Abstract
Molecular apocrine is a subtype of estrogen receptor-negative (ER.) breast cancer, which is characterized by a steroid-response gene signature that includes androgen receptor, FOXA1, and a high frequency of ErbB2 overexpression. In this study, we demonstrate that there is a strong association between the overexpression of FOXA1 and ErbB2 in ER- breast tumors. This has led us to identify a cross-regulation network between FOXA1 and ErbB2 signaling in ER- breast cancer. We present two mechanisms to explain the association between FOXA1 and ErbB2 overexpression in molecular apocrine cells. In one process, ErbB2 signaling genes CREB1 and c-Fos regulate FOXA1 transcription, and in another process, AP2α regulates the expression of both FOXA1 and ErbB2. Moreover, we demonstrate that FOXA1, in turn, regulates the transcription of ErbB2 signaling genes. This includes a core gene signature that is shared across two molecular apocrine cell lines. Importantly, the most upregulated (RELB) and downregulated (PAK1) genes in this signature are direct FOXA1 targets. Our data suggest that FOXA1 acts as a dual-function transcription factor and the repressive function of FOXA1 on RELB can be explained by the recruitment of its binding partner corepressor TLE3. It is notable that a group of FOXA1-regulated genes vary across molecular apocrine cell lines leading to the differences in the functional effects of FOXA1 on extracellular signal-regulated kinase phosphorylation and cell viability between these lines. This study demonstrates that there is a cross-regulation network between FOXA1 and ErbB2 signaling that connects FOXA1 to some of the key signaling pathways in ER- breast cancer.
Introduction
Estrogen receptor-negative (ER-) breast cancer is a heterogeneous disease that constitutes around 30% of all cases [1]. To develop effective targeted therapies for ER- breast cancer, there is need to better understand the biology of this disease. ER- breast cancer can be divided into molecular apocrine and basal subtypes based on expression microarray profiling [2]. Molecular apocrine subtype is characterized by a steroid-response gene signature that includes androgen receptor (AR), FOXA1, TFF3, and a high frequency of ErbB2 overexpression (ErbB2+) [2–4]. Recent studies have shown that AR expression is observed in approximately 50% of ER- breast tumors and more than 50% of these cases also have ErbB2 overexpression [5–7].
There is a growing body of evidence to support a significant role for the AR and ErbB2 signaling pathways in molecular apocrine breast cancer. Notably, there is a functional cross talk between the AR and ErbB2 signaling in molecular apocrine cells, which modulates cell proliferation and expression of steroid-response genes [8]. Moreover, we have recently identified a positive feedback loop between the AR signaling and extracellular signal-regulated kinase (ERK) pathways in molecular apocrine breast cancer [9]. In this feedback loop, AR regulates ERK phosphorylation through the mediation of ErbB2 and, in turn, ERK-CREB1 signaling regulates the transcription of AR in molecular apocrine cells [9]. Furthermore, it has been shown that AR mediates ligand-dependent activation of Wnt and ErbB2 signaling pathways through direct transcriptional induction of WNT7B and ErbB3 [10]. Importantly, AR signaling is a potential therapeutic target in ER-/AR+ breast cancer [10–13].
Another notable gene in the molecular apocrine signature is the transcription factor FOXA1 [2–4]. FOXA1 has emerged as a critical modulator of ER and AR function with a significant role in breast and prostate cancers [14–18]. In addition, recent studies suggest that FOXA1 has a complex regulatory function with the ability to both facilitate and restrict key transcription factors such as AR [15,19]. However, there are limited data available regarding the function of FOXA1 in ER- breast cancer. Moreover, although GATA3 and ERα are known transcriptional activators of FOXA1 in ER+ breast cancer [20,21], the regulation of FOXA1 in ER- tumors is poorly understood.
We have previously demonstrated that, in a subset of ER-/AR+ breast cancer cells, heregulin, which activates ErbB2 and ErbB3, induces FOXA1 expression [8]. Furthermore, gene expression analysis has revealed that FOXA1 is expressed in approximately 70% of ErbB2+ breast tumors [22]. These findings suggest that ErbB2 signaling may have a role in the regulation of FOXA1; however, the mechanism involved in this process is yet to be identified.
In this study, we investigate a cross-regulation between FOXA1 and ErbB2 signaling in molecular apocrine breast cancer. We demonstrate that ErbB2 signaling activates FOXA1 mediated through its downstream transcription factors and FOXA1, in turn, regulates a distinct group of ErbB2 signaling genes.
Materials and Methods
Tissue Microarray Cohort and Immunohistochemistry
Three sets of breast cancer tissue microarray (TMA) slides were obtained from Pantomics (http://www.pantomics.com/tissue-arrays/Systems.htm#Breast_disease_tissue_arrays; BRC1501-3, Richmond, CA) as they were published before [23]. The TMA slides constituted of a total of 79 ER- breast tumors with known ErbB2 status. Immunohistochemistry (IHC) staining was performed using EnVision+ System-HRP (DakoCytomation, Sydney, Australia). Antigen retrieval was carried out using Target Retrieval Solution (DakoCytomation). Primary antibody incubation was performed with FOXA1 rabbit polyclonal antibody (ab23738) from Abcam (Cambridge, United Kingdom) at 1:200 dilution [24,25]. TMA cores were examined at 60x magnification using a light microscope (Nikon Instruments, Inc, Tokyo, Japan). A total of 1000 nuclei per each TMA slide were assessed, and the average score for two replicate slides was used for the analysis.
Scoring for FOXA1 was carried out based on a previously described semiquantitative scoring system [26]. On the basis of this system, percentage (P: score of 1 for each 10% positive tumor nuclei such as 10 = 91%–100%) and intensity (I: 1+ to 3+) of nuclear expression were multiplied to generate a numerical score (S = P x I), which could vary between 0 and 30 [26]. In our study, FOXA1 overexpression (FOXA1+) was defined as S > 15 (e.g., intense nuclear staining [3+] in >50% of cells). Using the median score as a cutoff has been previously used to assess protein overexpression in immunohistochemistry studies [27,28].
Cell Culture
Breast cancer cell lines MDA-MB-453, HCC-1954, and MCF-7 were obtained from the American Type Culture Collection (Manassas, VA). All tissue culture media were obtained from Invitrogen (Melbourne, Australia). MDA-MB-453 and HCC-1954 cell lines were cultured in L15 medium, 10% fetal bovine serum, and RPMI 1640 medium, respectively. The MCF-7 cell line was cultured in Dulbecco modified Eagle medium/F12 medium and 10% fetal bovine serum. Treatment with MEK inhibitor, CI-1040 (PD184352; Selleck Chemicals, Houston, TX) at 10 µM concentration, was carried out for 24 hours in full media.
Western Blot Analysis
FOXA1 rabbit polyclonal antibody was obtained from Abcam. Rabbit monoclonal ERK1/2 antibody and rabbit monoclonal phospho-ERK1/2 (Thr202/Tyr204) were obtained from Cell Signaling (Danvers, MA). Western blots were carried out at 1:1000 dilution of each primary antibody using 10 and 20 µg of cell lysates for the total and phospho-proteins, respectively. Protein concentrations from the cell isolates were measured using the BCA Protein Assay Kit (Thermo Scientific, Melbourne, Australia). Rabbit polyclonal α-tubulin antibody (Abcam) was used as the loading control. Analysis of band densities was performed using Bio-Profil Densitometer Software (Vilber Lourmat, Eberhardzel, Germany). All fold changes in band densities were measured relative to the control groups. Western blots were done in two biologic replicates, and the average fold change was shown for each set of experiments.
