Abstract
Background
The efficacy of pegylated IFN-α and ribavirin (pegIFN/RBV) in the treatment of Hepatitis C infection is limited by psychiatric adverse effects (IFN-PE). Our study examined the ability of differential gene expression patterns prior to therapy to predict emergent IFN-PE among 28 HIV/HCV co-infected patients treated with pegIFN-α2b/RBV.
Methods
Patients dually infected with HIV and HCV were evaluated at baseline and during treatment by board-certified psychiatrists who classified patients into 2 groups: those who developed IFN-PE and those who did not (IFN-NPE). Gene expression analysis (Affymetrix HG-U133A) was performed using PBMCs before and after initiation of treatment. ANOVA, post hoc analysis based on pair-wise comparisons and functional annotation analysis identified differentially expressed genes within and between groups. Prediction Analysis for Microarrays was used to test the predictive ability of selected genes.
Results
Twenty-four genes (16 up- and 8 down-regulated) that were differentially expressed at baseline in patients who subsequently developed IFN-PE compared to the IFN-NPE group showed the ability to predict IFN-PE with an accuracy of 82%. In 16 patients with IFN-PE, 135 genes (117 up-; 18 down-regulated) were significantly modulated following treatment. Of these, 10 genes have already been shown to be associated with neuropsychiatric illnesses and were significantly modulated only in patients who experienced IFN-PE.
Conclusions
We describe a novel molecular diagnostic biomarker panel to predict emergent IFN-PE in HIV/HCV-co-infected patients undergoing pegIFN/RBV treatment, which may improve the identification of patients at greatest risk for IFN-PE and suggest candidate therapeutic targets for preventing or treating IFN-PE.
Keywords: HIV/HCV, peg-Interferon, psychiatric toxicities, gene expression, prediction
Introduction
Chronic co-infection with hepatitis C virus (HCV) is documented in one-third of all HIV-infected persons in the United States, an estimated 250,000 people, and is associated with increased morbidity and mortality relative to mono-infection with either virus 1. Due to shared routes of transmission, HCV co-infection is observed in an even larger proportion of HIV-infected intravenous drug users with prevalence rates up to 90% 1,2. Since the advent of antiretroviral therapy (ART), AIDS-associated opportunistic infections have declined considerably. However, HIV/HCV co-infected individuals demonstrate lower rates of sustained virologic response (SVR) to HCV therapy as well as increased rates of viral relapse among those who have achieved end-of treatment response (ETR) 3–6. Furthermore, HIV/HCV co-infected patients often have high rates of illicit drug use and psychiatric disorders, rendering them more difficult to manage during pegylated IFN-α and ribavirin (pegIFN/RBV) therapy 7,8.
Approximately one-half of all patients receiving IFN-α-based therapy for HCV infection experience specific IFN-related psychiatric toxicities 7, which remain a major cause of dose reduction and treatment discontinuation in patients undergoing therapy for HCV 9,10. Moreover, psychiatric history and baseline symptomatology are not reliable predictors of subsequent development of IFN-related psychiatric toxicities in HCV-infected subjects undergoing therapy 10,11. In this regard, a recent study has suggested that emergence of IFN-related psychiatric toxicities correlate with HCV virologic response, strongly arguing for improved diagnosis and management of these toxicities 11. Besides early identification of emerging IFN-related psychiatric toxicities and active psychiatric management are critical in ensuring patient safety and enhancing the chance of eradication of HCV in this population. In this study, we developed molecular techniques to predict the emergence of serious psychiatric adverse events during treatment with pegIFN/RBV. Such an approach for screening and monitoring HIV/HCV co-infected individuals could help improve the tolerability of pegIFN-α treatment, thereby decreasing the discontinuation rates for toxicities and enhancing the SVR rates while further elucidating genetic factors underlying the development of psychiatric symptoms.
Materials and Methods
Study subjects
Thirty two patients with stable HIV disease (with or without ART) and chronic HCV infection were enrolled in IRB-approved studies at the National Institute of Allergy and Infectious Diseases (NIAID) in Bethesda, MD for treatment of HCV infection with peg-interferon-alpha-2b (1.5μg/Kg/wk) and ribavirin 1000–1200mg/day (pegIFN/RBV). All patients signed an NIAID IRB-approved protocol informed consent document. Four patients were taken off study (one had a manic episode while on study medications; two were lost to follow up and one dropped out for social issues after 1 dose of medications). 28 patients on medication for at least 4 weeks were categorized into two groups (defined below): those who experienced no psychiatric toxicity and those who experienced psychiatric toxicity while receiving pegIFN/RBV treatment
Study Subjects Selection
HIV/HCV co-infected patients were eligible if they were older than 18 years, had a CD4 count >100 cells/mm3, HCV viral load >2000 copies/ml, histologic evidence of chronic HCV infection and stable HIV disease being managed according to current HIV guidelines. Exclusion criteria included severe liver decompensation, active and severe psychiatric illness, active substance abuse or dependence (except nicotine) at baseline, severe cardiopulmonary illness, renal disease, a hemoglobinopathy or a retinopathy. Participants with active psychiatric illness were treated and stabilized prior to enrollment. If participants’ psychiatric illness could not be stabilized following mental health evaluation and treatment, they were excluded from the study. (psychiatric evaluations are detailed below)
Psychiatric Evaluations
All patients underwent standard pretreatment psychiatric evaluations conducted by board-certified psychiatrists from the National Institute of Mental Health. Patients were assessed for current and past psychiatric and substance abuse diagnoses via semi-structured clinical interview. Patients were seen in regular clinical psychiatric follow up to assess response to treatment and control of mental health symptoms. Based upon the determination of NIMH Psychiatry, they had a minimum of three months of remitted symptoms prior to receiving study medications. This clinical assessment was corroborated by consistent BDI-II scores of less than or equal to 9 at the time of interferon treatment initiation. The majority of patients screened had a history of substance use, but only patients with active substance abuse or dependence (within the preceding six months) were excluded from the study. Patients who presented with active mood, anxiety, or psychotic symptoms were treated clinically and stabilized prior to study enrollment; three patients were given new prescriptions for citalopram, one was prescribed a dose increase of escitalopram, and one was given risperidone. Four patients (including two of those given citalopram) increased their frequency of psychotherapy visits prior to study entry. During the study, depression was evaluated by examination for cause, standard diagnostic criteria (DSM-IV), and the Beck Depression Inventory (BDI)—the latter at every clinic visit (ranging from weekly to monthly, depending upon the demands of the IFN treatment schedule) 12. Patients who received at least 4 weeks of pegIFN/RBV treatment were classified into two groups, those with (IFN-PE) and without (IFN-NPE) psychiatric toxicity after initiation of therapy (Box 1). Categorization of patients into these two groups was performed at the conclusion of the study in a blinded fashion prior to analysis of gene expression. Study participation was discontinued for patients in whom severe psychiatric toxicity developed and could not be safely managed during ongoing pegIFN/RBV treatment.
