Abstract
Epstein-Barr virus (EBV) establishes a life-long latent infection in humans. In proliferating latently infected cells, EBV genomes persist as multiple episomes that undergo one DNA replication event per cell cycle and remain attached to the mitotic chromosomes. EBV nuclear antigen 1 (EBNA-1) binding to the episome and cellular genome is essential to ensure proper episome replication and segregation. However, the nature and regulation of EBNA-1 interaction with chromatin has not been clearly elucidated. This activity has been suggested to involve EBNA-1 binding to DNA, duplex RNA, and/or proteins. EBNA-1 binding protein 2 (EBP2), a nucleolar protein, has been proposed to act as a docking protein for EBNA-1 on mitotic chromosomes. However, there is no direct evidence thus far for EBP2 being associated with EBNA-1 during mitosis. By combining video microscopy and Förster resonance energy transfer (FRET) microscopy, we demonstrate here for the first time that EBNA-1 and EBP2 interact in the nucleoplasm, as well as in the nucleoli during interphase. However, in strong contrast to the current proposed model, we were unable to observe any interaction between EBNA-1 and EBP2 on mitotic chromosomes. We also performed a yeast double-hybrid screening, followed by a FRET analysis, that led us to identify HMGB2 (high-mobility group box 2), a well-known chromatin component, as a new partner for EBNA-1 on chromatin during interphase and mitosis. Although the depletion of HMGB2 partly altered EBNA-1 association with chromatin in HeLa cells during interphase and mitosis, it did not significantly impact the maintenance of EBV episomes in Raji cells.
INTRODUCTION
Epstein-Barr virus (EBV) is a lymphocryptovirus member of the Gammaherpesvirinae subfamily that infects >90% adults worldwide. Paradoxically, EBV has been associated with numerous tumors whose incidence increases in immunosuppressed patients (55). Although primary EBV infection usually occurs at an early age in an asymptomatic form, it can also result in infectious mononucleosis when it occurs during adolescence or later in life (43). The virus is mainly transmitted by saliva, and primary infection occurs in oropharyngeal cells. Then, EBV establishes a persistent latent infection mainly in the memory B-cell compartment (39). Latent EBV infection has been associated with several malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's lymphomas, T-cell lymphomas, gastric adenocarcinoma, and others (62, 73).
In latently infected B cells EBV genomes persist as multicopy covalently closed double-stranded circular episomes (28, 34). In proliferating cells these episomes undergo one DNA replication event per cycle and are efficiently partitioned during cell division, enabling the viral genomes to be maintained at a constant average copy number per cell (1, 70). Two viral components only, the latent origin of DNA replication, oriP, and the EBNA-1 protein, are required for episome maintenance in proliferating cells (32, 69, 71). oriP is composed of two elements: a dyad symmetry (DS) element that contains four EBNA-1 binding sites and a family of repeats (FR) containing 20 EBNA-1 recognition sites (4, 50). EBNA-1 is a 641-amino-acid (aa) protein in the B95.8 strain. Two functions of EBNA-1 are required for episome maintenance. First, by concomitantly binding to DS (19, 57), to replication cellular factors (13), and to cellular chromatin (24), EBNA-1 activates episome replication during the S phase. Second, EBNA-1 simultaneously associates with the FR element and cellular chromosomes during mitosis, therefore ensuring an efficient coupling between cellular and viral genome segregation (29, 38, 41). In addition, EBNA-1 modulates the expression of several viral (3, 52, 69) and numerous cellular promoters (10, 14, 35). Thus, EBNA-1 activities are intimately associated with its ability to interact with chromatin throughout the cell cycle. EBNA-1 interaction with chromatin during interphase has been evaluated by various biochemical fractionations procedures with controversial conclusions. Indeed, while Kanda et al. showed that EBNA-1 copurified with the chromatin, Daikoku et al. and Ritzi et al. found only minor amounts of EBNA-1 in the chromosomal pellet from interphase cells (12, 24, 51). During mitosis EBNA-1 binding to chromosomes is linked to the presence of three independent chromosome-binding sites (CBS), namely, CBS-1 (aa 72 to 84), CBS-2 (aa 328 to 365), and CBS-3 (aa 8 to 54) (16, 18, 36). However, the molecular mechanism responsible for EBNA-1 interaction with mitotic chromosomes has been much debated, and several non-mutually-exclusive hypotheses have been proposed. Sears et al. identified an AT-hook structure in the N terminus of EBNA-1 that could bind to AT-rich DNA (54). CBS-2 and CBS-3 also contain RGG motifs that have been identified within several RNA-binding proteins. Since Braco-19, a molecule that disrupts G-quadruplex RNA, also disrupted EBNA-1 interaction with metaphase chromosomes, it was suggested that EBNA-1 binding to RNA may be required to load EBNA-1 on chromosomes or to stabilize this interaction during mitosis (44). Finally, EBNA-1 may interact with chromatin through direct protein-protein interactions. Thus far, EBNA-1 binding protein 2 (EBP2) is the only cellular protein that has been shown to be required for EBNA-1 binding on mitotic chromosomes. EBP2 was initially isolated from a double-hybrid screen with EBNA-1 (58). EBP2 and EBNA-1 were further shown to copurify from insect cells coexpressing both proteins (58). EBP2 binds to a Gly/Arg-rich region of EBNA-1 that has been mapped between aa 325 and 376 (58), which closely mapped to CBS2. A truncation form of EBNA-1 mutant lacking EBP2 binding site (EBNA-1Δ325-376) was shown to promote the replication of oriP containing plasmids but was defective in supporting their long-term maintenance in human cells (58), as well as in yeast cells (27). EBNA-1Δ325-376 is still nuclear but fails to attach to the cellular chromosomes in cells artificially blocked in metaphase (67). Moreover, the apparent colocalization of EBNA-1 and EBP2 on mitotic chromosomes (42, 67) and the loss of EBNA-1 localization onto chromosome in cells where EBP2 expression has been silenced or where EBP2 has been dissociated from chromosome by an Aurora kinase (26) suggested that EBP2 could act as an docking protein for EBNA-1 on mitotic chromosomes.