Real-time Polymerase Chain Reaction Analysis
Total RNA extraction was performed as described before [29]. Real-time polymerase chain reaction (RT-PCR) to assess the expression levels of FOXA1 (assay ID: Hs00270129_m1), RELB (assay ID: Hs00232399_m1), and TLE3 (assay ID: Hs00183222_m1) was carried out using TaqMan Gene Expression Assays (Applied Biosystems, Melbourne, Australia) as instructed by the manufacturer. Housekeeping gene RPLP0 (Applied Biosystems) was used as a control. Relative gene expression = gene expression in the knockdown group / average gene expression in the control group. All experiments were performed in three biologic replicates.
FOXA1 Knockdown in Cell Lines
FOXA1 knockdown (KD) was carried out as described before [30] in MDA-MB-453 and HCC-1954 cells using the following small interfering RNA (siRNA) oligos (duplex; Sigma-Aldrich, Sydney, Australia): D1, 5′CACACAAACCAAACCGUCA; D2, 5′UGACGGUUUGGUUUGUGUG. TLE3 KD was carried out using the following two sets of siRNA oligos (duplex; Sigma-Aldrich): set 1: D1, 5′GUCAUCUACUAAACAAGAA; D2, 5′UUCUUGUUUAGUAGAUGAC; and set 2: D1, 5′GUCAUCUACUAAACAAGAA; D2, 5′UUCUUGUUUAGUAGAUGAC. Transfection of siRNA oligos using Lipofectamine RNAiMAX (Invitrogen) was carried out with a reverse transfection method as instructed by the manufacturer. The final siRNA duplex concentration was 20 nM for the KD experiments. Cells transfected with siRNA Universal Negative Control No. 1 (Sigma-Aldrich) were used as controls. In all experiments, the effects of KDs were assessed 72 hours after the siRNA transfections.
Epidermal Growth Factor Pathway Arrays
TaqMan Human Epidermal Growth Factor (EGF) Pathway Arrays constituting of 96 genes were obtained from Applied Biosystems (Part No: 4418774, the layout of this array is available under the following link: http://www3.appliedbiosystems.com/cms/groups/portal/documents/web_content/dev_043045.gif). Transfections with FOXA1-siRNA oligos and control-siRNA were carried out in MDAMB-453 and HCC-1954 cell lines as described previously. Seventy-two hours after transfections, total RNA was extracted for RT-PCR analysis. The efficiency of FOXA1-KD was validated using RT-PCR as described previously. A total of eight arrays were applied for two biologic replicate experiments with each of the FOXA1-siRNA and controlsiRNA oligos in MDA-MB-453 and HCC-1954 cell lines (four arrays per cell line).
After RT-PCR experiments using human EGF pathways arrays, cycle threshold (CT) values in each array plate were normalized to the mean CT values of four housekeeping genes on the plates (18S, GAPDH, HPRT1, and GUSB) to obtain ΔCT values for each gene. Next, the average ΔCT values for the control-siRNA plates in each cell line were deducted from the ΔCT values of the corresponding genes on FOXA1-siRNA plates to obtain ΔΔCT values. Finally, relative gene expressions were calculated using 2-ΔΔCT formula as described before [31].
Luciferase Reporter Assays
Full-length complementary DNA (cDNA) clones for CREB1, c-Jun, c-Fos, and AP2α were obtained from Open Biosystems (Thermo Scientific). Full-length cDNA clone for Elk1 was obtained from GeneCopoeia (Rockville, MD). The clones were validated by restriction digestion/sequencing and then subcloned into a pcDNA3.1 vector (Invitrogen) to generate expression constructs. The sequence of 1.5-kb promoter region of FOXA1 was obtained using Ensembl Genome Browser (http://www.ensembl.org/index.html) and PCR generated using the following primer set: forward, GCGCGGTACCCGCAGTGCAGATGCGTTCCC; reverse, GCGCGAGCTCACCTCCTGCGTGTCTGCGTAGT. Subsequently, FOXA1 promoter was cloned into a pGL3 luciferase reporter vector (Promega, Sydney, Australia) and validated by restriction digestion/sequencing.
To carry out the reporter assays, MCF-7 cells were cotransfected with the FOXA1 reporter vector and each of the transcription factors using ExGen 500 reagent (Fermentas Life Sciences, St Leon-Rot, Germany). The Renilla pRL-TK vector (Promega) was used as an internal control reporter. Cotransfection with FOXA1 reporter vector and an empty pcDNA vector was used as a control. Fortyeight hours after the transfections, reporter activities were measured using Dual-Glo Luciferase Assay System (Promega) in an Orion II Microplate Luminometer (Berthold Detection Systems, Pforzheim, Germany). The response ratios for transcription factors and control were measured relative to the internal control reporter (relative response ratio). All reporter assays were carried out in four biologic replicates.
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) assays were performed in MDA-MB-453 cell line using ChIP Assay Kit (USB Corporation, Cleveland, OH) as instructed by the manufacturer [32]. ChIP-grade rabbit monoclonal CREB1 (ChIPAb+ CREB Kit; Millipore, Melbourne, Australia), rabbit polyclonal AP2α (Abcam), rabbit polyclonal c-Fos (Abcam), rabbit polyclonal FOXA1 (Abcam), and rabbit polyclonal TLE3 (Proteintech, Chicago, IL) antibodies were applied at 4 µg per assay.
Two primer sets for each of the FOXA1, RELB, and PAK1 promoters were used for the end point RT-PCR amplification using SYBR green method (Applied Biosystems). FOXA1 promoter primers included the following: forward primer set 1, TCACTTGGCTCGGCTGACT (start: -960); reverse primer set 1, AGCCCCACTTTTGCTTCGT (start: -902); forward primer set 2, CTCCCCATTTCCCTCTTTCC (start site: -565); and reverse primer set 2, TGTTTGCAAAGCAGTGTAATTGG (start site: -505). RELB promoter primers included the following: forward primer set 1, CGCCCGGCCTCCAA (start site: -714); reverse primer set 1, GACCAGCGCCTGGAACAC (start site: -664); forward primer set 2, GGTAAGGCGTGATGGCTCTAAG (start site: -381); and reverse primer set 2, CGTCGCTGCCCATTGC (start site: -319). PAK1 promoter primers included the following: forward primer set 1, CTCTTGAGCCCTGGAGTTTGA (start site: -877); reverse primer set 1, CAGGCTGGAGTGCAGTGGTA (start site: -815); forward primer set 2, TGAATGGGTGTGGCTGTGTT (start site: -567); and reverse primer set 2, TGGACTGTATCAAGGGTCAGGAA (start site: -494). Amplification of input chromatin before immunoprecipitation at a dilution of 1:50 was used as a positive control. ChIP assays using nonspecific antibody (rabbit IgG) served as a negative control. The assays were carried out in three replicates and copy number changes were calculated as -Log2 value for each experimental set.
Cell Viability Assay
FOXA1-KD in MDA-MB-453 and HCC-1954 cells was carried out using reverse transfection as described previously. A total of 10,000 cells transfected with either FOXA1-siRNA or control-siRNA were seeded per each well of a 96-well plate. Seventy-two hours after transfections, cell viability was assessed using Vybrant MTT Proliferation Assay Kit (Invitrogen) as instructed by the manufacturer. MTT assays were performed in eight biologic replicates, and absorbance at 570 nm was measured using a plate reader.