| Classification of peg-IFN/RBV-related Psychiatric Toxicity |
|---|
| A. Emergent psychiatric symptoms (patients without psychiatric symptoms at baseline, regardless of psychiatric history) |
At any time during treatment:
|
|
|
| B. Worsening psychiatric symptoms (patients with baseline psychiatric symptoms) |
At any time during treatment:
|
Patients meeting criterion A and/or B were classified as experiencing IFN-PE.
DNA microarray analysis
Peripheral blood mononuclear cells (PBMCs) were collected pre-treatment and at the end of treatment with pegIFN/RBV (48 weeks unless stopped earlier for adverse events or non response to HCV therapy) and used for DNA microarray analysis. (Figure 1) The second sample was obtained within five days of the final dose of pegIFN-α 2b in each case. Total RNA and cRNA synthesis, labeling, and hybridization to the Affymetrix U133A human microarray chips (Affymetrix Inc. Santa Clara, CA) were performed according to the manufacturer’s recommended protocol. Gene expression values (log2) were determined by GC-Robust Multi-Array algorithm (Z. J. Wu and R. Irizarray; available at: http://www.bioconductor.org) followed by a Loess normalization using an R package (available at: http://www.elwood9.net/spike).
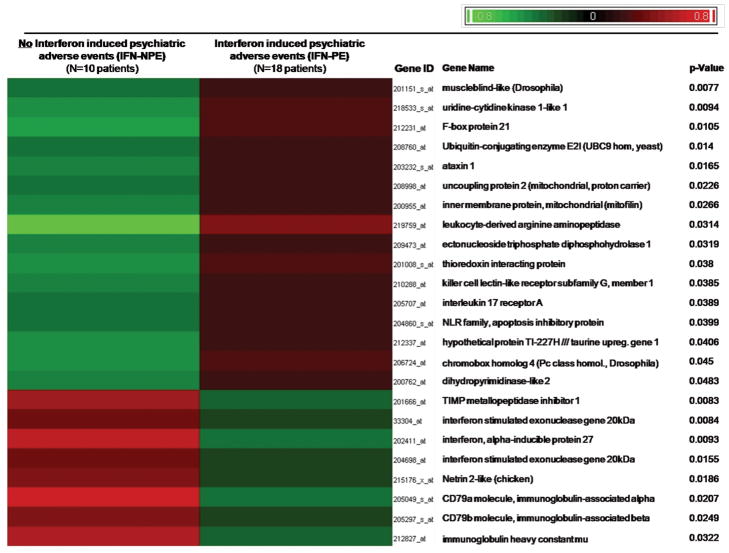
Figure 1. Differential Gene Expression between IFN-PE and IFN-NPE.
DNA microarray analysis was performed and at baseline, a significant association (P <0.05; log2MFD >0.58) has been found between specific PBMC expression profiles of 24 genes and the emergence of two different clinical phenotypes (IFN-PE: Interferon induced psychiatric adverse events vs. IFN-NPE: absence of such events) in 28 patients who underwent pegIFN-α2b/RBV therapy. Mean expression levels for each of these 24 genes were considered for supervised clustering after shift-mean normalization to highlight differences (across samples) in gene expression between patients who developed IFN-PE and those who did not.
Baseline Gene Expression Analysis
One-way Analysis of Variance (ANOVA, PARTEK Genomics Suite) was performed, followed bypair-wise post-hoc statistical comparisons based on assigning patients into 2 groups (IFN-PE vs. IFN-NPE). At baseline, 24 genes were selected for use in prediction analysis on the basis of having a significant group comparison P of <0.05 and an absolute Mean Fold Difference (log2MFC) of > 0.58.
