Background: The Na+/H+ antiporter NhaA undergoes conformational changes, with only a pH-locked, cytoplasmic-facing structure available.
Results: Based on pseudo-symmetry in the crystal structure, we modeled a periplasmic-facing conformation, supported by normal mode analysis and cross-linking experiments.
Conclusion: The model appears to represent an elevated pH conformation, revealing novel features of cation transport.
Significance: The periplasm-facing conformation is useful for understanding homologous human transporters.
Keywords: Computer Modeling, Protein Dynamics, Protein Structure, Sodium Proton Exchange, Transporters, NhaA, Cation Proton Antiporters, Ion Transport, Model-Structure, Normal Mode Analysis
Abstract
The Escherichia coli NhaA antiporter couples the transport of H+ and Na+ (or Li+) ions to maintain the proper pH range and Na+ concentration in cells. A crystal structure of NhaA, solved at pH 4, comprises 12 transmembrane helices (TMs), arranged in two domains, with a large cytoplasm-facing funnel and a smaller periplasm-facing funnel. NhaA undergoes conformational changes, e.g. after pH elevation to alkaline ranges, and we used two computational approaches to explore them. On the basis of pseudo-symmetric features of the crystal structure, we predicted the structural architecture of an alternate, periplasm-facing state. In contrast to the crystal structure, the model presents a closed cytoplasmic funnel, and a periplasmic funnel of greater volume. To examine the transporter functional direction of motion, we conducted elastic network analysis of the crystal structure and detected two main normal modes of motion. Notably, both analyses predicted similar trends of conformational changes, consisting of an overall rotational motion of the two domains around a putative symmetry axis at the funnel centers, perpendicular to the membrane plane. This motion, along with conformational changes within specific helices, resulted in closure at the cytoplasmic end and opening at the periplasmic end. Cross-linking experiments, performed between segments on opposite sides of the cytoplasmic funnel, revealed pH-dependent interactions consistent with the proposed conformational changes. We suggest that the model-structure and predicted motion represent alkaline pH-induced conformational changes, mediated by a cluster of evolutionarily conserved, titratable residues, at the cytoplasmic ends of TMs II, V, and IX.
Introduction
Conserved Na+/H+ antiporters preserve a delicate balance of sodium concentration and pH conditions in cells in all organisms (1). In Escherichia coli, the Na+/H+ transporter NhaA is responsible for the maintenance of this basic homeostasis, coupling the antiport of H+ with that of Na+ (2, 3). Studies of bacterial NhaA enhance our understanding of the human transporters to which they are evolutionarily related (4–7).
The crystal structure of NhaA has been determined at pH 4, facilitating the molecular level interpretation of preceding biochemical data (Fig. 1C) (8). This cytoplasm-facing structure contains 12 transmembrane helices (TMs)2 forming a large cytoplasmic funnel and a small periplasmic funnel. NhaA is made of two TM domains containing pseudo-symmetric segments (8, 9): the first domain consists of TMs III-V and X-XII, and the second domain consists of TMs I-II and TMs VI-IX. The two opposite-topology TMs IV and XI, referred to as the TM IV-XI assembly, are interrupted by unwound segments that cross each other at the membrane midplane. Notably, Asp-163 and Asp-164 of TM V face the TM IV-XI assembly and have been implicated in cation and proton binding (8, 10–12).
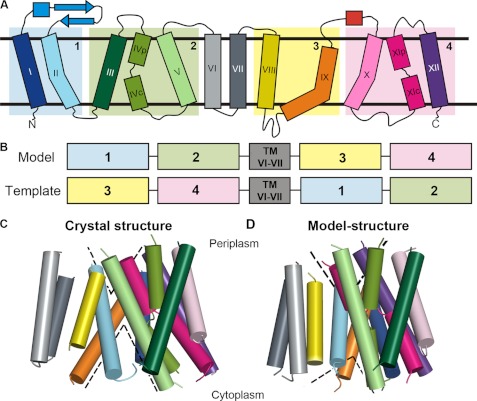
FIGURE 1.
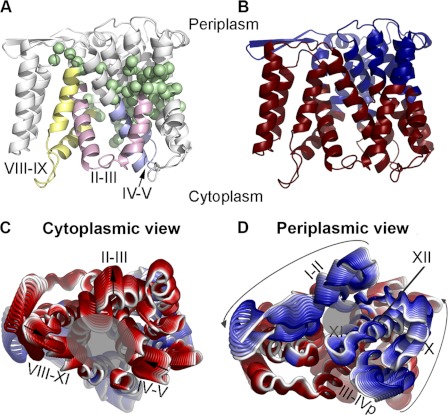
Modeling of a periplasm-facing state. A, schematic representation of the NhaA structure (8). Helices are marked with rectangles, and β-strands by arrows. The two pairs of pseudo-symmetric regions are shaded and marked by numbers. B, scheme of the homology modeling approach, displaying the fragments used as templates. C and D, side views of the crystal structure and periplasm-facing model. Both models are shown in cylinder representation, and colored according to panel A. The loops were omitted for clarity. The dashed lines mark the approximate locations of the cytoplasmic and periplasmic funnels.
In physiological conditions, the antiporter is pH-dependent, active at alkaline pH (pH 6.5–8.5) but not at acidic pH (1, 2), with many indications of pH-induced conformational changes (13–20). In particular, cryo-electron microscopy (cryoEM) difference maps have shown two distinct conformational changes: the first occurs in the funnel region in response to pH elevation, and the second was observed in the TM IV-IX assembly upon addition of Na+ or Li+ at alkaline pH conditions (17). Recently, site-directed tryptophan fluorescence revealed a pH-induced conformational change at F136W (helix IVc) and a ligand-induced conformational change at F339W (in the unwound segment of TM XI) at alkaline pH (21). These important data, however, cannot reveal the molecular events leading to the conformational changes. Here we try to fill this gap with the aid of two computational modeling techniques combined with biochemical cross-linking experiments.
Forrest et al. (22) previously utilized pseudo-symmetry in the outward-facing conformation of the LeuT transporter to model the transporter's inward-facing conformation; their results were supported by structural data (23). The approach was successfully applied to three other transporters with distinct folds (24–26). Following the same computational approach (22, 24, 25), we modeled a periplasmic-open state of NhaA based on the pseudo-symmetry in the low pH structure. We also studied the normal modes of motion in NhaA using elastic network models, an effective technique to elucidate functional dynamics of proteins (27), including transporters (28). By depicting the protein structure as a network of springs, the Gaussian Network Model (GNM) (29, 30) and Anisotropic Network Model (ANM) (31) can be used to explore the relative magnitudes and directions of slow and/or large-scale modes of motion.
Encouragingly, both computational approaches predicted similar structural transitions, further supported by biochemical cross-linking experiments that we performed. These data allowed us to provide an atomistic description of pH-induced conformational-changes in the NhaA antiporter within the context of an alternating-access mechanism of transport, in which a single binding site is alternately exposed to either side of the membrane (32, 33).
EXPERIMENTAL PROCEDURES
Structural Modeling
We obtained a sequence alignment between TMs III-V and TMs X-XII from a structural alignment via SKA (34), and manually aligned the sequences of TMs I-II and TMs VIII-IX. The conformation of TM VI-VII in the model was retained from the crystal structure. We adjusted the sequence alignment to obtain a model that is consistent with two generic traits of membrane proteins (35): the hydrophobicity pattern (36, 37) and evolutionary conservation profile (38, 39), that is, hydrophobic and variable positions face the lipids, while conserved and/or strongly hydrophilic residues are embedded in the interior, mediating helix-helix and substrate interactions (supplemental Figs. S1 and S9). Constraints were applied to maintain the helicity of the helical segments of the crystal structure. We selected the lowest energy model from 2,000 candidate conformations, generated using Modeler, version 9v8, which uses the CHARMM22 force field (40).
