Abstract
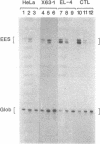
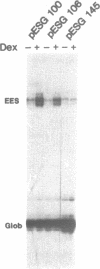
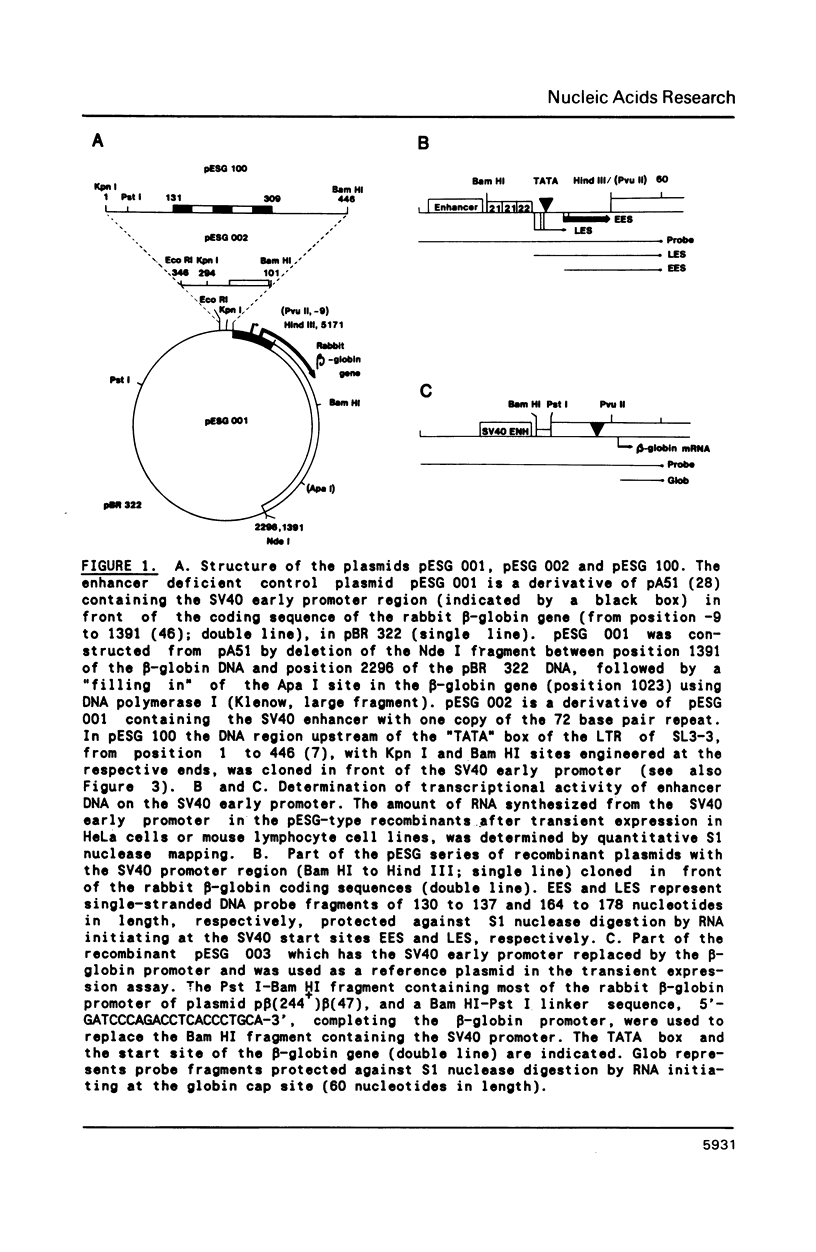
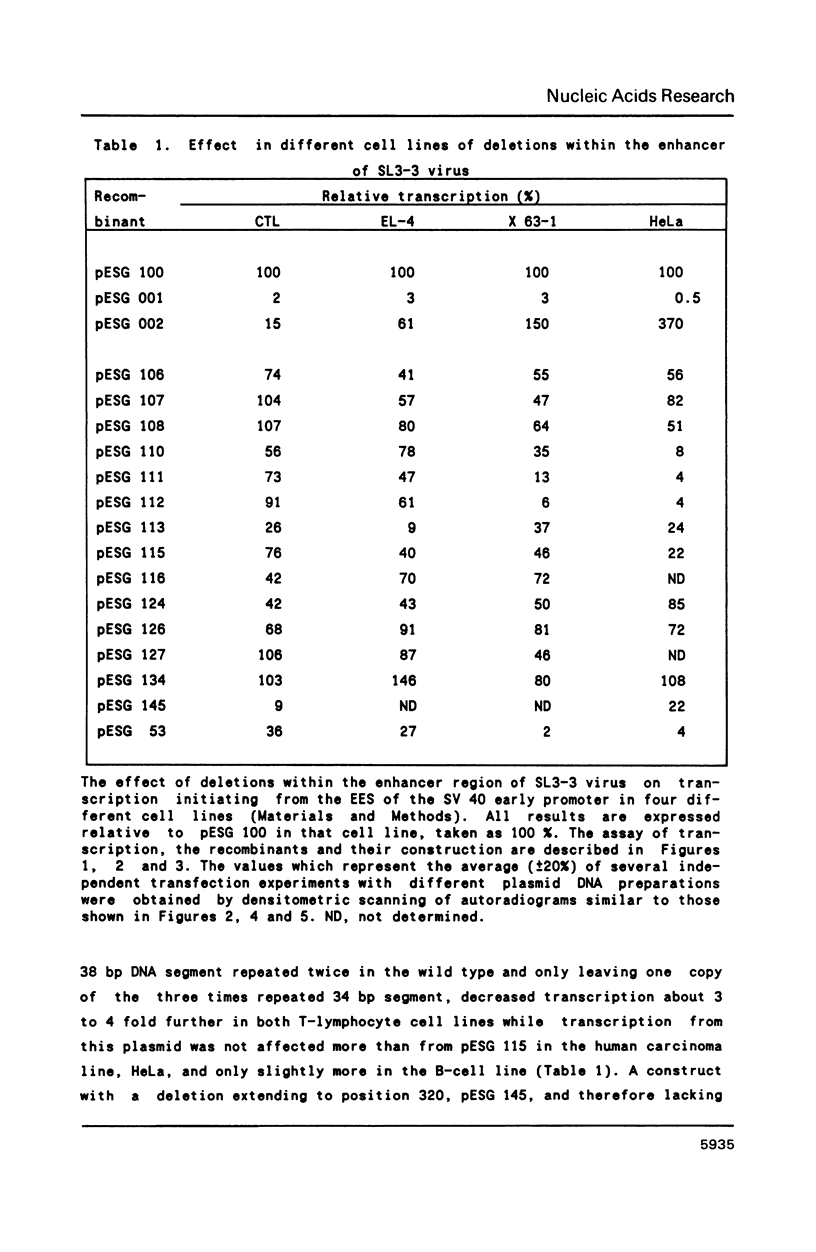
The long terminal repeat (LTR) of the retrovirus SL3-3 determines its tropism for T-lymphocytes and its ability to induce T-cell lymphomas in mice. We have studied the ability of different DNA sequences located upstream of the "TATA" box in the LTR of SL3-3 to enhance transcription in T-lymphocyte cell lines and other cell lines, employing a transient assay and quantitative S1 nuclease mapping. The enhancer was found to be composed of many DNA domains which determines different activities in different cell lines. We find enhancer sequence motifs with a high T-lymphocyte specificity in the DNA repetitions of the LTR, and other enhancer motifs active in a broader range of cells in the surrounding DNA segments. The localization of sequences preferentially active in T-cells within the repeated sequences containing differences between SL3-3 and the very closely related Akv virus, which is without the T cell tropism and leukaemogenicity of SL3-3, supports the notion that the enhancer sequence motifs with T-cell preferences are primary determinants of these properties.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Baker P. E., Gillis S., Smith K. A. Monoclonal cytolytic T-cell lines. J Exp Med. 1979 Jan 1;149(1):273–278. doi: 10.1084/jem.149.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Baty D., Barrera-Saldana H. A., Everett R. D., Vigneron M., Chambon P. Mutational dissection of the 21 bp repeat region of the SV40 early promoter reveals that it contains overlapping elements of the early-early and late-early promoters. Nucleic Acids Res. 1984 Jan 25;12(2):915–932. doi: 10.1093/nar/12.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Glucocorticoid regulation of murine leukemia virus transcription elements is specified by determinants within the viral enhancer region. J Virol. 1987 Feb;61(2):269–275. doi: 10.1128/jvi.61.2.269-275.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- GORER P. A. Studies in antibody response of mice to tumour inoculation. Br J Cancer. 1950 Dec;4(4):372–379. doi: 10.1038/bjc.1950.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. V., Bich-Thuy L. T., Stafford J., Queen C. Synergism between immunoglobulin enhancers and promoters. Nature. 1986 Jul 24;322(6077):383–385. doi: 10.1038/322383a0. [DOI] [PubMed] [Google Scholar]