Prediction Analysis for Microarrays
We performed Prediction Analysis for Microarrays (PAM) in order to evaluate the utility of baseline expression profiles to predict the probability of a HIV/HCV co-infected individual developing IFN-PE. The list of the 24 differentially expressed genes selected after the baseline analysis described above was tested for its predictive power in classifying patient samples according to IFN-PE using Prediction Analysis for Microarrays (PAM) software (version 2.0; Stanford University). The standardized difference, and thereby the number of relevant genes, was chosen by minimizing the prediction error using 10-fold balanced, leave-10%-out cross-validation within the training set. The threshold value that returned the lowest classification error with the fewest genes was deemed optimal. The same method can be used to predict classes (IFN-PE vs. IFN-NPE) for new samples 13.
Pre-Post Gene Expression Analysis
To gain insight into the types of neurobiological pathways that might mediate IFN-induced psychiatric toxicity in HIV/HCV co-infection, samples from 25 (out of 28 patients who received pegIFN/RBV) were successfully (hybridized to microarrays and) subjected to “post treatment” DNA microarray analysis. In patients who developed IFN-PE, genes were identified as being significantly modulated by antiviral treatment using the same statistical criteria as for the baseline analysis above (P <0.05; absolute Mean Fold Change (log2MFC) of >0.58). Supervised clustering of relative log2 gene expression values was performed to highlight changes in expression profiles following pegIFN/RBV therapy. Functional annotation analysis (DAVID 2.1) of the modulated genes was used to identify those already known to be involved in neurological and/or psychiatric disorders.
Real time PCR Validation
Total RNA was isolated from peripheral blood mononuclear cells (PBMCs), and 150 ng was used for reverse transcription (RT) analysis, using the manufacturer’s recommend protocol (ABI reverse transcription kit). One-fifteenth of the RT reaction mix was used for real-time polymerase chain reaction (PCR; in triplicate), using ABI_ Taqman gene expression master mix and the ABI 7900HT fast real-time PCR system. NRGN (ABI’s assay ID: Hs00382922_m1), GLUL (ABI’s assay ID: Hs00365928_g1, SNCA (ABI’s assay ID: Hs01103383_m1), TIMP1 (ABI’s assay ID: Hs00171558_m1) and GAPDH primers and probes were ordered through ABI’s premade Taqman gene expression assays system: Probe sets and GAPDH (ABI assay ID:Hs01003716_m1). Each gene expression data were normalized to GAPDH expression levels and expressed as relative log2 values.
Results
Study subjects and classification of toxicity
28 HIV/HCV co-infected patients were enrolled in this study. [Table 1] Post treatment samples from 3 patients were excluded from the analysis because of failed chip quality control tests; (2 developed IFN-PE.) Based on criteria described above, 18 (64%) patients developed psychiatric toxicity suggesting that IFN-PE is common in the HIV/HCV co-infected patient. Thirteen patients experienced a major depressive episode, four developed first-onset mixed mood/anxiety states and one had an exacerbation of pre-existing bipolar disorder attributable to IFN treatment. Twelve of the patients received new or augmented antidepressant medication therapy during the study (mirtazapine, sertraline, or paroxetine), two received atypical antipsychotics (risperidone), and at least six enhanced their engagement in psychotherapy. Baseline characteristics of those who developed psychiatric toxicity and patients who did not were similar with regard to age, sex, race, IL28B genotype, HCV viral load, HIV viral load, CD4+ T cell counts, HCV genotype, concurrent ART and baseline psychiatric history (Table 1). BDI screening results were consistent with other criteria for assigning toxicity in each case. The more detailed clinical assessments for a variety of psychiatric symptoms were sufficient thus the BDI data are not presented here.
Table 1.
Patient demographics
| Baseline Characteristic | IFN NPE n =10 | IFN PE n =18 | P values* |
|---|---|---|---|
|
| |||
| Age (yr) | 49.5 (41, 51) | 47 (43, 49.5) | 0.5 |
|
| |||
| Male gender (%) | 9 (90%) | 17 (94%) | 0.661 |
|
| |||