Bioinformatics and Statistical Analysis
Analysis of gene expression correlations. Expressions of FOXA1, AP2α, and ErbB2 in a total of 30 ER- breast cancer cell lines were extracted from the microarray data published by Neve et al. [33]. Expression data were normalized to the median expression of each gene across the cohort of ER- cell lines. Correlation coefficients between the normalized expression values of FOXA1 with those of ErbB2 and AP2α were obtained using the Pearson method.
Promoter analysis. Sequences of the 1-kb promoter region of FOXA1, RELB, and PAK1 genes were obtained using Ensembl Genome Browser (http://www.ensembl.org/index.html). Identification of putative binding sites in the promoter regions was carried out using PATCH public 1.0 software (http://www.gene-regulation.com/cgibin/pub/programs/patch/bin/patch.cgi).
Bioinformatics and statistical analysis. Heat map was generated using Spotfire DecisionSite for Functional Genomics (TIBCO, Somerville, MA). Biostatistical analysis was carried out using IBM SPSS Statistics 20 (Armonk, NY). The Mann-Whitney U test was applied for the comparison of nonparametric data. All error bars depict ±2 SEM.
Results
FOXA1 Is Frequently Overexpressed in ErbB2+/ER- Breast Tumors
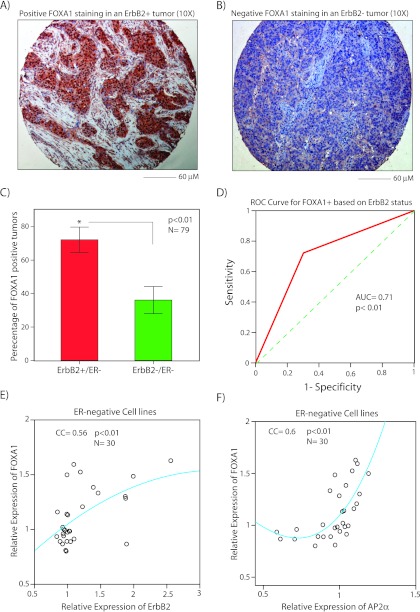
To assess the association between FOXA1 and ErbB2 overexpression in ER- breast cancer, we carried IHC staining for FOXA1 protein expression in a TMA cohort of 79 ER- tumors with known ErbB2 status as described in the Materials and Methods. This cohort constituted of a total of 36 ErbB2+ tumors defined as 3+ IHC staining for ErbB2 and 43 ErbB2- tumors. After IHC scoring of FOXA1 staining, we compared the percentage of FOXA1+ tumors, as defined by S > 15, between ErbB2+ and ErbB2- groups. Notably, we observed that ErbB2+ tumors had approximately 2.5-fold higher frequency of FOXA1 overexpression at 72% ± 7% compared with that of ErbB2- tumors at 30% ± 7% (P < .01; Figure 1, A–C). In addition, we analyzed the data using a cutoff of S > 24 as applied by Mehta et al. [34]. This approach also demonstrated a higher level of FOXA1 expression in ErbB2+ compared with ErbB2- tumors (Figure W1).
Figure 1.
The association between FOXA1 and ErbB2 expression in ER-negative breast tumors and cell lines. FOXA1 IHC in a breast tumor with ErbB2 overexpression (ErbB2+; A) and in an ErbB2- breast tumor (B). Magnifications, x10. (C) Percentage of FOXA1+ staining using IHC in ErbB2+/ER- and ErbB2-/ER- breast tumors. *P < .01 is for ErbB2+ versus ErbB2- groups. Error bars, ±2 SEM. (D) Receiver operating characteristic analysis to test ErbB2 status as a predictor for FOXA1 overexpression (FOXA1+) in ER- breast tumors. Dashed line is a diagonal reference line. (E) Scatterplot to demonstrate the correlation between the relative expression of FOXA1 and ErbB2 in ER- cell lines. Regression fitted line using the quadratic method is depicted. (F) Scatterplot to demonstrate the correlation between relative expression of FOXA1 and AP2a as described in E.
We next tested whether FOXA1 overexpression could be predicted based on ErbB2 status using receiver operating characteristic analysis. This method demonstrated an area under the curve (AUC) of 0.71 for predicting FOXA1+ tumors based on ErbB2 status, indicating that ErbB2 status can classify FOXA1 expression pattern in ER. breast tumors (P < .01; Figure 1D). Altogether, these findings suggest that FOXA1 overexpression is associated with ErbB2+ status in ER- breast tumors.
FOXA1 Expression Correlates with ErbB2 and AP2α Transcript Levels in ER- Cell Lines
We further assessed the association between FOXA1 and ErbB2 expression patterns using the expression microarray data available from a cohort of thirty ER- breast cancer cell lines [33]. In addition, we examined a possible association between the expression of FOXA1 and AP2α using this data set. AP2α is a transcription factor with an established role in the regulation of ErbB2 expression and is also a top-ranking gene in the steroid-response gene signature [3,35–40]. Therefore, a possible association between FOXA1 and AP2α expression would have potential biologic significance in ErbB2+/ER- breast cancer.
The expression data for FOXA1, ErbB2 and AP2α were extracted and normalized as described in the Materials and Methods (Table W1). We next measured Pearson correlation coefficients (CCs) between the expression of FOXA1 and that of ErbB2 and AP2α. Notably, FOXA1 expression had significant correlations with the expression of both ErbB2 (CC = 0.56, P < .01; Figure 1E) and AP2α (CC = 0.6, P < .01; Figure 1F). These data further confirm a robust association between FOXA1 and ErbB2 expression patterns in ER- breast cancer. In addition, AP2α and ErbB2 expressions have a similar level of correlation with that of FOXA1 in ER- cell lines.
ERK Regulates FOXA1 Expression
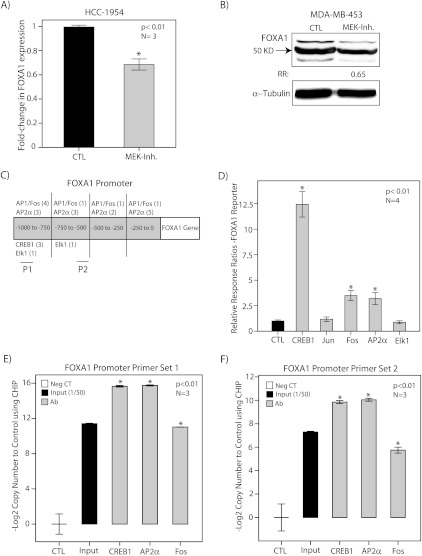
We next investigated the underlying mechanism for the observed overexpression of FOXA1 in ErbB2+/ER- breast tumors. A possible mechanism for this association is the regulation of FOXA1 transcription by the ErbB2 signaling pathway. To assess this possibility, we first examined the effect of ERK, an important downstream mediator of ErbB2 signaling, on FOXA1 expression using MEK inhibitor CI-1040 [9]. MDA-MB-453 and HCC-1954 cell lines were treated with CI-1040 at 10 µM concentration for 24 hours, and vehicle (DMSO)-treated cells were used as controls. FOXA1 expression was then assessed using RT-PCR and Western blot analysis. We observed that MEK inhibition reduced the level of FOXA1 transcript and protein levels by approximately 35% (Figure 2, A and B). These findings suggest that ERK signaling regulates FOXA1 expression in molecular apocrine cells.
Figure 2.