- Gronostajski R. M., Adhya S., Nagata K., Guggenheimer R. A., Hurwitz J. Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol Cell Biol. 1985 May;5(5):964–971. doi: 10.1128/mcb.5.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundström T., Zenke W. M., Wintzerith M., Matthes H. W., Staub A., Chambon P. Oligonucleotide-directed mutagenesis by microscale 'shot-gun' gene synthesis. Nucleic Acids Res. 1985 May 10;13(9):3305–3316. doi: 10.1093/nar/13.9.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Jantzen H. M., Strähle U., Gloss B., Stewart F., Schmid W., Boshart M., Miksicek R., Schütz G. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987 Apr 10;49(1):29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Kappler J. W., Skidmore B., White J., Marrack P. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med. 1981 May 1;153(5):1198–1214. doi: 10.1084/jem.153.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney J. F., Radbruch A., Liesegang B., Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979 Oct;123(4):1548–1550. [PubMed] [Google Scholar]
- Kieny M. P., Lathe R., Lecocq J. P. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983 Dec;26(1):91–99. doi: 10.1016/0378-1119(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Lenz J., Crowther R., Klimenko S., Haseltine W. Molecular cloning of a highly leukemogenic, ecotropic retrovirus from an AKR mouse. J Virol. 1982 Sep;43(3):943–951. doi: 10.1128/jvi.43.3.943-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Haseltine W. A. Localization of the leukemogenic determinants of SL3-3, an ecotropic, XC-positive murine leukemia virus of AKR mouse origin. J Virol. 1983 Aug;47(2):317–328. doi: 10.1128/jvi.47.2.317-328.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Matthes H. W., Staub A., Chambon P. The segmented paper method: DNA synthesis and mutagenesis by rapid microscale "shotgun gene synthesis". Methods Enzymol. 1987;154:250–287. doi: 10.1016/0076-6879(87)54080-0. [DOI] [PubMed] [Google Scholar]