| Non-white race/ethnicity (%) | 8 (80%) | 7 (39%) | 0.055 |
|
| |||
| BMI | 27.3 (23, 30.9) | 25.7 (23.7, 27.05) | 0.281 |
|
| |||
| HCV Genotype 1 | 9 (90%) | 13 (72%) | 0.375 |
|
| |||
| IL28B CC genotype | 3 (30%) | 6 (30%) | 1 |
|
| |||
| Fibrosis | |||
| 0–2 | 7 (70%) | 10 (55.6%) | |
| 3–4 | 3 (30%) | 8 (44.4%) | 0.689 |
|
| |||
| Underlying psychiatric history | 4 (40%) | 6 (33%) | 0.725 |
|
| |||
| Baseline psychiatric meds | 2 (20%) | 4 (22%) | 1 |
|
| |||
| HCV RNA (log10 IU/mL) | 6.47 (5.97, 6.90) | 6.46 (6.25, 6.71) | 0.9 |
|
| |||
| HCV RNA >800,000 IU/mL (%) | 8 (80%) | 16 (89%) | 0.602 |
|
| |||
| CD4 cells/uL | 514 (404,700) | 606 (490, 816) | 0.194 |
|
| |||
| Baseline HIV RNA (IU/mL) | < 50 (< 50, <50) | < 50 (< 50, 699) | 0.458 |
|
| |||
| On ART | 9 (90%) | 13 (72%) | 0.375 |
|
| |||
| EFV-Based ART | 3 (30%) | 4 (22%) | 0.674 |
|
| |||
| HIV suppression | 8 (80%) | 11 (61%) | 0.417 |
|
| |||
| SVR | 0 | 8 (44.4%) | *0.025 |
Values are median (interquartile ranges) or (percentages). P values: χ2 or Mann Whitney tests
Abbreviations: BMI(basal metabolic index); HAART (Highly active antiretroviral therapy); SVR (sustained virologic response)
Differential gene expression profiles of study subjects (IFN-PE vs. IFN-NPE)
In order to identify markers that might predict IFN-PE, we analyzed the differential gene expression profiles from all 28 subjects at baseline and identified 24 genes that were differentially expressed (P <0.05; log2MFC >0.58) in the patients who experienced psychiatric toxicity after initiation of treatment compared to those who did not (Figure 1, Supplemental List 1). In patients with IFN-PE, 16 genes were expressed at higher levels and 8 genes were expressed at lower levels prior to pegIFN/RBV treatment. Interestingly, interleukin-17 receptor (IL17RA) expression was up- while interferon alpha-inducible protein 27 (IFI27) and interferon-stimulated exonuclease gene 20kDa (ISG20) were down-regulated compared to IFN-NPE.(comparative changes in the relative expression between groups)
Use of baseline gene expression profiles to predict the development of IFN-PE
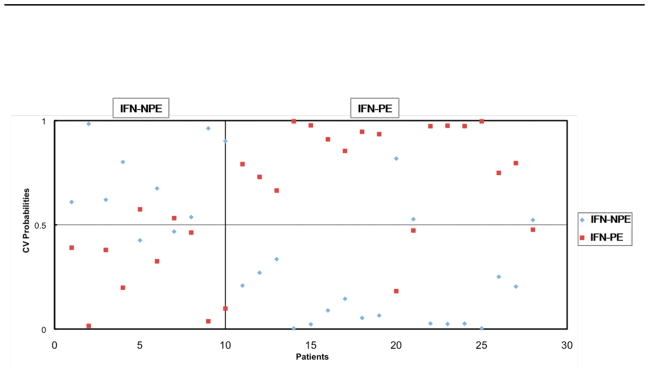
Using these 24 genes, the PAM model was able to accurately classify 23 (82%) into IFN-PE and IFN-NPE groups (Figure 2). The positive predictive value (PPV) was 84% (observed number of patients with IFN-PE out of the predicted number with IFN-PE) and the negative predictive value (NPV) was 80% (observed number of patients with IFN-NPE out of the predicted number with IFN-NPE).
Figure 2. Predictive Ability of 24 genes for psychiatric adverse events.
Based on 24 genes differentially regulated genes at baseline, prediction of psychiatric adverse events (IFN-PE) was performed with an accuracy of 23/28 (82.15%) using Prediction Analysis for Microarrays (PAM) software (version 2.0; Stanford University) in 28 patients who were about to initiate pegIFN-α2b/RBV treatment as discussed in the materials and methods section. (Those with IFN-PE are coded red while IFN-NPE is blue). Samples belonging to the known respective clinical phenotype of each patient are indicated across the upper X-axis. The 10× cross-validation probabilities of a given sample belonging to each patient clinical phenotype are indicated by the symbols on the graph: IFN-PE, development of psychiatric adverse effects vs. IFN-NPE, absence of such effects. The selected threshold was 0.05. The sum of probabilities for a given sample will equal 100%.
Identification of genes associated with emergence of psychiatric toxicity
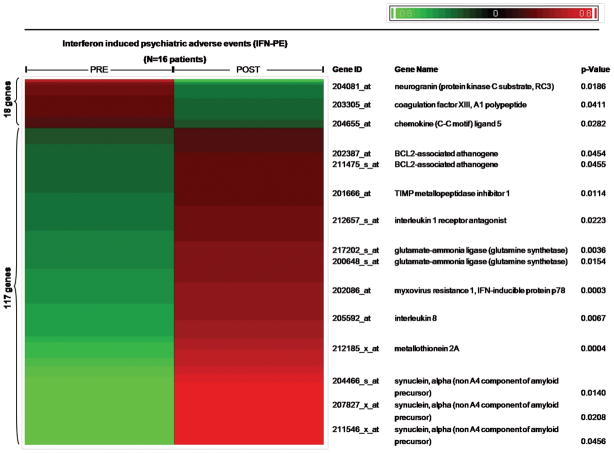
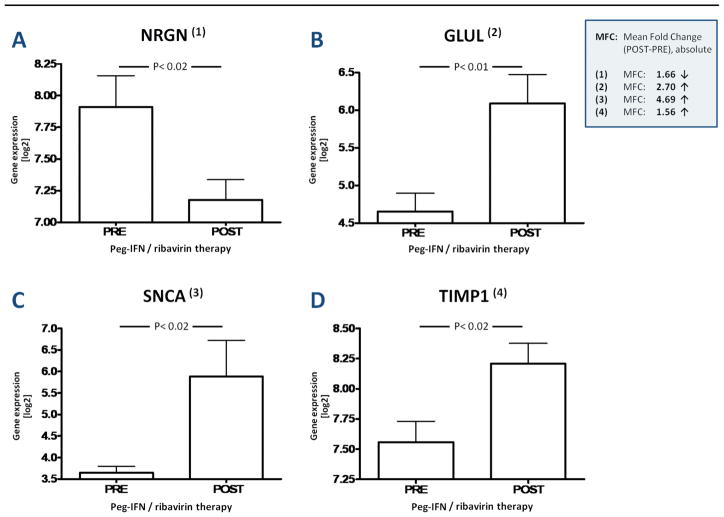
Rigorous statistical analysis of pre-to-post expression profiles identified 135 genes (P<0.05, log2MFC >0.58) that were modulated in the IFN-PE group (N=16 patients): 117 were up-regulated and 18 down-regulated (Figure 3) (Supplemental List 2). Using the same statistical cutoffs, only 32 genes were found to be significantly modulated in the IFN-NPE group (N=9 patients) (Supplemental List 3). A Venn diagram including these sets of 135 and 32 genes, respectively, revealed a subset of 13 genes that were significantly modulated (all up-regulated) in all 25 patients following pegIFN/RBV therapy; consequently, they are unlikely to be biologically relevant for the development of IFN-PE (Supplemental List 4). In contrast, 10 other genes (distinct from those 13 genes) with high degrees of both statistical and biological significance were identified by applying a systematic literature mining algorithm (DAVID 2.1) to all 135 significantly modulated genes within the IFN-PE group (Figure 3, Supplemental List 5). Of these, the expression of 3 genes (Neurogranin or Protein kinase C substrate (NRGN), Coagulation factor XIII/A1 polypeptide (F13A1), and Chemokine “C-C motif” ligand 5 (CCL5)) was down-regulated (Figure 3) in those who developed psychiatric toxicity. The expression of 7 genes (BCL2-associated athanogene (BAG1), Interleukin 1 Receptor Antagonist (IL1RN), Myxovirus resistance 1/interferon-inducible protein p78 mouse. (MX-1), Glutamate synthetase (GLUL), TIMP metallopeptidase inhibitor 1 (TIMP1), Metallothionein 2A (MT2A), and synuclein alpha (SNCA)) was induced by pegIFN/RBV in these individuals. IFN therapy resulted in the down-regulation of the expression of NRGN (P<0.02) and CCL5 (P<0.03), respectively, and in the up-regulation of the expression of MX-1 (P<0.001), MT2A (P<0.001), GLUL (P<0.01), SNCA (P<0.02) and TIMP1 (P<0.02) (Figure 4-I) which are highlighted based on existing biological significance. These genes were then validated by PCR. Thus, we identified PBMC gene expression profiles and biological pathways modulated by pegIFN/RBV therapy and specifically associated with the development of IFN-induced psychiatric toxicity in HIV/HCV co-infected patients.
Figure 3. Differential Expression of 10 important genes associated with psychiatric adverse events.
135 differentially expressed mRNAs (P <0.05; log2MFC >0.58) were identified in 16 patients who developed IFN-PE and whose samples were successfully hybridized to microarrays at the end of pegIFN-α2b/RBV treatment: 18 genes were down- and 117 genes were up regulated. Functional annotation analysis of all 135 genes by use of DAVID 2.1 resulted in the identification of 10 genes that were characterized by a high degree of biological significance for the development of neurological and/or psychiatric disorders. (Red: up-regulated; Green: down-regulated)
Supervised clustering was performed after shift-mean normalization of mean expression values for each of these 135 genes to highlight differences (across samples) in expression profiles at the end of pegIFN-α2b therapy relative to baseline.
Figure 4. I A–D: DNA Microarray Data On Selected Relevant Genes.
Significant induction (P <0.05; log2MFC >0.58**) of 4 genes by pegIFN-α2b/RBV therapy in 16 patients* that developed interferon associated psychiatric adverse effects (IFN-PE). The y-axis displays (log2) gene expression values normalized to GAPDH expression values.
* Samples of only 25 patients treated with pegIFN-α2b/RBV (16 IFN-PE; 9 IFN-NPE) were included in microarray analysis at the end of therapy vs. 28 patient samples (18 IFN-PE; 10 IFN-NPE) that were included in microarray analysis at baseline).
** Absolute log2MFC >0.58 equals an absolute MFC >1.5
Discussion
In this study, we have described a novel molecular diagnostic approach that accurately predicts the emergence of psychiatric toxicity associated with IFN therapy for chronic HCV in HIV/HCV coinfected individuals. First, these results might aid in optimizing therapy among patients undergoing HCV treatment using IFN and enhance opportunities to eradicate the virus. Second, these findings provide insights into the pathophysiology of emergent psychiatric complications with the possible association of neurogranin, glutamate synthetase, TIMP-1 and synuclein A gene expression changes with IFN-PE and thereby could potentially serve as biomarkers of IFN-PE. Earlier studies have shown that a pre-existing psychiatric diagnosis is not predictive of IFN-induced psychiatric events 11,14. Hence, there is a need to improve the prediction of which patients will develop toxicity prior to initiation of IFN therapy, thereby allowing physicians to perform risk assessments and institute pre-emptive therapy for susceptible subjects. Our previous observations have indicated that emergent serious adverse events are often seen with patients who respond virologically to pegIFN/RBV therapy, further emphasizing the potential utility of developing and employing such a tool 11.