The regulation of FOXA1 expression by ErbB2- ERK signaling. (A) Relative expression of FOXA1 using RT-PCR in HCC-1954 cell line after treatment with CI-1040 (MEK Inh.) at 10 µM concentration for 24 hours. Expression is relative to that of vehicle-treated cells (CTL). *P < .01 is for CI-1040 versus control. (B) FOXA1 protein levels using Western blot analysis in MDA-MB-453 cell line after treatment with CI-1040 as described in A. (C) Putative transcription factor binding sites for CREB1, c-Jun, c-Fos, AP2α, and Elk1 in 1-kb promoter region of FOXA1. P1 (primer set 1) and P2 (primer set 2) are regions of amplification for ChIP assays. (D) Luciferase reporter assay. The transcriptional activation of FOXA1 promoter by CREB1, c-Jun, c-Fos, AP2α, and Elk1 expression constructs was assessed using dual-luciferase assays in MCF-7 cells, and relative response ratios are reported. Cotransfection with the FOXA1 reporter vector and an empty pcDNA vector was used as a control. *Compared with the control group. (E) ChIP assay using CREB1, AP2α, and c-Fos antibodies and primer set 1 in MDA-MB-453 cell line. The results of end point RT-PCR amplification for ChIP assay are demonstrated with primer set 1 for FOXA1 promoter. Input indicates input chromatin at a dilution of 1:50; Neg. CT, nonspecific antibody. The relative copy number changes to control are shown as -Log2 values. *P < .01 is for each antibody versus Neg. CT. (F) ChIP assay using CREB1, AP2α, and c-Fos antibodies and primer set 2 as described in E. All error bars, ±2 SEM.
FOXA1 Is a Target of CREB1, AP2α, and c-Fos Transcription Factors
To further identify a mechanism for our observations, we studied whether FOXA1 is regulated by some of the key transcription factors in the ErbB2-ERK signaling pathway. In this respect, we examined CREB1, Elk1, c-Jun, and c-Fos, which are well-characterized downstream mediators of ErbB2-ERK signaling [41–46]. In addition, we investigated a potential role for AP2α in the regulation of FOXA1 because we observed a significant correlation between AP2α and FOXA1 expression (Figure 1F) as well as the fact that AP2α is a critical regulator of ErbB2 transcription [35–40].
Examination of 1-kb FOXA1 promoter region identified several putative binding sites for the previously mentioned transcription factors (Figure 2C). We subsequently used luciferase reporter assays to examine the effects of these predicted transcription factors on the regulation of FOXA1 promoter. Owing to a high degree of transfectability, MCF-7 cells were used for the reporter assay experiments [9,32]. MCF-7 cells were cotransfected with the FOXA1 reporter vector and each of the CREB1, c-Jun, c-Fos, AP2α, and Elk1 expression constructs. Cotransfection with the FOXA1 reporter vector and an empty pcDNA vector was used as a control. Forty-eight hours after the transfections, reporter activities were measured, and relative response ratios were calculated as described in Materials and Methods. We observed a marked increase in FOXA1 reporter activity with CREB1 by approximately 12-fold (P < .01; Figure 2D). In addition, there was an approximately three-fold increase in FOXA1 reporter activity with AP2α and c-Fos constructs (P < .01; Figure 2D). However, c-Jun and Elk-1 did not activate the FOXA1 promoter (Figure 2D).
To assess the binding of CREB1, AP2α, and c-Fos to FOXA1 promoter, we next carried out ChIP assays in MDA-MB-453 cell line. Two sets of primers for FOXA1 promoter in proximity to the predicted binding sites were used for the end point RT-PCR amplification (Figure 2C). Amplification of input chromatin at a dilution of 1:50 and ChIP using nonspecific antibody (rabbit IgG) were applied as positive and negative controls, respectively. Copy number changes were calculated as -Log2 value for each experimental set (Figure 2, E and F). Importantly, we observed a significant enrichment for FOXA1 promoter region with each of the CREB1, AP2α, and c-Fos antibodies using both primer sets (P < .01; Figure 2, E and F). Altogether, these data demonstrate that FOXA1 is a target gene of CREB1, AP2α, and c-Fos transcription factors.
FOXA1 Regulates a Gene Signature in the ErbB2 Signaling Pathway
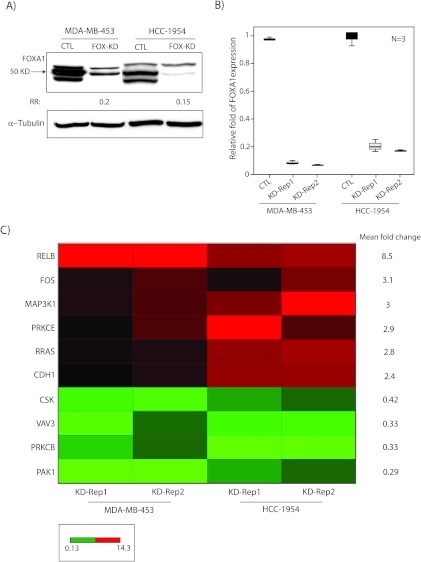
We next explored whether FOXA1 has a role in transcriptional regulation of the EGFR-ErbB2 signaling pathway. To investigate this possibility, we assessed the effect of FOXA1-KD on the expression of EGFR-ErbB2 signaling genes in MDA-MB-453 and HCC-1954 cell lines using Human EGF Pathway Arrays as described in Materials and Methods. FOXA1-KD was carried out using a siRNA duplex, and the efficiency of KD was evaluated by Western blot analysis and RT-PCR. We observed at least 80% reduction of FOXA1 transcript and protein levels after FOXA1-KD (Figure 3, A and B). It is notable that RT-PCR validations of FOXA1-KD were carried out in all replicate experiments applied for Human EGF Pathway Arrays (Figure 3B).
Figure 3.
FOXA1-regulated gene signature in ErbB2 signaling. (A) Western blot analysis to show FOXA1 protein level after FOXA1-KD (FOX-KD) in MDA-MB-453 and HCC-1954 cell lines. Fold changes (RR) in band densities were measured relative to the control (CTL). (B) RT-PCR to demonstrate FOXA1-KD efficiencies in MDA-MB-453 and HCC-1954 cell lines. FOXA1 expression after KD was assessed relative to nontargeting siRNA control (CTL), and fold change is shown for each cell line and replicate experiment. Rep1 indicates replicate 1; Rep2, replicate 2. (C) Heat map of FOXA1-regulated gene signature in ErbB2 signaling generated using RT-PCR data. Heat map is constituted of genes with more than two-fold change in expression after FOXA1-KD (KD) in both MDA-MB-453 and HCC-1954 cell lines. Mean fold change in expression is shown for each gene. Red and green depict up-regulation and down-regulation, respectively. Bar indicates the range of fold changes in gene expression.
We subsequently carried out RT-PCR using Human EGF Pathway Arrays and analyzed the expression data as described in Materials and Methods. We identified genes with more than two-fold change in expression between FOXA1-KD and control experiments. Importantly, there was a core gene signature consisting of ten genes that showed more than two-fold change in expression after FOXA1-KD in both MDA-MB-453 and HCC-1954 cell lines (Figure 3C). This FOXA1-regulated gene signature included six upregulated and four downregulated genes (Figure 3C). The upregulated genes were RELB (8.5-fold), c-Fos (3.1-fold), MAP3K1 (3-fold), PRKCE (2.9-fold), R-Ras (2.8-fold), and CDH1 (2.4-fold), with mean fold increase in their expression levels across two cells lines ranging from 2.4- to 8.5-fold (Figure 3C). The downregulated genes included PAK1 (0.29), PRKCB (0.33-fold), VAV3 (0.33-fold), and CSK (0.42-fold), with mean fold reduction in their expression levels across two cells ranging from 0.29- to 0.42-fold (Figure 3C).