- Matthes H. W., Zenke W. M., Grundström T., Staub A., Wintzerith M., Chambon P. Simultaneous rapid chemical synthesis of over one hundred oligonucleotides on a microscale. EMBO J. 1984 Apr;3(4):801–805. doi: 10.1002/j.1460-2075.1984.tb01888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Mohit B., Fan K. Hybrid cell line from a cloned immunoglobulin-producing mouse myeloma and a nonproducing mouse lymphoma. Science. 1971 Jan 8;171(3966):75–77. doi: 10.1126/science.171.3966.75. [DOI] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Nowock J., Borgmeyer U., Püschel A. W., Rupp R. A., Sippel A. E. The TGGCA protein binds to the MMTV-LTR, the adenovirus origin of replication, and the BK virus enhancer. Nucleic Acids Res. 1985 Mar 25;13(6):2045–2061. doi: 10.1093/nar/13.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen F. S., Crowther R. L., Tenney D. Y., Reimold A. M., Haseltine W. A. Novel leukaemogenic retroviruses isolated from cell line derived from spontaneous AKR tumour. Nature. 1981 Jul 9;292(5819):167–170. doi: 10.1038/292167a0. [DOI] [PubMed] [Google Scholar]
- Robb R. J., Munck A., Smith K. A. T cell growth factor receptors. Quantitation, specificity, and biological relevance. J Exp Med. 1981 Nov 1;154(5):1455–1474. doi: 10.1084/jem.154.5.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Haseltine W. A., Lenz J., Ruprecht R., Cloyd M. W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985 Sep;55(3):862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C., Westphal H. M., Carlson C., Bosshard H., Beato M. Molecular model of the interaction between the glucocorticoid receptor and the regulatory elements of inducible genes. DNA. 1986 Oct;5(5):383–391. doi: 10.1089/dna.1986.5.383. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Vigneron M., Matthes H., Wildeman A., Zenke M., Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986 Jan 9;319(6049):121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Matthes H., Wintzerith M., Chambon P. Transcription from the SV40 early-early and late-early overlapping promoters in the absence of DNA replication. EMBO J. 1983;2(9):1605–1611. doi: 10.1002/j.1460-2075.1983.tb01631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooyen A., van den Berg J., Mantei N., Weissmann C. Comparison of total sequence of a cloned rabbit beta-globin gene and its flanking regions with a homologous mouse sequence. Science. 1979 Oct 19;206(4416):337–344. doi: 10.1126/science.482942. [DOI] [PubMed] [Google Scholar]