-let-7 that is conserved in other Caenorhabditis species (Fig. 3a and Supplementary Fig. 5a,b). To test if let-7 miRNA mediates association of ALG-1 with primary let-7 transcripts, we performed RIP assays, using late L3 stage WT and let-7(n2853) mutant animals. The let-7(n2853) mutant animals harbour a point mutation (G→A) at the fifth nucleotide of the mature miRNA (Fig. 3a), which disrupts base-pairing to target mRNAs13–17. Primary let-7 immunopurified with ALG-1 in WT, but not in let-7(n2853) extracts (Fig. 3b and Supplementary Fig. 6a), indicating that the association of Argonaute with primary let-7 transcripts requires the mature let-7 miRNA.
Figure 3. Mature let-7 regulates its own maturation.
a, Base-pairing of mature let-7 to a site in pri-let-7 with the G→A mutation in let-7(n2853) indicated. b, Detection of the indicated transcripts by RIP of WT and let-7(n2853). c, Northern analysis of RNA from WT and let-7(n2853). d–f, Levels of pri-let-7 relative to 18S rRNA and pre- or mature let-7 relative to 5.8S rRNA in WT or let-7(n2853) quantified from c. g, Analysis of mature let-7(n2853) relative to 18S rRNA from the indicated strains or a 1:1 mix of RNA from let-7(n2853) and WT (mean ± s.e.m., n = 3, *, P < 0.05).
Northern blot analysis of primary let-7 in staged let-7(n2853) and WT animals during development from mid L3 until the mid-L4 stages showed similar or higher levels in let-7(n2853) compared to WT animals (Fig. 3c–d and Supplementary Fig. 6b–d). Despite the generally higher levels of primary let-7, precursor and mature let-7 were substantially reduced in let-7(n2853) compared to WT worms (Fig. 3c–f and Supplementary Fig. 6b–f).
Our cumulative data indicate that binding of ALG-1 via mature let-7 miRNA to pri-let-7 results in reduced transcript levels and increased mature levels, consistent with a role for ALG-1 in promoting the processing of primary let-7 transcripts. To further test this idea, we asked if introduction of WT let-7 would boost the levels of mature let-7 (n2853). Since the let-7(n2853) primary transcript has an intact LCS, the presence of WT mature let-7 miRNA is predicted to recruit ALG-1 to the transcript to promote processing and increase mature let-7(n2853) levels. Strains containing the WT let-7 gene integrated on chromosome II and the let-7(n2853) allele on the X chromosome [let-7(n2853); let-7(+)] were tested for effects on the levels of mature let-7(n2853) RNAs. Compared to the let-7(n2853) strain, the levels of mature let-7(n2853) were increased in the let-7(n2853); let-7(+) strain (Fig. 3g). This up-regulation is likely muted by destabilization of the mature miRNA attributed to the n2853 mutation, which impairs the target-mediated protection conferred to mature miRNAs by their target mRNAs15.
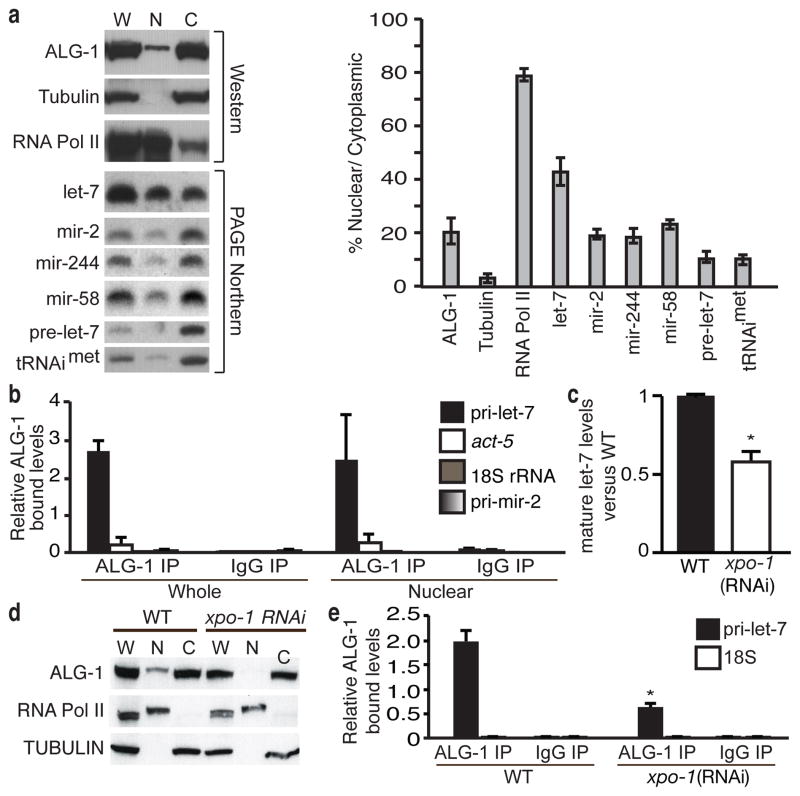
Since processing of miRNA primary transcripts typically occurs in the nucleus, we investigated the association of ALG-1 with pri-let-7 in this compartment. About 20% of ALG-1 protein localized to the nuclear fraction (Fig. 4a). Notably, almost half of the mature let-7 miRNA was associated with the nuclear fraction, which was double the amount of three other tested miRNAs (Fig. 4a). Using ALG-1 RIP assays of whole cell and nuclear fractions from WT animals, we observed strong and specific association of pri-let-7 with ALG-1 in both extracts (Fig. 4b).