Normal Mode Analysis using Elastic Network Models
In GNM and ANM (27, 29, 31, 41), the collective modes of motion are determined by the particular structural topology, represented by a contact matrix. The fluctuations of each residue and their inter-correlations in the slowest modes were obtained from GNM calculations, performed with an in-house program with the commonly used cutoff of 10 Å to indicate contact between Cα atoms. To predict the direction of the motion, we carried out ANM computations via the HingeProt webserver using the default cutoff of 15Å (42). We matched the corresponding GNM and ANM slow modes based on their mean-square fluctuations, to associate the size and direction of detected motions (supplemental Fig. S6). More details are available as supplemental data.
Intra-molecular Cross-linking at the Cytoplasmic End of NhaA in Situ
Site-directed intramolecular cross-linking was conducted in situ on membrane vesicles isolated from TA16 (43) cells expressing the various NhaA double-Cys replacements, as previously described (44). Membranes (2 mg of protein) were resuspended in a buffer (3 ml) containing one of the following homo-bifunctional cross-linkers, freshly prepared: 2 mm BMH (1,6-bis-maleimidohexane) or 1 mm pPDM (N,N′-p-phenylenedimaleimide) and processed at pH 6.5. Mutants expressing the double Cys replacements L138C/H256C and A142C/H256C were processed at pH 8.5 as well, whereas mutants expressing S146C/E252C were processed also at pH 9. The affinity-purified protein was treated with trypsin at pH 9 and resolved on SDS-PAGE to identify the proteolytic products according to (45). Because the only trypsin-cleavable site (Lys-249) is located between the two Cys replacements, trypsinolysis of untreated samples results in two tryptic peptides with faster mobility than that of the intact protein (17.24 and 32.5 kDa, designated high frequency and low frequency, respectively). On the other hand, intramolecular cross-linking results in one fragment whose mobility is equal to that of the intact protein (46). Each experiment was repeated at least twice with practically identical results. Notably, there was no intermolecular cross-linking that could have been recognized by a band of a mobility of the NhaA dimer.
RESULTS
Modeling the Periplasm-facing State of NhaA
In accordance with prior studies (8, 9), we postulated that NhaA consists of two pairs of pseudo-symmetric segments (Fig. 1A). This type of structural relationship can be used for modeling an alternate state if there is a distinct difference between the internal conformations of the pseudo-symmetric segments in the known state (22, 24, 25). While TMs III-V and TMs X-XII superimposed very well (Figs. 1A and supplemental Fig. S1A, referred to as segments 2 and 4), superimposing TMs I-II and TMs VIII-IX (Fig. 1A, segments 1 and 3, respectively) with the SKA program did not reveal a structural relationship suitable for modeling an alternate state (supplemental Fig. S1B). However, the comparison did reveal an intriguing difference; specifically, although TM II and TM IX are approximately of the same length, there is a difference in their conformations: TM II is only slightly kinked, while TM IX is more banana-shaped (Figs. 1A and supplemental Fig. S1D). Since in the crystal structure TMs II and IX line the cytoplasmic funnel, and TM II also contributes to the periplasmic funnel (8, 19), we reasoned that this conformational difference might contribute to the fact that in the crystal structure the funnel in the cytoplasmic side is larger than that in the periplasmic side. We also considered TMs I-II and III-V as one non-contiguous structural repeat, and TMs VIII-IX and X-XII as a second structural repeat, with each repeat containing five TM helices. It can be seen that these two repeats differ in the relative translation of the first two helices of each repeat with respect to the last three helices of the repeat (supplemental Fig. S1C).
In line with the aforementioned hypothesis, we were able to obtain a model-structure of NhaA in a distinct conformation by swapping the conformations of the structural repeats (Figs. 1B and supplemental Fig. S2). The two helical domains, the first consisting of TMs I, II, VIII, and IX and the second bundle including TMs III, IV, V, X, XI, and XII, are rotated relative to one another in comparison to their positioning in the crystal structure. This reflects the different relative positions of segments 1 or 3 with respect to segments 2 or 4, respectively (Figs. 1B and 2). This relative rotational change can be most clearly seen when superposing the two structures using only segments 2 and 4 (supplemental Fig. S3C) or only segments 1 and 3 (supplemental Fig. S3B). The remaining peripheral TMs VI and VII retained the same conformation in the model as in the crystal structure, and their exact positioning in the model is of lower confidence (supplemental text).
FIGURE 2.
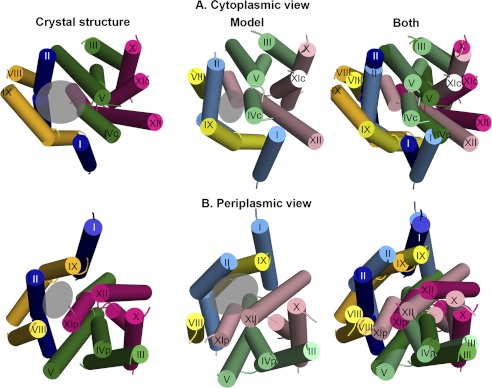
The crystal structure versus periplasm-facing model. The crystal structure and model are shown as cylinders, with the loops and TMs VI and VII excluded, and helical segments marked. The crystal and model are colored according to segments 1–4, specified in Fig. 1B, with the crystal colored in darker shades than the model. The approximate locations of the funnels are marked with transparent gray circles. Panels A and B show the crystal structure, model, and a superimposition of both from left to right, from the cytoplasm (A) and the periplasm (B). The crystal structure shows a large cytoplasmic funnel, whereas in the model the cytoplasmic funnel is closed, and the periplasmic funnel is more open. In addition, the model displays enlarged cytoplasmic and periplasmic entries to the TM IV-XI assembly (Fig. 3).
The primary consequence of the rotation of the two domains, and of the more local changes in the conformation of several helices, is that, in comparison to the crystal structure, the model is more open to the periplasm, whereas its cytoplasmic funnel is smaller in volume (Figs. 1, C and D and 2). In both conformations, the cytoplasmic funnel is lined by TMs II, IVc, V, and IX (Fig. 2A). In the model-structure, the domain rotation changed the positions of these TM segments relative to each other; this in turn results in closure of the cytoplasmic funnel. In particular, the cytoplasmic ends of TM II and TM IX pack up against IVc, displacing TM I, which now also packs against TM XII (Fig. 2A). Bending of TM II and straightening of TM IX may also contribute to the closure (Fig. 3). Interestingly, we also observe a decrease in the tilt of TMs IVc and V relative to the membrane normal, concurrent with an increased tilt of TM XII. As a result, a separate pathway from the cytoplasmic funnel on the cytoplasmic side, leading to the unwound region of the TM IV-XI assembly, is enlarged because of an increase in the distance between the cytoplasmic end of TM XII and those of TMs IVc and V (Figs. 2A and 3A). Notably, as TMs IV and V line both the cytoplasmic funnel and the pathway to the TM IV-XI assembly, the change in their relative orientations contributes to the closure of the funnel and also to the enlargement of the pathway at the model's cytoplasmic end.
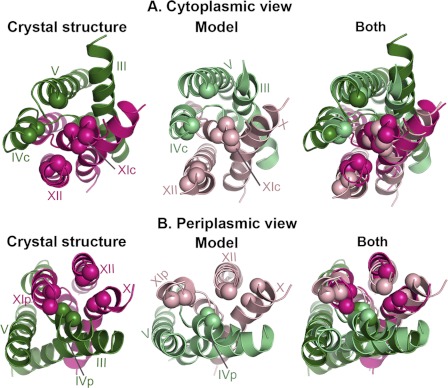
FIGURE 3.
Enlarged pathways to the TM IV-XI assembly. In all panels, the crystal structure, periplasm-facing model and their superimposition are shown in cartoon representation and colored as in Fig. 2, and TM helices are labeled. For clarity, only TMs III-V and TMs X-XII are displayed. In both model and crystal structure, Cα atoms of equivalent positions lining the cytoplasmic and periplasmic ends of the pathways to the TM IV-XI assembly are shown as spheres, demonstrating the changes in the pathways' conformations. It is apparent that both pathways are enlarged in the model compared with the crystal. A, cytoplasmic view. B, periplasmic view.