Our study used a unique bioinformatics-based methodology to determine biomarker profiles predictive of IFN-PE in HIV-infected subjects undergoing HCV treatment with pegIFN/RBV. Baseline expression of 24 genes predicted the occurrence of IFN-PE with an accuracy of 82%, which will be invaluable for clinical purposes if confirmed in larger trials. IFN-inducible genes (IFIGs) such as ISG20 and IFI27 were among the genes down regulated in the IFN-PE group at baseline compared to IFN-NPE. We have previously reported that HIV/HCV co-infected subjects who do not respond to treatment with pegIFN/RBV have an increased basal expression of IFIGs 15 and that serious IFN-related neuropsychiatric complications may be seen among sustained viral responders (SVR) and not among non-responders (NR) to anti-HCV therapy 11. Hence, the over expression of IFIGs seen with subjects who did not experience psychiatric toxicity further suggests associations between IFIGs, poor antiviral response rates and fewer adverse events. These results provide support for the use of a novel molecular diagnostic biomarker panel that may be useful in predicting and optimizing the management of HIV-infected individuals undergoing therapy for HCV. Further validation of these gene profiles in a prospective clinical trial is necessary to establish clinical utility of this observation.
In order to identify genes that were expressed/depressed in association with incipient IFN-PE, we compared before and after therapy gene expression profiles from 25 patients and identified ten genes related to numerous CNS functions that were induced exclusively in those who experienced IFN-PE. Of those, three genes were down-regulated and eight were up-regulated by pegIFN/RBV. While all ten gene products were previously associated with the occurrence of neuropsychiatric manifestations, NRGN, GLUL and SNCA confirmed by PCR were of particular interest. In the CNS, these genes participate in pathways implicated in the pathogenesis of psychiatric disorders through alterations of serotonin, dopamine and glutamate neurotransmitter systems and/or disruption of synaptic and neuronal plasticity.
The NRGN gene product neurogranin (Ng) is a post-synaptic IQ-motif-containing protein that accelerates Ca2+ dissociation from calmodulin (CaM), a key regulator of long-term potentiation and long-term depression in CA1 pyramidal neurons 16–18. Notably, altered Ng activity mediates the effects of NMDA receptor (N-methyl D-aspartate receptor) hypofunction that has been implicated in the pathogenesis of schizophrenia 19 and also appears to have a role in mood disorders, as well, so the affective, cognitive, and perceptual symptoms in our IFN-treated patients may all have some connection to this observed genetic modulation.
Glutamine synthetase (GS) is a ubiquitous enzyme that catalyzes the conversion of ammonia and glutamate to glutamine 20. GS is expressed from the GLUL gene throughout the brain, and it plays a central role in the detoxification of ammonia and in the metabolic regulation of the neurotransmitter glutamate 21. Since glutamate is the main physiologic activator of NMDA receptors, GS modulation may be related to IFN-PE. Furthermore, since GS activity influences astrocyte function and affects neuronal activity, it is plausible that up-regulation of GS is a compensatory response in cells of monocyte/macrophage lineage following an insult, which in our study may be the effects of IFN on the brain 22,23.
The third candidate gene, alpha-synuclein (SNCA) belongs to a family of vertebrate proteins, encoded by three different genes: α, β, and γ. The protein has generated great interest in the last few years after the discovery that a mutation in the α-synuclein gene is associated with familial autosomal-dominant early-onset forms of Parkinson’s Disease 24. Previous findings have suggested that α-synuclein plays a role in neurotransmitter release and in synaptic plasticity 24. As with the example of GS, we believe that up-regulation of SNCA may be a compensatory response in cells of monocyte/macrophage lineage that might contribute to IFN-induced psychiatric toxicity 24.
Since this microarray-generated gene signature for IFN-PE was identified using PBMC samples there may be emergent conditions linked to HIV/HCV co-infection-specific pathogenesis that might allow immune cells from the periphery to gain access into the CNS and, thus express and directly secrete in the brain parenchyma neuropsychiatric-modulating gene products in response to IFN-α. In this regard, it has been shown that HIV-infection is associated with increased blood-brain-barrier (BBB) disruption and increased leukocyte penetration into the CNS mediated by increased extracellular levels of Matrix Metallopeptidase 9 (MMP-9) released from monocytes and T-cells 25–27. HIV Tat, gp120 and IFN-α have been shown to up-regulate MMP-9 expression by inducing the production of tumor necrosis factor α (TNF-α) and interleukin 1 (IL-1) in mononuclear leukocytes 27,28. These cytokines participate in pathways reportedly involved in the development of psychiatric disorders through alterations in the CNS of serotonin, dopamine and glutamate neurotransmitter systems and/or disruption of synaptic and neuronal plasticity 29,30. Thus, IFN-α induced gene products expressed and/or secreted by immune cells may have a role in the development of psychiatric disorders in HIV/HCV co-infection. This would also explain how gene products secreted in response to IFN-α might be pathogenic factors that further enhance IFN-PE in HIV/HCV-co-infected patients as compared to HCV mono-infected patients 4. In this regard, IFN is a potent inducer of IL-1 and TNF-α. These cytokines are reportedly involved in the development of psychiatric disorders through various alterations in the CNS 29,30. Given recent studies suggesting that sufficient amounts of peripherally administrated IFN can cross the human BBB 31, we suggest that gene products secreted in response to IFN-α might be a pathogenic factor that further enhances IFN-PE in HIV/HCV-co-infected patients as compared to HCV mono-infected patients.