Figure 4. Association of ALG-1 with pri-let-7 in nuclear fractions.
a, Detection in whole cell (W), nuclear (N) or cytoplasmic (C) fractions of the indicated proteins or RNAs (mean ± s.e.m., n = 3). b, Levels of the indicated transcripts relative to their respective inputs analysed by RIP and detected by qRT-PCR (mean ± s.e.m., n = 2). c, Ratio of mature let-7 from xpo-1 relative to control RNAi after normalization to 18S rRNA (mean ± s.e.m., n = 7, *, P < 0.001). d, Detection of the ALG-1 protein in fractions from WT and xpo-1-depleted animals. e, Analysis of pri-let-7 or 18S rRNA relative to their respective inputs by RIP and qRT-PCR of control (WT) or xpo-1 RNAi (mean ± s.e.m., n = 4, *, P < 0.001).
Recently, the nuclear transport receptor Exportin-1 (XPO-1) was implicated in regulating primary miRNA processing through an unknown mechanism18. As previously reported, depletion of xpo-1 by RNAi resulted in a two-fold decrease in mature let-7 levels (Fig. 4c)18. Moreover, xpo-1(RNAi) reduced the nuclear localization of ALG-1 by a factor of 3.5 ± 0.1 (mean ± s.e.m., n = 5, P < 0.001), but did not affect total ALG-1 levels, compared to WT controls (Fig. 4d). Additionally, RIP assays revealed reduced association of ALG-1 with pri-let-7 in xpo-1 depleted animals (Fig. 4e). Although Exportin-1 is best characterized as a nuclear export factor, it has been shown to interact with Argonaute proteins in mammalian cells19 and, thus, it remains to be determined how direct the role is for XPO-1 in mediating nuclear interactions between ALG-1 and pri-let-7.
Our studies reveal a novel paradigm of ncRNAs as targets of miRNA induced silencing complexes. We find that Argonaute associates with let-7 miRNA primary transcripts in C. elegans and with a subset of pri-let-7 RNAs in human cells, indicating that transcripts other than mRNAs can also be miRNA targets and should be considered in target prediction endeavours. Argonaute is best characterized for its role in directing deadenylation and translational repression of bound mRNAs through incompletely resolved mechanisms4,5. Although the mechanism by which ALG-1 regulates let-7 primary transcripts is yet to be fully elucidated, it appears to depend on the subcellular localization of Argonaute, as regulated by XPO-1, and to be independent of LIN-28 (Supplementary Fig. 7). Recently, miR-709 was reported to inhibit the processing of pri-miR-15a/16-1 in mouse cells20. The discovery that the miRNA complex can regulate the processing of primary transcripts reveals a new role for Argonaute in the miRNA pathway. Moreover, this is the first example of a direct miRNA auto-regulatory loop, whereby mature let-7 binds and promotes processing of its own primary transcript. This amplification mechanism may be important for efficient production of mature let-7 from the oscillating levels of primary transcript substrates during C. elegans development. Our demonstration of direct miRNA regulation of let-7 biogenesis adds to the growing list of factors that impose post-transcriptional control on production of this important miRNA8,21–27. Since misregulation of let-7 leads to disease28, the let-7 positive feedback loop presents a new target for therapeutic interventions designed to restore appropriate miRNA levels.