On the periplasmic side, the funnel (Fig. 2B) expands due to the rotation of the domains and the altered conformations of TM II and TM IX in the model (supplemental Fig. S1D). The periplasmic funnel, lined mainly by TMs II and XIp in the crystal structure, is formed between TMs II, XIp, IX and XII in the model (Figs. 2B and 3B). In addition, a separate pathway to the periplasm, located within the TM VI-XI assembly, is enlarged somewhat by separation of TMs IVp and XIp (Figs. 2B and 3B). Since it lines both the periplasmic funnel and the putative pathway to the TM IV-XI assembly, TM XIp mediates both the opening at the periplasmic funnel and the expansion of the pathway toward the assembly.
Normal Mode Analysis of the Crystal Structure
We investigated the global, cooperative (correlated) dynamics of the crystal structure using normal mode analysis. We focused on the two slowest GNM modes, i.e. those with the most significant contribution to the overall motion (supplemental Fig. S4), and obtained the mean-square fluctuations of the residues and inter-residue cooperative dynamics. The directions of these motions were inferred from the corresponding ANM modes (supplemental Fig. S5).
The Slowest Mode of Motion
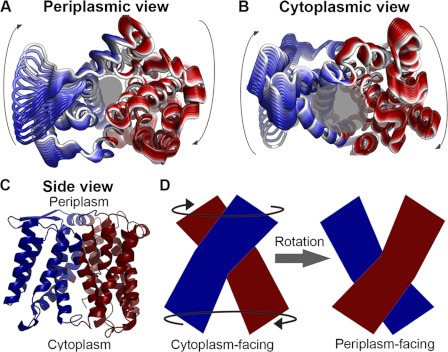
The slowest GNM mode (GNM1) revealed hinges that reside at, or in the vicinity of, evolutionarily conserved and functional residues, (supplemental Fig. S6A and Table S1), as is typical of hinges (47). Almost all the detected hinges were located along the putative axis that separates NhaA into its two structural domains (supplemental Fig. S6B). The GNM cooperative dynamics were concentrated around hinges situated at “inter-domain” connection points. The proposed motions within each helical bundle were positively correlated, while being anti-correlated to the proposed motions of the other bundle (Fig. 4C). The corresponding ANM mode (ANM1, supplemental Fig. S5A) manifested a rotational or twisting motion of the domains relative to one another, around the central axis (Fig. 4, A and B and supplemental Movie S1). These deformations altered the general shape of the two funnels, while preserving their volume. Accordingly, residues that originally lined the pore still retained their roles, albeit changed their exact positioning around the funnels.
FIGURE 4.
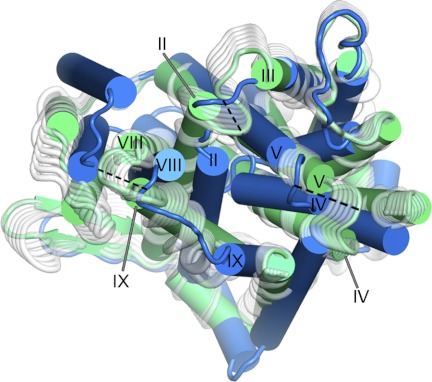
The slowest mode (GNM1/ANM1). Panels A and B, fluctuations predicted by ANM1, associated with GNM1 (supplemental Fig. S5), mapped onto the structure and viewed from the periplasm (A) or cytoplasm (B). The structural changes of ANM1 are colored in shades of red and blue according to their GNM1 dynamical correlation, with the direction of motion indicated by paling of the color tone from dark red/blue to white, and arrows for the general direction of motion. Transparent gray circles mark the approximated funnels, as in Fig. 2. C, inter-residue correlated dynamics of GNM1 mapped onto the NhaA crystal structure. The dynamics of the two separate helical bundles are oppositely correlated, with red and blue indicating opposite correlations. D, suggested implication of this motion. Schematic side views of NhaA in the cytoplasm-facing and periplasm-facing states, with arrows marking the direction of motion depicted by ANM1 (A and B, supplemental Movie S1). The rotational motion described by ANM1 could potentially contribute to alternating-access via separate yet simultaneous rotations of the two distinct helical bundles, colored red and blue as in panel C.
The Second Slowest Mode
The hinge regions in the second slowest GNM mode (GNM2, supplemental Fig. S6C) were also enriched with highly conserved residues that are important for function (supplemental Table S1). The hinges in the TM IV-XI assembly and flanking helices, along with TM IX, roughly in the membrane midplane, separate these TM segments into cytoplasmic and periplasmic rigid parts (Fig. 5A). As the modes are orthogonal, independent of one another (27, 29, 31, 41), the hinge plane in GNM2 (Fig. 5A) is distinct from that of GNM1 (supplemental Fig. S6A). The dynamics of the cytoplasmic rigid parts were positively correlated, while anti-correlated with the dynamics of the periplasmic ends (Fig. 5B). Three positively correlated, highly fluctuating, rigid segments lined the cytoplasmic funnel; these are the cytoplasmic ends of TMs II-III, TMs IVc-V, and TMs VIII-IX (Fig. 5, A and B). In the corresponding ANM2 mode (supplemental Fig. S5B) the TMs II-III segment moved away from the center of the funnel, and the cytoplasmic ends of TMs IVc-V and TMs VIII-IX approached one another at the center of the funnel (Fig. 5, C and D, supplemental Movies S2 and S3).
FIGURE 5.
The second-slowest mode (GNM2/ANM2). A, side view of the NhaA crystal structure in cartoon presentation, with the Cα atoms of the GNM2 hinge regions (supplemental Fig. S6C) shown as green spheres. Rigid segments lining the cytoplasmic funnel, consisting of parts of TMs II-III, TMs IV-V, and TMs VIII-IX, are colored pink, purple, and yellow, respectively. B, same as in panel A, with the structure colored according to the cooperative dynamics of GNM2, as in Fig. 4C. Panels C and D, ANM2 fluctuations, corresponding to GNM2 (supplemental Fig. S5), viewed from the cytoplasm and periplasm, respectively, with transparent gray circles indicating the funnels. The fluctuations are colored in tones of red and blue according to their cross-correlation values (B), as described for Fig. 4A. The directions of motion are marked with arrows, and the TM segments of interest specified with roman letters.
Agreement between the Two Computational Approaches
By superimposing the crystal structure and the periplasm-facing model, we were able to derive a rough estimate of the conformational changes leading to the transition between them (Fig. 2). We compared these trends of motion to the two slow modes of motion, keeping in mind that normal mode analysis is capable of revealing the relative magnitude of the fluctuations and their directions but not their precise amplitudes (31). Remarkably, we detected an overall qualitative agreement between our two independent computational results (Figs. 6, supplemental Fig. S7 and Movies S4–S6). According to both methods, the cytoplasmic ends of TMs IV-V and TMs VIII-IX approach each other near the center of the cytoplasmic funnel, effectively closing it off (Figs. 2A, 5C, and 6 and supplemental Movie S4). Notably, straightening of TM IX was predicted by both the second mode of motion and the periplasm-facing model (supplemental Fig. S7). Likewise, a rotational motion suggested by ANM1, which could be described as twisting around an axis running through the two funnels and separating the two domains (Fig. 4, A and B and supplemental Movie S1), can also be detected when examining the transition between the crystal structure and model (Fig. 2 and supplemental Movies S5 and S6). Still, two features of the periplasmic-facing model were not suggested by the normal mode analysis, namely, opening of the periplasmic funnel, and moderate opening at the cytoplasmic and periplasmic entrances of TMs IV-XI (Figs. 2 and 3). These differences may be attributed to the limitations of normal mode analysis, discussed below.
FIGURE 6.
Comparison of ANM2 to the transition between the crystal and model structures. Periplasmic view of the crystal structure and periplasm-facing model, shown in cartoon representation and colored green and blue, respectively. ANM2 fluctuations are displayed as white, semi-transparent, cartoons. Only one of the two directions of ANM motion is shown. The dashed lines indicate the transition between the crystal- and model-structure in helices of interest.