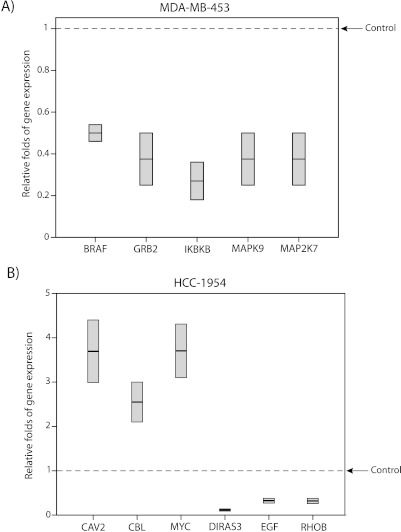
Moreover, we identified a separate group of genes, with more than a two-fold change in expression after FOXA1-KD, which were different between MDA-MB-453 and HCC-1954 cell lines (Figure 4, A and B). This differentially regulated gene list in MDA-MB-453 cell line included BRAF, GRB2, IKBKB, MAPK9, and MAP2K7 that were downregulated by 0.27- to 0.49-fold (Figure 4A). In HCC-1954 cell line, CAV2, CBL, and MYC were upregulated by 2.6- to 3.7-fold and DIRAS3, EGF, and RHOB were downregulated by 0.12- to 0.33-fold (Figure 4B).
Figure 4.
ErbB2 signaling genes that are differentially regulated by FOXA1 between molecular cell lines. (A) RT-PCR to demonstrate the relative folds of gene expression to control after FOXA1-KD in MDA-MB-453 cell line. Box plots show genes with more than two-fold change in expression after FOXA1-KD, which were unique to MDA-MB-453 cell line. (B) RT-PCR to demonstrate the relative folds of gene expression to control after FOXA1-KD in HCC-1954 cell line. Box plots to show genes with more than two-fold change in expression after FOXA1-KD, which were unique to HCC-1954 cell line.
These findings suggest that FOXA1 regulates the transcription of a gene signature in the ErbB2 signaling pathway. In addition, FOXA1 has both positive and negative regulatory effects on ErbB2 signaling genes as evidenced by down-regulation and up-regulation of these genes after FOXA1-KD, respectively.
Differences in FOXA1-Regulated Genes across Cell Lines Have Functional Implications
Because there are a group genes differentially regulated by FOXA1 between MDA-MB-453 and HCC-1954 cell lines (Figure 4, A and B), we next investigated whether these differences in FOXA1-regulated genes have any functional implications in molecular apocrine cells. To assess this, we examined the effect of FOXA1-KD on ERK phosphorylation and cell viability using Western blot analysis and MTT assay, respectively.
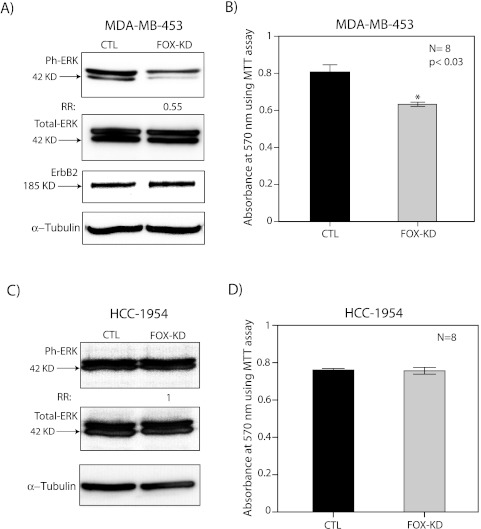
In MDA-MB-453 cell line, FOXA1-KD reduced the level of ERK phosphorylation by 45% compared with the control without any change in total ERK level (Figure 5A). Moreover, this effect was associated with a significant decrease in cell viability by approximately 25% (P < .03; Figure 5B). It is notable that ErbB2 level did not change after FOXA1-KD, indicating that ErbB2 expression is not regulated by FOXA1 in this cell line (Figure 5A). In contrast, FOXA1-KD did not affect the level of ERK phosphorylation and cell viability in HCC-1954 cell line (Figure 5, C and D). These findings suggest that FOXA1 expression is necessary for maintaining ERK phosphorylation and cell viability in MDA-MB-453 line as opposed to HCC-1954 line. Therefore, the functional effects of FOXA1 can vary across molecular apocrine cells.
Figure 5.
The effect of FOXA1 KD on ERK phosphorylation and cell viability. (A) Western blot analysis to measure the level of phosphorylated ERK (ph-ERK), total ERK, and ErbB2 after FOXA1-KD (FOX-KD) in MDA-MB-453 cell line. Fold change was assessed relative to nontargeting siRNA control (CTL). (B) MTT assay to measure cell viability after FOX-KD in MDA-MB-453 cell line. CTL indicates nontargeting siRNA control. *P < .03 is for FOX-KD versus CTL. (C) Western blot analysis to measure the level of phosphorylated ERK (ph-ERK) and total ERK after FOX-KD in HCC-1954 cell line. (D) MTT assay to measure cell viability after FOX-KD in HCC-1954 cell line. All error bars, ±2 SEM.
RELB and PAK1 Are FOXA1 Target Genes in ErbB2 Signaling
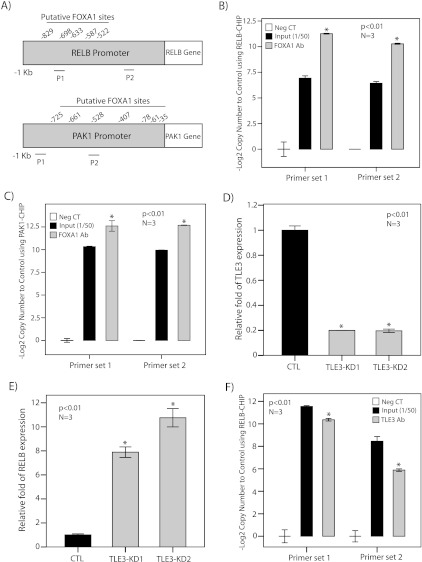
We subsequently assessed whether selective signature genes are direct targets of FOXA1. For this purpose, we examined FOXA1 binding to the promoters of RELB and PAK1, which were the most upregulated and downregulated genes in the gene signature, respectively (Figure 3C). It is notable that examination of 1-kb promoter regions of RELB and PAK1 identified several putative binding sites for FOXA1 (Figure 6A).
Figure 6.