METHODS SUMMARY
C. elegans worms were grown and synchronized by standard methods at 25°C. Polyacrylamide gel electrophoresis and Agarose Northern blotting methods were used to detect smaller and larger RNAs species, respectively7. For reverse transcription PCR (RT-PCR) assays, RNA was extracted and cDNA was synthesized using random hexamers8. For quantification of mature let-7 miRNA from the let-7(n2853) strain we used Taqman Small RNA Assays with a custom-made RT primer, specific for the mutant sequence (Applied Biosystems). RNA Immunopurification (RIP) assays in C. elegans were performed with anti-ALG-1 antibodies (Thermo Fisher Scientific) and in HeLa cells with antibodies that recognize Ago1–4 (4F9, sc-53521, Santa Cruz; 2e12/1c9, Sigma)8,29.
METHODS
Nematode culture and strains
C. elegans worms were synchronized by standard methods, cultured at 25°C and collected at the indicated time points. The wild-type strain used is N2 Bristol. The WT (PQ320) and Δalg-1 transgenic strains 1 & 2 (PQ402 & PQ404) express the WT pri-let-7 transcript or the Δalg-1 pri-let-7 transcript with the ALG-1 binding site deleted from single-copy transgenes integrated in Chromosome II using the MosSCI system30 along with the endogenous primary let-7 from chromosome X. The WT (PQ425) and Δalg-1 (PQ426) transgenic animals in the let-7(mn112) background were created by crossing PQ320 or PQ404 into let-7(mn112) animals, which do not express precursor or mature let-7. The let-7(n2853);let-7(+) transgenic animals express the WT primary let-7 transcript from a single-copy transgene integrated in Chromosome II and the endogenous let-7(n2853) transcript from the X Chromosome and were created by crossing PQ320 into the let-7(n2853) strain.
Northern Blotting
Polyacrylamide gel electrophoresis (PAGE) and Agarose Northern blotting methods were used to detect smaller and larger RNAs species, respectively7. The primers used for probe templates are described in Supplemental Table 1. To detect let-7 primary transcripts expressed only from transgenes in the let-7(mn112) background, the probe was limited to the 190 nt deletion missing in the let-7(mn112) allele. Equal amounts of probes complementary for the WT and the let-7(n2853) miRNAs were used in PAGE Northern analyses for the mature and precursor forms. Precursor levels in the WT and let-7(n2853) strains were also analysed with a probe specific for the loop region, which is identical in the WT and mutant let-7 RNAs. RNA bands were quantified using the ImageJ software package.
Reverse Transcription – Polymerase Chain Reaction (RT-PCR) assays
RNA was extracted using standard Trizol (Invitrogen) procedure and treated with RQ1 DNAse (Promega). cDNA synthesis was performed with Superscript II or III reverse transcriptase (Invitrogen) using random hexamers. Standard PCR was performed with the primers listed in Supplemental Table 1 and the products were resolved on agarose gels. Quantitative Real Time-PCR (qRT-PCR) was performed using SYBR Green or Fast SYBR Green (Applied Biosystems) on ABI Prism 7000 Real Time PCR or StepOne instruments. Quantification of wildtype mature let-7 or let-7(n2853) miRNA at the L4 stage was performed with Taqman Small RNA Assays (Applied Biosystems) following the manufacturer’s instructions using the hsa-let-7a RT primer and Taqman probe or a custom-made RT primer and Taqman probe specific for the let-7(n2853) sequence, respectively. Expression data were normalized to 18S rRNA. Levels of mature let-7(n2853) for the 1:1 mix of RNA from WT and let-7(n2853) animals were adjusted by a factor of 2 to compensate for the 2-fold dilution of the mature WT and let-7(n2853) miRNAs. Statistical analysis was performed with paired one- or two-tailed Student’s t-test.
RNA Immunopurification (RIP) assays
RNA immunopurification assays in C. elegans extracts were performed in L4 stage worms as described previously with minor modifications8. An aliquot (5% of total volume) was collected as RNA input control, the lysates were pre-cleared for 1 h at 4°C with Protein G Dynabeads and incubated overnight with 2–7ug of custom polyclonal anti-ALG-1 antibody (Thermo Fisher Scientific) or control IgG (total, Caltag Laboratories or Rat anti-GFP Clone 1A5, Santa Cruz) with gentle shaking at 4°C. Protein G Dynabeads were added the next day, incubated at 4°C for 1 h with gentle shaking, washed twice with lysis buffer for 10 min at 4°C and RNA was extracted and cDNA was synthesized with random primers using Superscript III.