Biochemical Study of Conformational Changes in Situ
Tests of accessibility to various SH-reagents of single Cys replacements and cross-linking between double Cys replacements, as a function of changes of various conditions, are widely-used to identify conformational changes of membrane proteins in situ (48, 49). For NhaA, such tests have been conducted in the physiological pH range (pH 6.5–8.5) when NhaA is activated, measuring response to pH level and/or different ligand concentrations, and the results have been compared with the pH 4 crystal structure.
Accessibility Measurements as a Function of pH
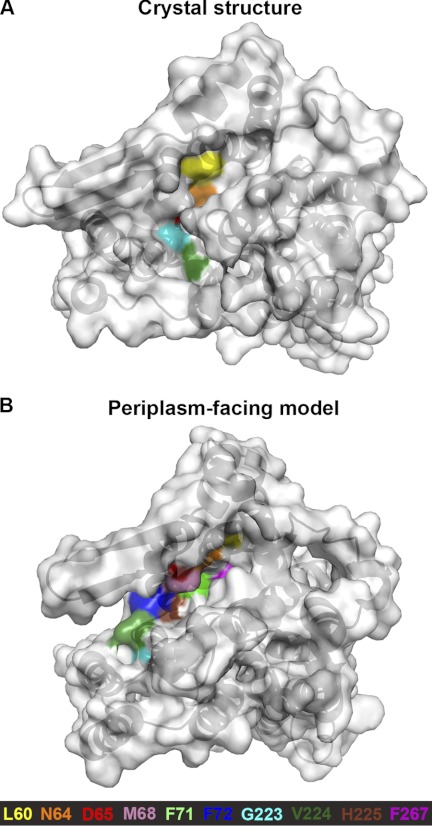
As reactivity of the membrane-permeant SH reagent N-ethyl maleimide (NEM) requires water, accessibility of Cys replacements to NEM within a protein implies the presence of a water-filled cavity connected directly or not with the periplasm or cytoplasm. The complementary reagents 2-sulfonatoethyl methanethiosulfonate (MTSES) and 2-(trimethyl ammonium)-ethylmethanethiosulfonate (MTSET) are charged, membrane-impermeable, and comparable in size to hydrated Na+ (19). Therefore, modification by these reagents indicates the presence of solvent-accessible funnels within the protein through which the reagents can reach the Cys replacement. Accessibility data for these reagents as a function of pH are available for Cys replacements on TMs II, IV, V, VIII, IX, X, and XI. The model agrees with most of the experimental data, as follows: (i) residues on TM II line the cation pathway (19); several positions accessible to the periplasmic side at physiological pH are not exposed in the crystal structure at pH 4, but are exposed in the periplasm-facing model (residues Asn-64, Asp-65, Met-68, Phe-71, and Phe-72) (Fig. 7). (ii) Except for the N-terminal residues of TM VIII (G223C, V224C, and H225C), no TM VIII Cys mutants were accessible to MTSES in intact cells, implying that this helix lines only the entrance to the periplasmic funnel (50). Furthermore, the accessibility to MTSES was not affected by pH. We observed that, in agreement with these data, no TM VIII positions, other than G233, V224, and H225, were significantly accessible from the periplasm, either in the model or in the crystal structure (Fig. 7). (iii) In TM IX, pH-dependent conformational changes were identified at the cytoplasmic side at Lys-249 and Glu-252, and the cytoplasmic funnel deepens at physiological pH (16), which should correspond to a cytoplasmic-facing, rather than a periplasm-facing, conformation, as depicted by the model. From the periplasmic end of TM IX, no MTSET-accessible residues were detected up to and including Phe-267 (16). Of these residues, only Phe-267 lines the periplasmic funnel in the model-structure (Fig. 7B). However, it is located at the bottom of the funnel, and in a narrow region, which would be inaccessible to the large reagent. (iv) Several studies examined the accessibility of residues in the TM IV-XI assembly and surrounding helices. Accessibility assessments of TM IV residues (51), as well as scans of TMs IV and V in HPNhaA, a close homologue from Helicobacter pylori (52, 53), showed that only the equivalents of Ile-128 and Asp-164 are accessible to the periplasmic side (52, 53). A few positions in the center of TM X were accessible to reagents from the cytoplasmic end at pH 7.5 (44). Last, cross-linking was obtained between several residue pairs in the proximal TM helices IV and XI (55) indicating that positions in TMs IV and XI within the membrane core are exposed as well. While none of these positions is directly accessible in the crystal or in the model, the cytoplasmic and periplasmic pathways to the assembly region are enlarged in the model compared with the crystal structure (Fig. 3A), potentially exhibiting a trend of motion toward the formation of accessible pathways.
FIGURE 7.
Accessibility measurements of Cys mutants in TM II (19), TM VIII (50), and TM IX (16), mapped on the crystal and model structures. Periplasmic view of the crystal structure (panel A) and periplasm-facing model (panel B) as in Fig. 2B, with the proteins shown in cartoon representation with mildly transparent surface. Cys mutants that reacted with at least one SH-reagent (16, 19, 50) are colored according to the legend. In TM II, all accessible positions are exposed in the model, while in the crystal structure only Leu-60 and Asn-64 are solvent-exposed. Indeed, L60C was reactive at both pH 6.5 and pH 8.5, whereas Asn-64 and Asp-65 were accessible to the large MTSES reagent only at pH 8.5 (19). The crevice in which Asn-64 resides in the crystal structure is narrower than that in the model-structure, which could explain the lack of reactivity with MTSES at pH 6.5. Overall, the model corresponds to TM II accessibility data produced at alkaline pH, whereas the crystal structure better depicts the measurements at pH 6.5. In TM VIII, accessible positions G223C, V224C, and H225C are exposed in the model, independently of pH conditions (50), while only the former two are exposed in the crystal. The other TM VIII residues (colored beige) were all inaccessible, and are indeed not exposed in either the structure or the model. This indicates that the model agrees with the experimental data for TM VIII. In TM IX, no MTSET accessibility was detected at physiological pH up to and including Phe-267 (16). This corresponds to both the crystal and model, with Phe-267 lining the narrow end of the periplasmic funnel in the latter, where it is unlikely to be exposed to the large MTSET reagent.
Cross-linking
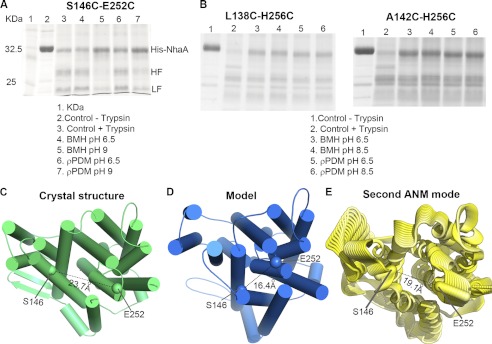
To examine the predicted conformational changes of TMs IVc-V and TM IX, we conducted cross-linking experiments at two pH conditions between three Cys pairs on TM IVc or the TMs IVc-V loop, and TM IX (L138C-H256C, A142C-H256C, and S146C-E252C). For the former two pairs, we conducted the experiments in pH 6.5 and 8.5, whereas for the latter pair the alkaline range was set to 9, as the E252C causes an alkaline shift in the pH activity range of NhaA (56). Two bifunctional cross-linking reagents were used: PDM and BMH, with spacing arms of 9.2–12.3 Å and 3.5–15.6 Å, respectively. According to the Cα-Cα distances of these residue pairs in the crystal structure, in the periplasm-facing model, and in the crystal structure after the most extreme deformation along the ANM2 motion, only one pair, namely S146C-E252C, is predicted to change sufficiently that the difference would be detected by cross-linking (supplemental Table S2 and Fig. 8, C–E). Indeed, S146C and E252C cross-linked at pH 9 but not 6.5, while the other pairs cross-linked at both pH levels (Fig. 8, A and B). These results suggest that upon pH change from 6.5 to alkaline range, TMs IVc and IXc approach each other, in agreement with both the suggested periplasm-facing model and ANM motion (Fig. 8, C--E), indicating that the conformational changes predicted herein are pH-induced.