A major limitation is that a majority of the study subjects carried a pre-existing psychiatric diagnosis. The influence of those illnesses and the medications used to treat them in this relatively small sample may have affected gene expression profiles. Specifically, about one-fifth of the patients were taking psychotropic medications at baseline (mostly SSRI antidepressants), and the majority who experienced IFN-PE had a medication change (most commonly the addition of mirtazapine or an SSRI). Although there is no known data regarding the effects of these medications on the genes identified through sampling of our patients at different stages of treatment, an interfering effect cannot be ruled out. Ideally it would be preferable to study a group of subjects without pre-morbid mental illness and then identify those who develop neuropsychiatric adverse events, which is highly improbable in clinic populations of dually infected patients. To offset potential bias, the binary classification of psychiatric toxicity was performed prior to genomic analysis in a blinded fashion with respect to viral response to treatment. Furthermore this study does not include any HCV monoinfected patients and as such the results are only applicable to HIV/HCV coinfected population. Future studies will address these concerns in ongoing clinical trials of HCV-infected subjects treated with direct acting antivirals (DAAs) in combination with pegIFN/RBV therapy.
In summary, IFN-induced neuropsychiatric adverse events are common in patients undergoing HCV treatment and threaten the prolonged course of therapy required for successful eradication of HCV infection. This study describes a novel molecular approach to aid the prediction of IFN-emergent toxicity based upon pre-treatment gene profiles. Moreover, IFN induces several candidate genes that may be causally related to the occurrence of psychiatric symptoms. These genes could serve as biomarkers or as novel therapeutic targets for IFN-associated psychiatric complications, and may elucidate more generally applicable mechanisms of psychopathology as well. Future studies will evaluate the roles of these genes and proteins in patients undergoing HCV treatment in a prospective manner.
Supplementary Material
1] Differential gene expression profiles of study subjects
2] List of 135 genes that were specifically modulated in the IFN-PE group as described in the materials and methods section.
3] List of 32 genes that were significantly modulated in the IFN-NPE group as described in the materials and methods section.
4] List of 13 genes that were significantly modulated in all patients as described in the materials and methods section.
5] List of 10 important biologic genes that were significantly modulated only in IFN-PE
Acknowledgments
R.J, K.S, P.M, R.D: study design, protocol writing, patient management and manuscript preparation. K.A, M.A, Y.J, L.R: Lab studies, data analysis and manuscript preparation. R.H, O.A, M.H, patient management and manuscript preparation
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E.
This research was supported in whole or in part by the Intramural Research Program of the NIH (National Institute of Allergy and Infectious Diseases).
Footnotes
Conflict of Interest Statement: None of the authors have any conflicts of interest to report.
Disclaimer
The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
References
- 1.Alter MJ, Kruszon-Moran D, Nainan OV, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999 Aug 19;341(8):556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 2.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with Human Immunodeficiency Virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis. 2002 Mar 15;34(6):831–837. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 3.Carrat F, Bani-Sadr F, Pol S, et al. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. Jama. 2004 Dec 15;292(23):2839–2848. doi: 10.1001/jama.292.23.2839. [DOI] [PubMed] [Google Scholar]
- 4.Laguno M, Murillas J, Blanco JL, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for treatment of HIV/HCV co-infected patients. AIDS. 2004 Sep 3;18(13):F27–36. doi: 10.1097/00002030-200409030-00003. [DOI] [PubMed] [Google Scholar]
- 5.Soriano V, Perez-Olmeda M, Rios P, Nunez M, Garcia-Samaniego J, Gonzalez-Lahoz J. Hepatitis C virus (HCV) relapses after anti-HCV therapy are more frequent in HIV-infected patients. AIDS Res Hum Retroviruses. 2004 Apr;20(4):351–353. doi: 10.1089/088922204323048096. [DOI] [PubMed] [Google Scholar]
- 6.Torriani FJ, Ribeiro RM, Gilbert TL, et al. Hepatitis C virus (HCV) and human immunodeficiency virus (HIV) dynamics during HCV treatment in HCV/HIV coinfection. J Infect Dis. 2003 Nov 15;188(10):1498–1507. doi: 10.1086/379255. [DOI] [PubMed] [Google Scholar]
- 7.Sulkowski MS, Thomas DL. Perspectives on HIV/hepatitis Cvirus co-infection, illicit drug use and mental illness. AIDS (London, England) 2005 Oct;19(Suppl 3):S8–12. doi: 10.1097/01.aids.0000192064.09281.48. [DOI] [PubMed] [Google Scholar]
- 8.Voigt E, Schulz C, Klausen G, et al. Pegylated interferon alpha-2b plus ribavirin for the treatment of chronic hepatitis C in HIV-coinfected patients. The Journal of infection. 2006 Jul;53(1):36–42. doi: 10.1016/j.jinf.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Zdilar D, Franco-Bronson K, Buchler N, Locala JA, Younossi ZM. Hepatitis C, interferon alfa, and depression. Hepatology (Baltimore, Md. 2000 Jun;31(6):1207–1211. doi: 10.1053/jhep.2000.7880. [DOI] [PubMed] [Google Scholar]
