Abstract
Summary: The database for annotation, visualization and integrated discovery (DAVID), which can be freely accessed at http://david.abcc.ncifcrf.gov/, is a web-based online bioinformatics resource that aims to provide tools for the functional interpretation of large lists of genes/proteins. It has been used by researchers from more than 5000 institutes worldwide, with a daily submission rate of ∼1200 gene lists from ∼400 unique researchers, and has been cited by more than 6000 scientific publications. However, the current web interface does not support programmatic access to DAVID, and the uniform resource locator (URL)-based application programming interface (API) has a limit on URL size and is stateless in nature as it uses URL request and response messages to communicate with the server, without keeping any state-related details. DAVID-WS (web service) has been developed to automate user tasks by providing stateful web services to access DAVID programmatically without the need for human interactions.
Availability: The web service and sample clients (written in Java, Perl, Python and Matlab) are made freely available under the DAVID License at http://david.abcc.ncifcrf.gov/content.jsp?file=WS.html.
Contact: xiaoli.jiao@nih.gov; rlempicki@nih.gov
1 INTRODUCTION
The database for annotation, visualization and integrated discovery (DAVID) bioinformatics resources (Dennis et al., 2003; Huang et al., 2009) consists of an integrated biological knowledge base (Sherman et al., 2007) and a comprehensive set of analytic tools (Hosack et al., 2003; Huang et al., 2007a, b, 2008) to extract biological themes from large gene/protein lists, such as those derived from genomic and proteomic studies. For any uploaded gene list, the DAVID bioinformatics resources provide not only the typical gene-term enrichment analysis but also tools that allow users to condense large gene lists into gene functional groups, visualize many-genes-to-many-terms relationships, cluster redundant and heterogeneous terms into groups, search for interesting and related genes or terms, dynamically view genes from their lists on biopathways and more. However, the current web interface does not support programmatic access to DAVID consequently constraining the user to the set workflows and data format options that are provided. Users have to manually upload their lists, set the background population, select the species and annotation categories and choose the tools for data analysis. The uniform resource locator (URL)-based application programming interface (API) allows users to access DAVID programmatically but because of the URL size limit and its stateless nature, users are allowed to submit only light-duty jobs (i.e. a gene list with no more than 400 genes for some browsers) and cannot change many default settings, such as species, background population, list selection, output format, etc. In a case where a user has a 1000 lists to be analyzed, each with a user-defined background, and would like to compare the output generated by DAVID. Due to the limitations of the current web interface and URL-based API, these types of tasks would not be feasible. However, the web services can be programmed to complete the task in a timely manner without human interactions. DAVID-WS is made stateful by keeping the state-related input of a user operation in a session context that can be accessed by subsequent user operations within the same session. Users can add lists, change background populations, select species and categories and reset functional parameters for data analysis, as well as query all tools within the same session and format output as desired. Our performance testing shows that it took about 6–9 s to generate the output for computationally intensive client tasks such as gene functional classification or functional annotation clustering with 2000 genes. The client code provided by DAVID-WS can be easily integrated into programs, work flows and interactive analysis tools as computational components. Compared with the current web interface and the URL-based API, DAVID-WS is a more efficient and flexible tool for using the resources of DAVID to foster user discovery.
2 SERVER IMPLEMENTATION
The implementation of DAVID-WS adheres to current W3C (http://www.w3.org/) standards including Extensible Markup Language (XML)-based languages such as Simple Object Access Protocol (SOAP) and Web Service Definition Language (WSDL).
The server is implemented in Java using Apache Axis2 (http://axis.apache.org/axis2/java/core) to call DAVID functional modules, such as gene functional classification, functional annotation clustering, functional annotation chart, functional annotation table, etc. (Fig. 1). More than 20 operations are currently available on the web page and can be categorized into four groups, as follows:
Add List: gene list/background population submission;
Lookup: check available conversion types, backgrounds, categories, species, tool options, etc.;
Select: select background populations, categories and species;
Report: generate output from DAVID modules/tools.