FIGURE 8.
In situ intra-molecular cross-linking in NhaA. Membrane vesicles were prepared from TA16 strain expressing the indicated double Cys-replacement mutants as described in “Experimental Procedures.” A and B, membranes were cross-linked, in situ, by the bi-functional cross linkers, BMH or p-PDM at various pHs, or not cross-linked (controls) as depicted on the figure. For analysis of the cross-linked products, the Ni2+-NTA affinity-purified protein was treated at pH 8.5 with trypsin and resolved by SDS-PAGE as described under “Experimental Procedures.” The experiments were repeated at least three times, and the results were essentially identical. HF and LF, are the tryptic fragments. The trypsin cleavable site Lys-249 is located between the two Cys replacements. Therefore, HF and LF are visible on the gel when cross linking does not take place (Variant S146C-E252C at pH 6.5, lanes 4 and 6). However, when cross-linking occurs (Variant S146C-E252C at pH 9, lanes 5 and 7), HF and LF remain linked and a single band similar in mobility to the native protein appears (lane 2). The cross linking of variants L138C-H256C and A142C-H256C was indifferent to pH. C–E, distance between Ser-146 and Glu-252 (the Cα atoms depicted as spheres) is shown on the crystal structure (cytoplasmic view, cylinder-presentation) (C), the periplasmic-facing model (D), and the second ANM mode (in cartoon representation) (E).
DISCUSSION
The NhaA transporter serves as a prototype for human cation proton antiporters (CPA) (4, 5, 7), but our molecular understanding of its function is limited to a single, cytoplasm-facing, conformation (8, 12). We used the pseudo-symmetry of NhaA to suggest a model of a periplasm-facing state of the transporter (Figs. 1, 2, and 3), and in parallel we used normal mode analysis to explore the predicted motion of the cytoplasm-facing crystal structure based on the extent of the contacts in the x-ray structure (Figs. 4, A–C and 5). Both methods predicted similar conformational changes that were validated by cross-linking experiments (Fig. 8, A and B) and previous biochemical data (Fig. 7).
Limitations of the Computational Methodologies
The major structural alteration predicted herein could probably not be accessed using molecular dynamics simulations, nor using other commonly used approaches that are based on all-atom description, because of the large size of the transporter, the restricted simulation time currently available, and the complexity of the conformational change (10, 57). Thus, we were compelled to use coarse-grained approximations that have their own limitations. The comparative modeling procedure used to construct the periplasm-facing conformation predicts the overall architecture of this state, at an approximate accuracy resolution of ∼3 Å (58). It also cannot take into account any additional structural rearrangements beyond those contained within the pseudo-symmetry. The GNM/ANM analyses predict the general trends of motion, while overlooking the details (28, 30, 31, 47). Moreover, the ANM allows examination of the individual modes in three-dimensional, but the integrated motion, consisting of a superimposition of all modes of motion, cannot be predicted, as this requires correctly combining each mode's phase and amplitude. Additionally, the motion revealed by normal mode analysis covers a limited scope of the conformational landscape; taking the NhaA structure as the energetic minimum, we survey its surrounding equilibrium dynamics, but cannot cross high energetic barriers (59). Lastly, although the two slowest modes appear to describe the transition well, some contribution of other modes may be required to accomplish the whole transition. Nevertheless, as several lines of experimental evidence support the validity of both analyses, we maintain that the computations are useful for deriving hypotheses to be tested in experiments.
Motion, Transition, and Alternating Access
Transporters alternate between inward- and outward-facing conformations, often illustrated as rocking of two independent domains (32, 33). In this study, we detected two types of motions. Directly consistent with this alternating-access concept, in the periplasm-facing model and in the ANM2 motion, segments lining the cytoplasmic funnel approach its center in a coordinated manner (Figs. 2A and 5C and supplemental Movies S2-S4), while the model-structure also shows simultaneous opening of the periplasmic funnel (Figs. 2B and 7). In GNM1, the two domains exhibit opposite dynamical correlations, each rotating around a putative symmetry axis in the corresponding ANM1 motion (Fig. 4, A and B and supplemental Movie S1), similar to the relative rotation of the two domains observed when comparing the periplasm-facing model with the crystal structure (supplemental Fig. S3, B and C). Interestingly, measuring the positioning of the two domains revealed a rotation of 22° in the model relative to the crystal structure. This is comparable to the helical rotations observed when comparing different structural conformations of LeuT (ranging between 9° and 45°, with most of the helices rotating ∼20°) (60), and those measured for the inter-domain rotations between the crystal structures and models of GltPh (37°, (24)) and LacY (51°, (25)). The division into two domains is reminiscent of the independent domains illustrated in classical alternating-access, and we thus suggest that the rotational motion could be related to switching between states as well. Although this might not be the intuitive motion one would imagine in order to switch between conformations, this could be an interesting alternative to a simple rocking (Fig. 4, C and D).
Structural Implications of the Periplasm-facing Scaffold
In the periplasm-facing model, the cytoplasmic funnel adopts a closed conformation (Figs. 1D and 2A), in agreement with cross-linking data (Fig. 8). While the periplasmic funnel in the model is larger than that in the crystal, it does not widen as much as the cytoplasmic funnel in the crystal structure (Figs. 2 and 7). Nevertheless, the model is in excellent agreement with TM II accessibility data, which show that Phe-71 and Phe-72 are accessible only to NEM (19), indicating that they reside in a narrower cavity than preceding residues, and that the periplasmic funnel ends at Phe-72 (19).
Interestingly, the structural model also predicts some opening of putative pathways on the cytoplasmic and periplasmic sides that lead into the TM IV-XI assembly (Figs. 2 and 3). The location of these pathways is consistent with observations from MD simulations, which suggest separate cation and proton pathways through the funnels and via the TM IV-XI assembly region, respectively (10). It is also consistent with the observation that several residues in the centers of TMs IV, X, and XI were exposed under physiological conditions (44, 51). Nevertheless, the opening of the periplasmic pathway in the model (Fig. 3B) is insufficient to expose the functionally important Asp-164 (8, 10–12) to the periplasm, although relatively small additional conformational changes, enlarging the pathway, would increase the periplasmic accessibility of this residue. Notably, the HpNhaA equivalent of Asp-164 was accessible to the periplasm upon Cys mutation (53). As no other TM V positions were accessible to SH-reagents from the periplasmic side upon Cys mutation, it is apparent that an entire face of TM V does not line the periplasmic funnel, an observation consistent with our periplasm-facing model (Figs. 1, C and D and 2). We thus propose that Asp164 could be exposed to the periplasmic side through pathway within the TM IV-XI assembly, suggesting how the residue may participate in cation binding.
pH-induced Conformational Changes?
Studies involving diverse experimental approaches have demonstrated that NhaA undergoes conformational changes in response to pH alteration (13–21). Two general views of this pH-dependent activation have been postulated, although they are not mutually exclusive. According to the first view, the apparent pH-induced activation results from direct competition between H+ and Na+ (or Li+) (61). According to the second view, the protons are not only ligands, but are also involved in pH regulation of NhaA's activity (13–21).