FOXA1 binding to RELB and PAK1 promoters and TLE3 regulation of RELB. (A) Putative FOXA1 binding sites in 1 kb promoter regions of RELB and PAK1. P1 (primer set 1) and P2 (primer set 2) are regions of amplification for ChIP assays. (B) ChIP assay for RELB promoter using FOXA1 antibody in the MDA-MB-453 cell line. The results of end point RT-PCR amplification for ChIP assay are shown with two sets of primers for RELB promoter. Input indicates input at a dilution of 1:50; Neg. CT, nonspecific antibody. The relative copy number changes to control are shown as -Log2 values. *P < .01 is for FOXA1 Ab versus Neg. CT. (C) ChIP assay for PAK1 promoter using FOXA1 antibody. The results of end point RT-PCR amplification for ChIP assay are shown with two sets of primers for PAK1 promoter as described in B. (D) RT-PCR to demonstrate TLE3-KD efficiencies with two sets of siRNA duplexes (KD1 and KD2) in MDA-MB-453 cell line. TLE3 expression after KD was assessed relative to nontargeting siRNA control (CTL), and the fold change is shown for each duplex. (E) RT-PCR to show RELB expression levels after TLE3-KD with two sets of siRNA duplexes as described in D. (F) ChIP assay for RELB promoter using TLE3 antibody. The results of end point RT-PCR amplification for ChIP assay are shown with two sets of primers for RELB promoter as described in B. *P < .01 is for TLE3 Ab versus Neg. CT. All error bars, ±2 SEM.
To assess FOXA1 binding to the promoters of RELB and PAK1, we carried out ChIP assays using FOXA1 antibody in MDA-MB-453 cell line. Two sets of primers in proximity to the predicted binding sites were used for each of the RELB and PAK1 promoters to assess the end point RT-PCR amplification (Figure 6A). Amplification of input chromatin at a dilution of 1:50 and ChIP using nonspecific antibody (rabbit IgG) were applied as positive and negative controls, respectively. Copy number changes were calculated as -Log2 value for each experimental set (Figure 6, B and C). ChIP assays demonstrated significant binding of FOXA1 to the promoters of RELB and PAK1 (P < .01; Figure 6, B and C). Moreover, the results were reproducible using both primer sets for each of the promoters (Figure 6, B and C). These findings indicate that RELB and PAK1 are FOXA1 target genes in the ErbB2 signaling pathway.
FOXA1 Binding Partner Corepressor TLE3 Regulates RELB
Our results indicate that FOXA1 negatively regulates a group of genes in the ErbB2 signaling pathway such as RELB. It has been shown that FOXA1 can act as a pioneer factor to recruit transcriptional corepressor TLE3 resulting in gene repression distal to the recruitment site [47,48]. To investigate whether this recruitment of TLE3 would provide a possible mechanism for the negative regulatory effects of FOXA1 observed in our study, we examined the role of TLE3 in the regulation of RELB as an example of a target gene repressed by FOXA1.
We first investigated the effect of TLE3-KD on RELB expression in MDA-MB-453 cell line. TLE3-KD was carried out using two sets of siRNA duplexes and the efficiency of KD experiments were assessed using RT-PCR. We observed approximately 80% reduction in the level of TLE3 expression with both siRNA duplexes (Figure 6D). We next measured RELB expression levels after TLE3-KD using RT-PCR. Notably, there was a significant increase in RELB expression by 8- to 11-fold after TLE3-KD that was reproducible with both sets of TLE3-siRNA duplexes (P < .01; Figure 6E).
We subsequently tested whether TLE3 can be recruited to the RELB promoter regions that bind FOXA1 using ChIP assays with the same primer sets applied for FOXA1-RELB ChIP experiments. Amplification of input chromatin at a dilution of 1:50 and ChIP using nonspecific antibody (rabbit IgG) were applied as positive and negative controls, respectively. Copy number changes were calculated as -Log2 value for each experimental set (Figure 6F). Importantly, we observed significant enrichment for RELB promoter region using TLE3 antibody with both primer sets (P < .01; Figure 6F). These results indicate that, in a similar fashion to FOXA1, TLE3 negatively regulates RELB and is recruited to the same promoter regions of this gene.
Discussion
Molecular apocrine is a subtype of breast cancer that is characterized by the overexpression of steroid-response genes such as AR and FOXA1 [2–4]. A key feature of this subtype is a high prevalence of ErbB2 overexpression, which is present in more than 50% of cases [2,3,5]. In fact, ErbB2 has a significant biologic role in molecular apocrine subtype, including a cross talk with the AR signaling pathway that regulates cell growth and survival [8–10]. FOXA1 is another notable gene in molecular apocrine signature that has recently gained much attention with respect to its role as a modulator of steroid receptors [14–18]. However, there are limited data available regarding the functional role and regulation of FOXA1 in ER- breast cancer.
In this study, we demonstrated that there is a strong association between the overexpression of FOXA1 and ErbB2 at both protein and transcription levels in ER- breast tumors (Figure 1). These findings are further supported by expression microarray studies that have shown FOXA1 expression is present in most ErbB2+ breast tumors and cell lines [22,49]. In addition, we observed that FOXA1 expression is induced by heregulin [8] and reduced by MEK inhibition in molecular apocrine cells (Figure 2, A and B). Altogether, these data suggest a role for ErbB2 signaling in the regulation of FOXA1 transcription. Therefore, we sought to identify an interaction between FOXA1 and ErbB2 signaling in ER- breast cancer.
Our findings suggest that ErbB2 signaling regulation of FOXA1 is mediated through transcription factors CREB1 and c-Fos (Figure 2, D–F). CREB1 is a critical downstream mediator of the EGFR-ErbB2 pathway, which is activated by both Akt and ERK signaling [42–44,50]. It is notable that we have previously identified AR as another CREB1 target gene [9]. Moreover, CREB1 strongly activates both FOXA1 and AR promoters by more than 10-fold (Figure 2D and Chia et al. [9]). These findings suggest that CREB1 acts as a key transcription factor in the regulation of steroid-response genes. C-Fos is the second transcription factor in the ErbB2 signaling that activates FOXA1. C-Fos is a part of AP1 family of transcription factors that is activated by ERK-mediated phosphorylation and increase in mRNA expression [46,51]. Interestingly, c-Fos is also one of the upregulated genes following FOXA1-KD, suggesting that c-Fos expression itself is negatively regulated by FOXA1 (Figure 3C).
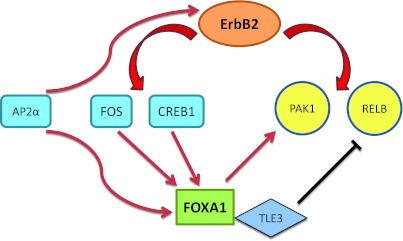
Another underlying mechanism that explains the association between FOXA1 and ErbB2 overexpression is the role of transcription factor AP2α in regulating both of these genes. AP2α is an established transcriptional regulator of ErbB2 that contributes to the overexpression of this gene in breast tumors [35–40]. In addition, we have demonstrated that FOXA1 is an AP2α target gene, and there is a significant correlation between the expression patterns of these two genes in a cohort of 30 ER- cell lines (Figures 2, D–F, and 1F). The expression data used for this correlation analysis was derived from the largest genomics data set available for ER-negative cell lines [33]. It is notable that AP2α itself is a member of steroid-response signature [3], suggesting that a network of interacting transcription factors including AR, FOXA1, and AP2α plays a significant role in the biology of molecular apocrine breast cancer. Altogether, we have identified two layers of transcriptional regulation to explain the association between FOXA1 and ErbB2 overexpression in molecular apocrine cells. In one layer, ErbB2 signaling genes CREB1 and c-Fos regulate FOXA1 transcription and in another layer, AP2α regulates the expression of both FOXA1 and ErbB2 genes (Figure 7).
Figure 7.
Schematic diagram of a cross-regulation network between FOXA1 and the ErbB2 signaling pathway in ER-negative breast cancer. Red and black arrows denote stimulatory and inhibitory effects, respectively.