- 10.Dieperink E, Willenbring M, Ho SB. Neuropsychiatric symptoms associated with hepatitis C and interferon alpha: A review. Am J Psychiatry. 2000 Jun;157(6):867–876. doi: 10.1176/appi.ajp.157.6.867. [DOI] [PubMed] [Google Scholar]
- 11.Osinusi A, Rasimas JJ, Bishop R, et al. HIV/Hepatitis C virus-coinfected virologic responders to pegylated interferon andribavirin therapy more frequently incur interferon-related adverse events than nonresponders do. J Acquir Immune Defic Syndr. 2010 Mar 1;53(3):357–363. doi: 10.1097/QAI.0b013e3181c7a29d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnau RC, Meagher MW, Norris MP, Bramson R. Psychometric evaluation of the Beck Depression Inventory-II with primary care medical patients. Health Psychol. 2001 Mar;20(2):112–119. doi: 10.1037//0278-6133.20.2.112. [DOI] [PubMed] [Google Scholar]
- 13.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieperink E, Ho SB, Thuras P, Willenbring ML. A prospective study of neuropsychiatric symptoms associated with interferon-alpha-2b and ribavirin therapy for patients with chronic hepatitis C. Psychosomatics. 2003 Mar-Apr;44(2):104–112. doi: 10.1176/appi.psy.44.2.104. [DOI] [PubMed] [Google Scholar]
- 15.Lempicki RA, Polis MA, Yang J, et al. Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. The Journal of infectious diseases. 2006 Apr 15;193(8):1172–1177. doi: 10.1086/501365. [DOI] [PubMed] [Google Scholar]
- 16.Gaertner TR, Putkey JA, Waxham MN. RC3/Neurogranin and Ca2+/calmodulin-dependent protein kinase II produce opposing effects on the affinity of calmodulin for calcium. J Biol Chem. 2004 Sep 17;279(38):39374–39382. doi: 10.1074/jbc.M405352200. [DOI] [PubMed] [Google Scholar]
- 17.Putkey JA, Kleerekoper Q, Gaertner TR, Waxham MN. A new role for IQ motif proteins in regulating calmodulin function. J Biol Chem. 2003 Dec 12;278(50):49667–49670. doi: 10.1074/jbc.C300372200. [DOI] [PubMed] [Google Scholar]
- 18.Huang KP, Huang FL, Jager T, Li J, Reymann KG, Balschun D. Neurogranin/RC3 enhances long-term potentiation and learning by promoting calcium-mediated signaling. J Neurosci. 2004 Nov 24;24(47):10660–10669. doi: 10.1523/JNEUROSCI.2213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai G, Coyle JT. Glutamatergic mechanisms in schizophrenia. Annu Rev Pharmacol Toxicol. 2002;42:165–179. doi: 10.1146/annurev.pharmtox.42.082701.160735. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto H, Konno H, Yamamoto T, Ito K, Mizugaki M, Iwasaki Y. Glutamine synthetase of the human brain: purification and characterization. J Neurochem. 1987 Aug;49(2):603–609. doi: 10.1111/j.1471-4159.1987.tb02906.x. [DOI] [PubMed] [Google Scholar]
- 21.Khelil M, Rolland B, Fages C, Tardy M. Glutamine synthetase modulation in astrocyte cultures of different mouse brain areas. Glia. 1990;3(1):75–80. doi: 10.1002/glia.440030110. [DOI] [PubMed] [Google Scholar]
- 22.Zamora AJ, Cavanagh JB, Kyu MH. Ultrastructural responses of the astrocytes to portocaval anastomosis in the rat. J Neurol Sci. 1973 Jan;18(1):25–45. doi: 10.1016/0022-510x(73)90018-x. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto H. Hyperammonemia, increased brain neutral and aromatic amino acid levels, and encephalopathy induced by cyanide in mice. Toxicol Appl Pharmacol. 1989 Jul;99(3):415–420. doi: 10.1016/0041-008x(89)90150-6. [DOI] [PubMed] [Google Scholar]
- 24.Paxinou E, Chen Q, Weisse M, et al. Induction of alpha-synuclein aggregation by intracellular nitrative insult. J Neurosci. 2001 Oct 15;21(20):8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persidsky Y, Zheng J, Miller D, Gendelman HE. Mononuclear phagocytes mediate blood-brain barrier compromise and neuronal injury during HIV-1-associated dementia. J Leukoc Biol. 2000 Sep;68(3):413–422. [PubMed] [Google Scholar]
- 26.Ghorpade A, Persidskaia R, Suryadevara R, et al. Mononuclear phagocyte differentiation, activation, and viral infection regulate matrix metalloproteinase expression: implications for human immunodeficiency virus type 1-associated dementia. J Virol. 2001 Jul;75(14):6572–6583. doi: 10.1128/JVI.75.14.6572-6583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafrenie RM, Wahl LM, Epstein JS, Yamada KM, Dhawan S. Activation of monocytes by HIV-Tat treatment is mediated by cytokine expression. J Immunol. 1997 Oct 15;159(8):4077–4083. [PubMed] [Google Scholar]
- 28.Capuron L, Miller AH. Cytokines and psychopathology: lessons from interferon-alpha. Biol Psychiatry. 2004 Dec 1;56(11):819–824. doi: 10.1016/j.biopsych.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004 Jun 24;429(6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 30.Schaefer M, Schwaiger M, Pich M, Lieb K, Heinz A. Neurotransmitter changes by interferon-alpha and therapeutic implications. Pharmacopsychiatry. 2003 Nov;36(Suppl 3):S203–206. doi: 10.1055/s-2003-45131. [DOI] [PubMed] [Google Scholar]
- 31.Raison CL, Borisov AS, Majer M, et al. Activation of central nervous system inflammatory pathways by interferon-alpha: relationship to monoamines and depression. Biol Psychiatry. 2009 Feb 15;65(4):296–303. doi: 10.1016/j.biopsych.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1] Differential gene expression profiles of study subjects
2] List of 135 genes that were specifically modulated in the IFN-PE group as described in the materials and methods section.
3] List of 32 genes that were significantly modulated in the IFN-NPE group as described in the materials and methods section.
4] List of 13 genes that were significantly modulated in all patients as described in the materials and methods section.
5] List of 10 important biologic genes that were significantly modulated only in IFN-PE