Consistent with the latter view, recent studies have shown two distinct conformational changes: one in response to pH, and the other, at alkaline pH, in response to ligand addition (17, 21). CryoEM projection maps of two-dimensional crystals of NhaA indicated conformation changes induced by pH change (4–8) at the funnel region, and other ligand-induced changes at alkaline pH in the TM IV-XI assembly region, ascribed to TM IVp (17). Correspondingly, a site-directed fluorescence study showed that TM IVc is affected by the pH-induced conformational change, and TM XI (at the active site) by ligand-induced changes (21). We suggest that the periplasm-facing model and the deformations depicted by the slowest modes of motion correspond to the pH-induced conformational change observed in these prior studies. This suggestion is supported by several lines of evidence. First, the extent of structural changes at the cytoplasmic funnel of the model-structure is larger than in the periplasmic funnel (Figs. 2 and 7), which can account for the increase in density detected by the cryoEM maps upon increasing pH (see Fig. 7 in Ref. 17). Second, site-directed Trp fluorescence revealed a pH-induced conformational change at TM IVc (21), in agreement with the model-structure (Figs. 1, C and D and 2A) and second mode of motion (Fig. 5C). Third, our cross-linking data were pH-dependent (Fig. 8A), indicating closure at the cytoplasmic funnel at pH 9. Further supporting evidence includes the accessibility of TM II residues at alkaline pH (19), which is consistent with the periplasm-facing model (Fig. 7). Moreover, mutations in various residues in TMs II and IX shifted the pH dependence profile (16, 19), as did intermolecular cross-linking of V254C of TM IX (62). The fact that TMs II and IX undergo substantial structural changes in our model further implies that the modeled conformation corresponds to pH-induced alterations. Finally, Lys-249 is the only cleavable site of all putative sites in NhaA, and cleavage was shown to be pH-sensitive, occurring in alkaline pH and not in acidic pH (13, 46). The extensive rearrangements predicted at the cytoplasmic ends of the helices by our analysis might also explain the pH-dependent sensitivity of Lys-249 of TM IX to trypsin cleavage (13, 46): although the accessibility of the residue itself remains similar in both the crystal structure and model, the extensive rearrangements at the cytoplasmic end may potentially affect the ability of trypsin to bind to the cleavage site.
Previous experimental data indicate that solvent-accessible pathways are formed around the TM IV-XI assembly at physiological pH (16, 51–53, 55), possibly creating ion-accessible pathways (these pathways have also been observed in MD simulations (10)). The experimental data can be grouped into three types: (i) accessibility to SH-reagents of some residues in TMs IV (51, 52) and V (53) in NhaA and in its homologue from Helicobacter pylori; most of these residues are situated at the periplasmic ends and accessible from the periplasmic side; (ii) accessibility to SH-reagent of a few residues in TM X in NhaA (16); (iii) cross-linking between residues from TMs IV and XI, indicating also that these residues are exposed (55). The ligand-induced conformational changes detected in the TM IV-XI assembly region by cryoEM (17) and by site-directed tryptophan fluorescence (21) can potentially account for these accessibility measurements. While the conformational changes in the periplasm-facing model were larger in the funnels than in the TM IV-XI assembly and flanking helices, we did observe structural rearrangements in the latter region as well (Figs. 2 and 3). Although the current conformation cannot account for the above mentioned accessibility of residues in TMs IV, V, X, and XI (16, 51–53, 55), it does suggest a clear trend of opening toward the assembly region from both sides of the membrane (Fig. 3). Assuming that ligand-induced changes indeed induce solvent-accessible pathways within the assembly region, we conclude that the model is essentially compatible with these data.
Previous studies have suggested that a “pH sensor” perceives the pH signal (a change in intra-cellular pH) that is then transduced to activate NhaA (64). Based on the crystal structure (8), computation (65), and experimental data (reviewed in Ref. 12), the pH sensor was identified at the cytoplasmic half of TM IX, loop VIII-IX, and the cytoplasmic end of TM II. Consistent with this suggestion, the cytoplasmic ends of TMs II and IX indeed undergo pronounced changes in their internal structural conformation in the model compared with the crystal structure. Our cross-linking data indicate that the modeled structural changes are indeed pH-induced.
On the Transport Mechanism
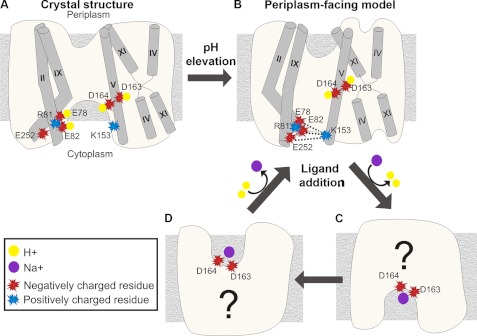
Computational studies have previously suggested that the evolutionarily conserved residues Glu-78 and Glu-82 of TM II are involved in pH sensing (57, 65). Furthermore, the mutation to Cys of each of these acidic residues, as well as of the conserved Glu-252 of TM IX, increased the apparent Km for Na+ and caused an alkaline shift in the activity profile (19, 56). A previous computational study calculated the pKa of charged residues in the crystal structure, predicting values of 5.4, 10.4, >15, >15 and 2.7 for the acidic Glu-78, Glu-82, Asp-163, Asp-164, and Glu-252, respectively, along with values of 15.4 and 9.3 for the basic Arg-81 and Lys-153, respectively (65). Interestingly, in contrast to the crystal structure, the modeling procedure placed the funnel-lining residues Glu-78, Glu-82, and Glu-252 in close proximity to the highly conserved Lys-153 of TM V, which faces into the funnel from the other side (supplemental Fig. S8). Electrostatic interactions among these residues could stabilize the conformation depicted by the model-structure. In light of these structural features, in conjunction with prior computational and experimental data, we propose a working hypothesis for the pH-induced conformational changes in NhaA within the context of an alternating-access mechanism (Fig. 9). At acidic pH, one or more of the conserved Glu-78, Glu-82, and Glu-252 residues are presumably protonated, potentially forming hydrogen bonds with the highly conserved Arg-81 of TM II (supplemental Figs. S8A and 9A). The model predicts that upon pH elevation, TMs II and IX change their conformations; concurrently, the glutamates are expected to become charged, attracting in turn Lys-153 (supplemental Figs. S8B and 9B). In this conformation, which would correspond to the periplasm-facing model, Asp-163 and Asp-164 are buried in the protein core, presumably remaining protonated. A second, ligand-induced change could expose one or both aspartates to the cytoplasm via motion of TM IVp, inducing rapid exchange of protons for Na+ (Fig. 9C), followed by swapping into an Na+-bound, periplasm-facing conformation (Fig. 9D). Upon the exchange of Na+ for protons, the antiporter could return to the partly-occluded periplasm-facing state (Fig. 9B). Overall, this mechanism connects the large conformational changes of TMs II and IX, predicted by our calculations, to the evidence indicating a response of specific residues in these TM helices to pH changes (19, 56, 57, 65), and it also connects these changes to pH-induced conformational changes in TM IV, and to the ligand-induced response detected at the TM IV-XI assembly (17, 21). The actual mechanism may be more complex and include more states.
FIGURE 9.
Hypothetical mechanism of pH-induced conformational changes and alternate access. The panels give schematic side views of the NhaA structure (or model), with the cytoplasm below. Helices of interest are marked with cylinders. Charged residues and ions are displayed according to the legend. A, crystal structure at acidic pH. One or more acidic residues at the cytoplasmic end would be protonated, as well as Asp-163 and Asp-164. The cluster of titratable residues at TMs II and IX does not interact with Lys-153 of TM V. B, upon pH elevation, the transporter undergoes a conformational change and shifts to the periplasm-facing model. We suggest that the cluster of titratable residues at TMs II and IX becomes negatively charged and associates electrostatically with Lys-153 of TM V. In this state, the ligand-binding residues Asp-163 and Asp-164 of TM V are buried in the protein core and would still be protonated. C and D, in alkaline pH, ligand addition induces a second conformational change, predicted to occur at the TM IV-XI assembly region (17, 21). This enables the exchange of Na+ for protons, presumably involving the exposure of Asp-163 and Asp-164 (C). Flipping of the antiporter to a periplasm-facing state (D) could allow the exchange of Na+ with two other H+ ions, and a return to state B.