Moreover, our data suggest that FOXA1, in turn, regulates the transcription of ErbB2 signaling genes inmolecular apocrine cells (Figures 3 and 4). FOXA1-regulated genes include a core gene signature that is shared across two molecular apocrine cell lines. Importantly, FOXA1-KD results in both down-regulation and up-regulation of ErbB2 signaling genes, suggesting that FOXA1 has both positive and negative transcriptional activities in molecular apocrine cells. Furthermore, the most upregulated (RELB) and downregulated (PAK1) genes in the signature are direct FOXA1 targets (Figure 6, B and C). These findings indicate that FOXA1 acts as a dual-function transcription factor with both activating and repressive activities (Figure 7). This pattern of gene regulation is in agreement with a recent study that showed FOXA1-KD results in a similar number of downregulated and upregulated genes in prostate cancer cells [15]. In fact, the repressive function of FOXA1 on RELB can be explained by the recruitment of its binding partner corepressor TLE3 (Figure 6, D–F). In this process, FOXA1 recruitment of TLE3 leads to gene repression distal to the recruitment site [47]. In addition, the removal of FOXA1 as a pioneering factor by KD can result in transcriptional reprogramming mediated through other transcription factors [15,19].
It is notable that a group of FOXA1-regulated genes vary across molecular apocrine cell lines leading to the differences in the functional effects of FOXA1 between these lines (Figures 4 and 5). In MDA-MB-453 cells, FOXA1 expression is necessary for ERK phosphorylation and maintaining cell viability. These effects can be explained by the fact that most of FOXA1-regulated genes in this cell line, including key ERK signaling mediators GRB2 and BRAF, are positively regulated by this gene (Figures 3C and 4A). It is known that GRB2, which mediates Ras activation, and BRAF, which is a Ras effector, are both required for full activation of ERK [52,53]. Therefore, down-regulation of these genes can explain the reduction in ERK phosphorylation and cell viability after FOXA1-KD in MDA-MB-453 cells. In contrast, FOXA1 expression is not necessary to maintain ERK phosphorylation and cell viability in HCC-1954 cell line, and this corresponds with the fact that FOXA1-KD results in a similar number of positively and negatively regulated genes in this cell line, which does not include GRB2 and BRAF (Figures 3C and 4B). These observations point out to the diversity of FOXA1 functional activities in molecular apocrine cells.
We identified RELB as a FOXA1 target gene that is markedly repressed by FOXA1 and TLE3 expression in molecular apocrine cells (Figures 3C and 6). RELB is a member of NF-κB family that can be activated by the EGFR-ErbB2 signaling pathway [54–56]. In addition, we observed that FOXA1 regulates another NF-κB-related gene IKBKB (IκB kinase B) in MDA-MB-453 cells (Figure 4A). Most widely studied mechanisms for the regulation of NF-κB activity involve changes in the phosphorylation levels of IκBα and RELA/p65 proteins [57]. Our results suggest that FOXA1-mediated transcriptional regulation of RELB may provide an alternative mechanism for the regulation of NF-κB pathway. It is notable that the NF-κB family of proteins has a significant role in the biology of ErbB2+/ER- breast cancer [55]. In addition to being involved in the ErbB2 signaling, some of the FOXA1-regulated genes such as RELB and IKBKB are also part of other signaling pathways such as NF-κB. Therefore, FOXA1 may play a regulatory function in interactions between some of the key signaling pathways such as AR, ErbB2, and NF-κB in molecular apocrine cells.
Furthermore, PAK1 (p21-activated kinase 1) represents a direct target of FOXA1 that is positively regulated by this gene (Figures 3C and 6C). PAK1 interacts with the GRB2 adapter protein, and this association is enhanced by the induction of EGFR signaling [58]. Considering that FOXA1 positively regulates GRB2 in one of the molecular apocrine lines (Figure 4A), this combined regulation of GRB2 and PAK1 can significantly modulate the downstream ERK signaling pathway.
In summary, we have identified a cross-regulation network between FOXA1 and the ErbB2 signaling pathway (Figure 7). Importantly, this cross-regulation connects FOXA1 to some of the key signaling elements in ER- breast cancer such as ERK-CREB1 axis, NF-κB genes, and transcription factor AP2α. Therefore, this novel signaling network has a critical function in the biology of ER- breast cancer and may provide a potential therapeutic target in this disease.
Supplemental Materials and Methods
Footnotes
This study is funded by grants from Princess Alexandra Hospital Private Practice Trust Fund, Cancer Collaborative Group, and Cancer Council Queensland.
This article refers to supplementary materials, which are designated by Table W1 and Figure W1, and are available online at www.neoplasia.com.
References
- 1.Putti TC, El-Rehim DM, Rakha EA, Paish CE, Lee AH, Pinder SE, Ellis IO. Estrogen receptor-negative breast carcinomas: a review of morphology and immunophenotypical analysis. Mod Pathol. 2005;18:26–35. doi: 10.1038/modpathol.3800255. [DOI] [PubMed] [Google Scholar]
- 2.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 3.Teschendorff AE, Naderi A, Barbosa-Morais NL, Caldas C. PACK: profile analysis using clustering and kurtosis to find molecular classifiers in cancer. Bioinformatics. 2006;15:2269–2275. doi: 10.1093/bioinformatics/btl174. [DOI] [PubMed] [Google Scholar]
- 4.Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, Gerald WL. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. doi: 10.1038/sj.onc.1209415. [DOI] [PubMed] [Google Scholar]
- 5.Park S, Koo J, Park HS, Kim JH, Choi SY, Lee JH, Park BW, Lee KS. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2010;21:488–492. doi: 10.1093/annonc/mdp510. [DOI] [PubMed] [Google Scholar]
- 6.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses' Health Study. Mod Pathol. 2011;24:924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niemeier LA, Dabbas DJ, Beriwal S, Striebel JM, Bhargava R. Androgen receptor in breast cancer: expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol. 2009;23:205–212. doi: 10.1038/modpathol.2009.159. [DOI] [PubMed] [Google Scholar]
- 8.Naderi A, Hughes-Davies L. A functionally significant cross-talk between androgen receptor and ErbB2 pathways in estrogen receptor negative breast cancer. Neoplasia. 2008;10:542–548. doi: 10.1593/neo.08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chia KM, Liu J, Francis GD, Naderi A. A feedback loop between androgen receptor and ERK signaling in estrogen receptor-negative breast cancer. Neoplasia. 2011;13:154–166. doi: 10.1593/neo.101324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, Rimm DL, Liu XS, Brown M. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naderi A, Chia KM, Liu J. Synergy between inhibitors of androgen receptor and MEK has therapeutic implications in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13:R36. doi: 10.1186/bcr2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naderi A, Liu J. Inhibition of androgen receptor and Cdc25A phosphatase as a combination targeted therapy in molecular apocrine breast cancer. Cancer Lett. 2010;298:74–87. doi: 10.1016/j.canlet.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Gucalp A, Traina TA. Triple-negative breast cancer: role of the androgen receptor. Cancer J. 2010;16:62–65. doi: 10.1097/PPO.0b013e3181ce4ae1. [DOI] [PubMed] [Google Scholar]