Axis2 supports four types of service scopes. They are request, transport session, SOAP session and application. DAVID-WS is configured to be stateful at the transport session level. When a client establishes a connection to the service, the server will create a new session for the client. The state-related input, such as the current list, species, backgrounds and annotation categories, can be stored in the session context and used by proceeding operations within the same session. Users can select and group operations into workflows and execute them in batch to automate analysis tasks. In Fig. 1, a typical DAVID-WS client can go through the following: (1) add gene/protein list; (2) lookup background/species/categories; (3) select background/species/categories and (4) get reports.
Fig. 1.
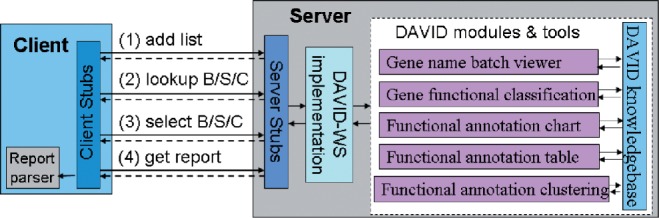
DAVID web services invocation
3 SAMPLE CLIENTS
Four light-weight ready-to-use clients (Java, Perl, Python and Matlab) are provided for users to connect to the server and consume the web services. A list of gene identifiers is the only input required from users in most situations. However, multiple lists can be supplied for more complex analysis tasks and users can reset the argument values in many ways, including changing the background, selecting categories and species or something similar. In addition, the Java and Perl clients were equipped with parsers to transform the Axis2 output into tab-delimited text format for the following reports, which are requested most often by users:
Functional annotation chart report.
Functional annotation table report.
Gene functional classification report.
Functional annotation clustering report.
The client code, with easy-to-follow instructions, can be freely downloaded from the web site. Users can simply replace the example gene list in the client code with their own gene lists to start analysis. The same example gene list was used in Huang et al. (2009) to illustrate the use of DAVID for browser clients. For more information about the client-generated reports and the usage of DAVID modules/tools, users may refer to Huang et al. (2009). Users may also enlist other programming languages and platforms to write their own client-side applications to send input and receive output in XML through Representational State Transfer, XML Remote Procedure Call or SOAP.
4 PERFORMANCE TESTING
We used Apache Jmeter (http://jakarta.apache.org/jmeter/) to configure test plans and perform load testing for DAVID-WS. A number of threads were started to simulate multiple users sending requests to the server, and the time each task consumes was documented and analyzed. We used a PC with 3 GB of memory, a 3.16-GHz CPU and a 100-Mbps internet connection and a laptop with 1.5 GB of memory, a 1.86-GHz CPU and a 54-Mbps WiFi Internet connection both with Windows XP operating systems to run Jmeter at different time schedules including normal working hours and off hours. Our simulation and performance testing showed that it took 1–3 s to generate the output for a typical gene list analysis task with l<1000 genes. For the most computationally intensive tasks, such as gene functional classification and functional annotation clustering with >2000 genes, the reports were generated and received by clients in 6–9 s with settings of 5–20 users/threads in the system. The performance shown by these results is quite acceptable for users and does not cause a heavy load on the DAVID server.
ACKNOWLEDGEMENTS
We thank our users, especially those who contributed their comments and questions to improve DAVID-WS during the beta testing.
Funding: This project has been funded by the National Institute of Allergy and Infectious Diseases (NIAID), National institutes of Health (NIH), under Contract No. NOI-CO-56000. The contents do not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Conflict of Interest: none declared.
Footnotes
† The authors wish it to be known that, in their opinion, the first 2 authors should be regarded as joint First Authors.
REFERENCES
- Dennis G., Jr., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Hosack D.A., et al. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007a;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007b;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., et al. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Sherman B.T., et al. DAVID knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]