CONCLUSIONS
This is the fourth example in which pseudo-symmetric inverted-repeats have been used to generate a new conformation based on a known structure of a TM transporter. Two of the previous cases (22, 24) were supported strongly by structural data, and we are hopeful that such will be the case here as well. Significantly, the ASBT transporter, which adopts a strikingly similar fold to that of NhaA, is also composed of pseudo-symmetric segments (26); it has been pointed out that changing the relative positioning of the two helical domains would result in an outward-facing conformation, as presented herein for NhaA (Figs. 2 and 4). Overall, these examples suggest that this general principle has emerged over and over again in the evolution of transporters (9, 66). The EmrE transporter also adheres to the same principle (54, 63, 66–69), although in this small homodimeric transporter the two subunits appear to alter their orientation with respect to each other in the dimer, while in the other cases two domains change their orientation within the same subunit. In this respect, the alternating-access mechanisms proposed for NhaA and the three other large transporters (22, 24, 25) are all variations of the theme proposed for the small EmrE transporter (67). The predicted cytoplasm-facing conformation could be useful for future studies of the ion translocation mechanism, driving novel structure-based hypotheses for NhaA. Moreover, it may further our understanding of the behavior of related human transporters of the CPA family in health and disease.
This work was supported in part by Grant 1331/11 from the Israel Science Foundation (to N. B-T.), and by NATO Traveling Grant No. CBP.MD.CLG 983009 (to T. H. and N. B-T.). This work was also supported in part by a fellowship from the Edmond J. Safra Center for Bioinformatics at Tel-Aviv University (to M. S.); by EDICT (European Drug Initiative on Channels and Transporters) Grant Number: EU EP7, and The USA-Israel Binational Science Foundation Grant Number: BSF 20050130 (to E. P.); by the DFG (German Research Foundation), Collaborative Research Center 807 “Transport and Communication across Biological Membranes” (to L. R. F.); and by Grant CBD.MD.CL6.984340 (to T. H.).
The model has been deposited in the Protein Model Database (PMDB) as accession number PM0078072.

This article contains supplemental Tables S1 and S2, Figs. S1–S9, and Movies S1–S6.
- TM
- transmembrane
- cryoEM
- cryo-electron microscopy
- GNM
- Gaussian Network Model
- ANM
- Anisotropic Network Model
- MTSES
- 2-sulfonatoethyl methanethiosulfonate
- MTSET
- 2-(trimethyl ammonium)-ethylmethanethiosulfonate.
REFERENCES
- 1. Padan E., Bibi E., Ito M., Krulwich T. A. (2005) Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717, 67–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Padan E., Tzubery T., Herz K., Kozachkov L., Rimon A., Galili L. (2004) NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim. Biophys. Acta 1658, 2–13 [DOI] [PubMed] [Google Scholar]
- 3. West I. C., Mitchell P. (1974) Proton/sodium ion antiport in Escherichia coli. Biochem. J. 144, 87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schushan M., Xiang M., Bogomiakov P., Padan E., Rao R., Ben-Tal N. (2010) Model-guided mutagenesis drives functional studies of human NHA2, implicated in hypertension. J. Mol. Biol. 396, 1181–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Landau M., Herz K., Padan E., Ben-Tal N. (2007) Model structure of the Na+/H+ exchanger 1 (NHE1): functional and clinical implications. J. Biol. Chem. 282, 37854–37863 [DOI] [PubMed] [Google Scholar]
- 6. Schushan M., Landau M., Padan E., Ben-Tal N. (2011) Two conflicting NHE1 model structures: compatibility with experimental data and implications for the transport mechanism. J. Biol. Chem. 286, le9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brett C. L., Donowitz M., Rao R. (2005) Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288, C223–C239 [DOI] [PubMed] [Google Scholar]
- 8. Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. (2005) Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 9. Boudker O., Verdon G. (2010) Structural perspectives on secondary active transporters. Trends Pharmacol. Sci. 31, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arkin I. T., Xu H., Jensen M. O., Arbely E., Bennett E. R., Bowers K. J., Chow E., Dror R. O., Eastwood M. P., Flitman-Tene R., Gregersen B. A., Klepeis J. L., Kolossváry I., Shan Y., Shaw D. E. (2007) Mechanism of Na+/H+ antiporting. Science 317, 799–803 [DOI] [PubMed] [Google Scholar]
- 11. Inoue H., Noumi T., Tsuchiya T., Kanazawa H. (1995) Essential aspartic acid residues, Asp-133, Asp-163, and Asp-164, in the transmembrane helices of a Na+/H+ antiporter (NhaA) from Escherichia coli. FEBS Lett. 363, 264–268 [DOI] [PubMed] [Google Scholar]
- 12. Padan E., Kozachkov L., Herz K., Rimon A. (2009) NhaA crystal structure: functional-structural insights. J. Exp. Biol. 212, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 13. Rothman A., Gerchman Y., Padan E., Schuldiner S. (1997) Probing the conformation of NhaA, a Na+/H+ antiporter from Escherichia coli, with trypsin. Biochemistry 36, 14572–14576 [DOI] [PubMed] [Google Scholar]
- 14. Venturi M., Rimon A., Gerchman Y., Hunte C., Padan E., Michel H. (2000) The monoclonal antibody 1F6 identifies a pH-dependent conformational change in the hydrophilic NH(2) terminus of NhaA Na(+)/H(+) antiporter of Escherichia coli. J. Biol. Chem. 275, 4734–4742 [DOI] [PubMed] [Google Scholar]
- 15. Karasawa A., Tsuboi Y., Inoue H., Kinoshita R., Nakamura N., Kanazawa H. (2005) Detection of oligomerization and conformational changes in the Na+/H+ antiporter from Helicobacter pylori by fluorescence resonance energy transfer. J. Biol. Chem. 280, 41900–41911 [DOI] [PubMed] [Google Scholar]
- 16. Tzubery T., Rimon A., Padan E. (2008) Structure-based functional study reveals multiple roles of transmembrane segment IX and loop VIII-IX in NhaA Na+/H+ antiporter of Escherichia coli at physiological pH. J. Biol. Chem. 283, 15975–15987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Appel M., Hizlan D., Vinothkumar K. R., Ziegler C., Kühlbrandt W. (2009) Conformations of NhaA, the Na+/H+ exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J. Mol. Biol. 388, 659–672 [DOI] [PubMed] [Google Scholar]
- 18. Dzafić E., Klein O., Screpanti E., Hunte C., Mäntele W. (2009) Flexibility and dynamics of NhaA Na+/H+-antiporter of Escherichia coli studied by Fourier transform infrared spectroscopy. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 72, 102–109 [DOI] [PubMed] [Google Scholar]
- 19. Herz K., Rimon A., Olkhova E., Kozachkov L., Padan E. (2010) Transmembrane segment II of NhaA Na+/H+ antiporter lines the cation passage, and Asp-65 is critical for pH activation of the antiporter. J. Biol. Chem. 285, 2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vinothkumar K. R., Smits S. H., Kühlbrandt W. (2005) pH-induced structural change in a sodium/proton antiporter from Methanococcus jannaschii. EMBO J. 24, 2720–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozachkov L., Padan E. (2011) Site-directed tryptophan fluorescence reveals two essential conformational changes in the Na+/H+ antiporter NhaA. Proc. Natl. Acad. Sci. U.S.A. 108, 15769–15774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forrest L. R., Zhang Y. W., Jacobs M. T., Gesmonde J., Xie L., Honig B. H., Rudnick G. (2008) Mechanism for alternating access in neurotransmitter transporters. Proc. Natl. Acad. Sci. U.S.A. 105, 10338–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. (2008) The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Crisman T. J., Qu S., Kanner B. I., Forrest L. R. (2009) Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc. Natl. Acad. Sci. U.S.A. 106, 20752–20757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Radestock S., Forrest L. R. (2011) The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J. Mol. Biol. 407, 698–715 [DOI] [PubMed] [Google Scholar]