- 14.Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. EMBO J. 2011;30:3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, Sankila A, Turunen JP, Lundin M, Konsti J, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, Carroll JS. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakshatri H, Badve S. FOXA1 in breast cancer. Exp Rev Mol Med. 2009;11:e8. doi: 10.1017/S1462399409001008. [DOI] [PubMed] [Google Scholar]
- 18.Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the cover: location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sircoulomb F, Bekhouche I, Finetti P, Adelaide J, Ben Hamida A, Bonansea J, Raynaud S, Innocenti C, Charafe-Jauffret E, Tarpin C, et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer. 2010;10:539. doi: 10.1186/1471-2407-10-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naderi A, Liu J, Francis GD. A feedback loop between BEX2 and ErbB2 mediated by c-Jun signaling in breast cancer. Int J Cancer. 2012;130:71–82. doi: 10.1002/ijc.25977. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, Badve S. Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol. 2008;61:327–332. doi: 10.1136/jcp.2007.052431. [DOI] [PubMed] [Google Scholar]
- 27.Tomozawa S, Tsuno NH, Sunami E, Hatano K, Kitayama J, Osada T, Saito S, Tsuruo T, Shibata Y, Nagawa H. Cyclooxygenase-2 overexpression correlates with tumour recurrence, especially haematogenous metastasis, of colorectal cancer. Br J Cancer. 2000;83:324–328. doi: 10.1054/bjoc.2000.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camilleri-Broet S, Hardy-Bessard AC, Le Tourneau A, Paraiso D, Levrel O, Leduc B, Bain S, Orfeuvre H, Audouin J, Pujade-Lauraine E. HER-2 overexpression is an independent marker of poor prognosis of advanced primary ovarian carcinoma: a multicenter study of the GINECO group. Ann Oncol. 2004;15:104–112. doi: 10.1093/annonc/mdh021. [DOI] [PubMed] [Google Scholar]
- 29.Naderi A, Teschendorff AE, Beigel M, Cariati M, Ellis IO, Brenton JD, Caldas C. BEX2 is overexpressed in a subset of primary breast cancers and mediates nerve growth factor/nuclear factor-κB inhibition of apoptosis in breast cancer cell lines. Cancer Res. 2007;67:6725–6736. doi: 10.1158/0008-5472.CAN-06-4394. [DOI] [PubMed] [Google Scholar]
- 30.Naderi A, Liu J, Bennett IC. BEX2 regulates mitochondrial apoptosis and G1 cell cycle in breast cancer. Int J Cancer. 2010;126:1596–1610. doi: 10.1002/ijc.24866. [DOI] [PubMed] [Google Scholar]
- 31.Ma XL, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Naderi A, Liu J, Hughes-Davies L. BEX2 has a functional interplay with c-Jun/JNK and p65/RelA in breast cancer. Mol Cancer. 2010;19:111. doi: 10.1186/1476-4598-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta RJ, Jain RK, Leung S, Choo J, Nielsen T, Huntsman D, Nakshatri H, Badve S. FOXA1 is an independent prognostic marker for ERpositive breast cancer. Breast Cancer Res Treat. 2012;131:881–890. doi: 10.1007/s10549-011-1482-6. [DOI] [PubMed] [Google Scholar]
- 35.Vernimmen D, Gueders M, Pisvin S, Delvenne P, Winkler R. Different mechanisms are implicated in ERBB2 gene overexpression in breast and in other cancers. Br J Cancer. 2003;89:899–906. doi: 10.1038/sj.bjc.6601200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernimmen D, Begon D, Salvador C, Gofflot S, Grooteclaes M, Winkler R. Identification of HTF (HER2 transcription factor) as an AP-2 (activator protein-2) transcription factor and contribution of the HTF binding site to ERBB2 gene overexpression. Biochem J. 2003;370:323–329. doi: 10.1042/BJ20021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perissi V, Menini N, Cottone E, Capello D, Sacco M, Montaldo F, De Bortoli M. AP-2 transcription factors in the regulation of ERBB2 gene transcription by oestrogen. Oncogene. 2000;19:280–288. doi: 10.1038/sj.onc.1203303. [DOI] [PubMed] [Google Scholar]
- 38.Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, Winkler R, Boniver J, Delvenne P, Begon DY. The combined immunodetection of AP-2α and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Begon DY, Delacroix L, Vernimmen D, Jackers P, Winkler R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J Biol Chem. 2005;280:24428–24434. doi: 10.1074/jbc.M503790200. [DOI] [PubMed] [Google Scholar]
- 40.Bosher JM, Williams T, Hurst HC. The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA. 1995;92:744–747. doi: 10.1073/pnas.92.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adiseshaiah P, Li J, Vaz M, Kalvakolanu DV, Reddy SP. ERK signaling regulates tumor promoter induced c-Jun recruitment at the Fra-1 promoter. Biochem Biophys Res Commun. 2008;371:304–308. doi: 10.1016/j.bbrc.2008.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 43.Xing J, Ginty D, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 44.De Cesare D, Jacquot S, Hanauer A, Sassone-Corsi P. Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc Natl Acad Sci USA. 1998;95:12202–12207. doi: 10.1073/pnas.95.21.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Prywes R. Activation of the c-fos enhancer by the Erk MAP kinase pathway through two sequence elements: the c-fos AP-1 and p62TCF sites. Oncogene. 2000;19:1379–1385. doi: 10.1038/sj.onc.1203443. [DOI] [PubMed] [Google Scholar]
- 47.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eeckhoute J, Lupien M, Meyer CA, Verzi MP, Shivdasani RA, Liu XS, Brown M. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 2009;19:372–380. doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi N, Ito E, Azuma S, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Tatsuta K, Inoue J, et al. FoxA1 as a lineagespecific oncogene in luminal type breast cancer. Biochem Biophys Res Commun. 2008;365:711–717. doi: 10.1016/j.bbrc.2007.11.064. [DOI] [PubMed] [Google Scholar]
- 50.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 51.Monje P, Hernandez-Losa J, Lyons RJ, Castellone MD, Gutkind JS. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J Biol Chem. 2005;280:35081–35084. doi: 10.1074/jbc.C500353200. [DOI] [PubMed] [Google Scholar]
- 52.Xu TR, Vyshemirsky V, Gormand A, von Kriegsheim A, Girolami M, Baillie GS, Ketley D, Dunlop AJ, Milligan G, Houslay MD, et al. Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species. Sci Signal. 2010;3:ra20. [PubMed] [Google Scholar]
- 53.Nagata Y, Honda Y, Matsuda R. FGF2 induces ERK phosphorylation through Grb2 and PKC during quiescent myogenic cell activation. Cell Struct Funct. 2010;35:63–71. doi: 10.1247/csf.09024. [DOI] [PubMed] [Google Scholar]
- 54.Lu L, Wang LL, Li T, Wang J. NF-κB subtypes regulate CCCTC binding factor affecting corneal epithelial cell fate. J Biol Chem. 2010;285:9373–9382. doi: 10.1074/jbc.M109.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Laere SJ, Van der Auwera I, Van den Eynden GG, van Dam P, Van Marck EA, Vermeulen PB, Dirix LY. NF-κB activation in inflammatory breast cancer is associated with oestrogen receptor downregulation, secondary to EGFR and/or ErbB2 overexpression and MAPK hyperactivation. Br J Cancer. 2007;97:659–669. doi: 10.1038/sj.bjc.6603906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biswas DK, Cruz AP, Gansberger E, Pardee AB. Epidermal growth factor-induced nuclear factor κB activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci USA. 2000;97:8542–8547. doi: 10.1073/pnas.97.15.8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 58.Puto LA, Pestonjamasp K, King CC, Bokoch GM. p21-activated kinase 1 (PAK1) interacts with the Grb2 adapter protein to couple to growth factor signaling. J Biol Chem. 2003;278:9388–9393. doi: 10.1074/jbc.M208414200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.