RNA Immunopurification (RIP) in HeLa cells
RNA immunopurification in HeLa cell extracts were performed as described previously29 with minor modifications. Following centrifugation the supernatant was incubated with 150 ul of Dynal My1 Streptavidin-coated magnetic beads (Invitrogen) coupled to 7.5 ug of biotinylated Ago-specific 4F9 antibody, which recognizes Ago1–4, (sc-53521, Santa Cruz) or coupled to biotinylated control anti-CD4+ antibody (L3T4, eBiosciences), which were previously equilibrated with lysis buffer. Immunopurified Ago proteins were visualised with the 2e12/1c9 antibody (Sigma). Biotinylation of the 4F9 antibody was performed as described29. Supernatant and beads were incubated for 2 h with gentle shaking at 4°C and then washed twice with ice-cold lysis buffer for 5 min each with gentle shaking. RNA was extracted with Trizol and the miRNeasy RNA extraction kit (Qiagen) or standard phenol/chloroform extraction followed by DNAse treatment.
Subcellular fractionation of C. elegans
Fractionation of whole C. elegans worms was performed according to a protocol provided by the Mello Lab, University of Massachusetts, with certain modifications. L4 staged worms were harvested, flash-frozen and lysed in ice-cold isotonic lysis buffer (ILB) [(25 mM HEPES, 10 mM KCl, 5% v/v glycerol, 0.5 mM DTT, protease inhibitor cocktail (Complete Mini) and 24 U/ml RNAsin], using a glass tissue grinder with pestle. Lysates were centrifuged at 500 g for 30 sec at 4°C and 1/3 of the supernatant volume was collected (whole cell fraction) while the rest was centrifuged at 2,000g for 5 min at 4°C. The supernatant (cytoplasmic fraction) was subjected to 2 further centrifugations (2,000g for 5 min at 4°C) while the pellet (nuclear fraction) was washed twice with ILB (2,000g for 5 min at 4°C). Nuclear fractions were resuspended in a volume of ILB equilvalent to the cytoplasmic fractions.
Western blot analysis
Western blot analysis was performed as described previously14 with mouse monoclonal antibodies against tubulin (Sigma) and RNA polymerase II (Santa Cruz) or custom rabbit polyclonal antibody against a peptide in ALG-1 (Thermo Fisher Scientific). Goat-anti-mouse IgG and goat-anti-rabbit IgG secondary antibodies conjugated to horseradish peroxidase (Jackson Immunochemicals) were used along with ECL Plus chemiluminescence reagents (GE Healthcare) followed by exposure to MS Film (Kodak).
RNAi Treatments
One generation RNAi treatments on wildtype animals were performed, using the xpo-1 clone (JA:ZK742.1; primers: CAACGATTCCTCACCTGGAT and TTTTCGAGTTCATGCACGAG) from the commercially available C. elegans RNAi library (Source BioScience LifeSciences).
Supplementary Material
Acknowledgments
We thank J. Lykke-Andersen and members of the Pasquinelli lab for critical reading of the manuscript and D. Hogan for discussions. We thank F. Slack for originally pointing out the LCS in pri-let-7, the M. David lab for sharing their real time PCR machine, P. Van Wynsberghe for the Δalg-1 primary let-7 plasmid, C. Mello for the worm fractionation protocol, E. Moss for LIN-28 antibodies and A. Gorin, H. Jenq and S. Verma for technical assistance. Funding was provided by a Lymphoma and Leukemia Society Special Fellow Award 3611-11 (D.G.Z.), NIH CMG and NIH/NCI T32 CA009523 Training Grants (Z.S.K), the Swedish Board of Study Support (R.K.C.) and the US National Institutes of Health (GM071654), Keck, and Peter Gruber Foundations (A.E.P.).
Footnotes
AUTHOR CONTRIBUTIONS
A.E.P, D.G.Z and Z.S.K. designed the project and wrote the paper; D.G.Z (Figs. 1b–c, 2a, 3b,g, 4b–e and Supplementary Figs. 2, 3a–c, 4a, 6a), Z.S.K (Figs. 1d, 2c,d, 3c and Supplementary Figs. 3d, 4c,d,g, 6b), R.K.C. (Figs. 2b, 3g, 4a and Supplementary Fig. 4b), A.E.P (Fig. 4a and Supplementary Fig. 7) performed the experiments and analysed the data; A.E.P. supervised the studies.
The authors declare no competing financial interests.
References
- 1.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138:1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 5.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 6.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Wynsberghe PM, et al. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 10.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 11.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 12.Tan GS, et al. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 14.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee S, Fasler M, Bussing I, Grosshans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell. 2011;20:388–396. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 17.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bussing I, Yang JS, Lai EC, Grosshans H. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. Embo J. 2010;29:1830–1839. doi: 10.1038/emboj.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci U S A. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang R, et al. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res. 2011 doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Lehrbach NJ, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 26.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS One. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frokjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.