- 26. Hu N. J., Iwata S., Cameron A. D., Drew D. (2011) Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bahar I., Rader A. J. (2005) Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct. Biol. 15, 586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bahar I., Lezon T. R., Bakan A., Shrivastava I. H. (2009) Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem. Rev. 110, 1463–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bahar I., Atilgan A. R., Erman B. (1997) Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 30. Haliloglu T., Bahar I., Erman B. (1997) Gaussian dynamics of folded proteins. Physical Review Letters 79, 3090–3093 [Google Scholar]
- 31. Atilgan A. R., Durell S. R., Jernigan R. L., Demirel M. C., Keskin O., Bahar I. (2001) Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 80, 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jardetzky O. (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 33. Kaback H. R. (2005) Structure and mechanism of the lactose permease. C. R. Biol. 328, 557–567 [DOI] [PubMed] [Google Scholar]
- 34. Petrey D., Honig B. (2003) GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 374, 492–509 [DOI] [PubMed] [Google Scholar]
- 35. Kessel A., Ben-Tal N. (2010) Proteins: Structure, Function and Motion. CRC Press, Taylor and Francis Group [Google Scholar]
- 36. Illergård K., Kauko A., Elofsson A. (2011) Why are polar residues within the membrane core evolutionary conserved? Proteins 79, 79–91 [DOI] [PubMed] [Google Scholar]
- 37. Kessel A., Ben-Tal N. (2002) Free Energy Determinants of Peptide Association with Lipid Bilayers. Current Topics in Membranes: Peptide-Lipid Interactions (Simon S., McIntosh T., eds.), Vol. 52, pp. 205–253, Academic Press, San Diego, CA [Google Scholar]
- 38. Fleishman S. J., Ben-Tal N. (2006) Progress in structure prediction of α-helical membrane proteins. Curr. Opin. Struct. Biol. 16, 496–504 [DOI] [PubMed] [Google Scholar]
- 39. Adamian L., Liang J. (2006) Prediction of transmembrane helix orientation in polytopic membrane proteins. BMC Struct. Biol. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 41. Bahar I., Kaplan M., Jernigan R. L. (1997) Short-range conformational energies, secondary structure propensities, and recognition of correct sequence-structure matches. Proteins 29, 292–308 [DOI] [PubMed] [Google Scholar]
- 42. Emekli U., Schneidman-Duhovny D., Wolfson H. J., Nussinov R., Haliloglu T. (2008) HingeProt: automated prediction of hinges in protein structures. Proteins 70, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 43. Taglicht D., Padan E., Schuldiner S. (1991) Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J. Biol. Chem. 266, 11289–11294 [PubMed] [Google Scholar]
- 44. Kozachkov L., Herz K., Padan E. (2007) Functional and structural interactions of the transmembrane domain X of NhaA, Na+/H+ antiporter of Escherichia coli, at physiological pH. Biochemistry 46, 2419–2430 [DOI] [PubMed] [Google Scholar]
- 45. Rimon A., Tzubery T., Galili L., Padan E. (2002) Proximity of cytoplasmic and periplasmic loops in NhaA Na+/H+ antiporter of Escherichia coli as determined by site-directed thiol cross-linking. Biochemistry 41, 14897–14905 [DOI] [PubMed] [Google Scholar]
- 46. Gerchman Y., Rimon A., Padan E. (1999) A pH-dependent conformational change of NhaA Na(+)/H(+) antiporter of Escherichia coli involves loop VIII-IX, plays a role in the pH response of the protein, and is maintained by the pure protein in dodecyl maltoside. J. Biol. Chem. 274, 24617–24624 [DOI] [PubMed] [Google Scholar]
- 47. Bahar I. (2010) On the functional significance of soft modes predicted by coarse-grained models for membrane proteins. J. Gen. Physiol. 135, 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guan L., Kaback H. R. (2006) Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35, 67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guan L., Kaback H. R. (2007) Site-directed alkylation of cysteine to test solvent accessibility of membrane proteins. Nat. Protoc. 2, 2012–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Diab M., Rimon A., Tzubery T., Padan E. (2011) Helix VIII of NhaA Na(+)/H(+) antiporter participates in the periplasmic cation passage and pH regulation of the antiporter. J. Mol. Biol. 413, 604–614 [DOI] [PubMed] [Google Scholar]
- 51. Galili L., Rothman A., Kozachkov L., Rimon A., Padan E. (2002) Transmembrane domain IV is involved in ion transport activity and pH regulation of the NhaA-Na(+)/H(+) antiporter of Escherichia coli. Biochemistry 41, 609–617 [DOI] [PubMed] [Google Scholar]
- 52. Kuwabara N., Inoue H., Tsuboi Y., Nakamura N., Kanazawa H. (2004) The fourth transmembrane domain of the Helicobacter pylori Na+/H+ antiporter NhaA faces a water-filled channel required for ion transport. J. Biol. Chem. 279, 40567–40575 [DOI] [PubMed] [Google Scholar]
- 53. Kuwabara N., Inoue H., Tsuboi Y., Mitsui K., Matsushita M., Kanazawa H. (2006) Structure-function relationship of the fifth transmembrane domain in the Na+/H+ antiporter of Helicobacter pylori: Topology and function of the residues, including two consecutive essential aspartate residues. Biochemistry 45, 14834–14842 [DOI] [PubMed] [Google Scholar]
- 54.70 Bowie J. U. (2006) Flip-flopping membrane proteins. Nat. Struct. Mol. Biol. 13, 94–96 [DOI] [PubMed] [Google Scholar]
- 55. Galili L., Herz K., Dym O., Padan E. (2004) Unraveling functional and structural interactions between transmembrane domains IV and XI of NhaA Na+/H+ antiporter of Escherichia coli. J. Biol. Chem. 279, 23104–23113 [DOI] [PubMed] [Google Scholar]
- 56. Tzubery T., Rimon A., Padan E. (2004) Mutation E252C increases drastically the Km value for Na+ and causes an alkaline shift of the pH dependence of NhaA Na+/H+ antiporter of Escherichia coli. J. Biol. Chem. 279, 3265–3272 [DOI] [PubMed] [Google Scholar]
- 57. Olkhova E., Padan E., Michel H. (2007) The influence of protonation states on the dynamics of the NhaA antiporter from Escherichia coli. Biophys. J. 92, 3784–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Forrest L. R., Tang C. L., Honig B. (2006) On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys. J. 91, 508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cui Q., Bahar I. (2006) Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems, Boca Raton, Chapman & Hall/CRC [Google Scholar]
- 60. Krishnamurthy H., Gouaux E. (2012) X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature 481, 469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mager T., Rimon A., Padan E., Fendler K. (2011) Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from Escherichia coli: an electrophysiological study. J. Biol. Chem. 286, 23570–23581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gerchman Y., Rimon A., Venturi M., Padan E. (2001) Oligomerization of NhaA, the Na+/H+ antiporter of Escherichia coli in the membrane and its functional and structural consequences. Biochemistry 40, 3403–3412 [DOI] [PubMed] [Google Scholar]
- 63. Morrison E. A., DeKoster G. T., Dutta S., Vafabakhsh R., Clarkson M. W., Bahl A., Kern D., Ha T., Henzler-Wildman K. A. (2012) Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 481, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Padan E. (2008) The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem. Sci. 33, 435–443 [DOI] [PubMed] [Google Scholar]
- 65. Olkhova E., Hunte C., Screpanti E., Padan E., Michel H. (2006) Multiconformation continuum electrostatics analysis of the NhaA Na+/H+ antiporter of Escherichia coli with functional implications. Proc. Natl. Acad. Sci. U.S.A. 103, 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pornillos O., Chang G. (2006) Inverted repeat domains in membrane proteins. FEBS Lett. 580, 358–362 [DOI] [PubMed] [Google Scholar]
- 67. Fleishman S. J., Harrington S. E., Enosh A., Halperin D., Tate C. G., Ben-Tal N. (2006) Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J. Mol. Biol. 364, 54–67 [DOI] [PubMed] [Google Scholar]
- 68. Chen Y. J., Pornillos O., Lieu S., Ma C., Chen A. P., Chang G. (2007) X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. U.S.A. 104, 18999–19004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rapp M., Seppälä S., Granseth E., von Heijne G. (2007) Emulating membrane protein evolution by rational design. Science 315, 1282–1284 [DOI] [PubMed] [Google Scholar]