Surprisingly, there is still no evidence for a direct interaction between EBNA-1 and EBP2 in living mitotic cells. In the present study, we reexamined the dynamics of interaction between EBNA-1 and EBP2 in living cells, using a combination of live-cell imaging approaches. This study led us to conclude that EBP2 interacted with EBNA-1 during interphase but not during mitosis. Subsequent experiments identified HMGB2 (high-mobility group box 2), a well-characterized chromatin-bound protein, as a new partner for EBNA-1 on chromatin both during interphase and mitosis.
MATERIALS AND METHODS
Recombinant plasmids.
Plasmids pEGFP-N1, pEGFP-C1, pDsRed1-N1, and pDsRed1-C1 (BD Biosciences Clontech, Palo Alto, CA) encoded either a variant of the green fluorescent protein with enhanced fluorescence (EGFP) or a red fluorescence protein (DsRed).
Human EBP2 was expressed as a fusion protein to the N and C termini of EGFP and the N and C termini of DsRed, respectively, in pEBP2-EGFP, pEGFP-EBP2, pEBP2-DsRed, and pDsRed-EBP2. EBP2 cDNA was cloned from HeLa cells. For this purpose, total RNA was extracted with a High-Pure RNA isolation kit (Roche Diagnostics, Mannheim, Germany) and subjected to a reverse transcription by using a Ready-to-Go You Prime First-Strand Beads kit according to the manufacturer's recommendations (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom). The region encoding human EBP2 was subsequently amplified from the cDNA by PCR with the EBP2 upstream primer (5′-CCCAAGCTTGGGCCACCATGGACACTCCCCCGCTCTCG-3′) and the EBP2 downstream primer 5′-CGCGGATCCCGGTGTGTTCTGTTCTTCATCTTCTC-3′. The resulting PCR product was digested by HindIII and BamHI, gel purified, and cloned into pEGFP-N1, pEGFP-C1, pDsRed1-N1, and pDsRed1-C1.
pDsRed-N1-EBP2-oriP was obtained by inserting oriP sequence upstream of the cytomegalovirus (CMV) promoter into pDsRed-N1-EBP2. The region containing oriP was amplified from pCEP4 (Invitrogen) by PCR with the oriP upstream primer 5′-CGCCATGCATTAGTTATTAATGCCTTTATGTGTAACTCTTG-3′ and the oriP downstream primer 5′-CCGTAATTGATTACTATTAATAGTAATCAATTACGG-3′. Upstream and downstream primers were designed to have 21 bases (boldface letters) with homology to the linear ends of AseI-cleaved pDsRed-N1-EBP2 plasmid. The resulting PCR product was purified by affinity chromatography and cloned into pDsRed-N1-EBP2 by using a cold fusion cloning kit (SBI, Mountain View, CA).
The EBNA-1 ΔGlyAla deletion construct was obtained by using the pEGFP-C1-EBNA-1 plasmid containing the entire EBNA-1 coding sequence (aa 8 to 641) described previously (36), from which the central Gly-Ala repeated region (aa 93 to 325) was removed by using a two-step PCR overlapping procedure. Briefly, the DNA sequence encoding EBNA-1 N-terminal part (aa 8 to 90) (part A) was fused in frame to DNA encoding the EBNA-1 C-terminal part (aa 326 to 641) that we can separate into two parts (aa 326 to 486 for part B and aa 487 to 641 for part C). In the first PCR step, primers 1 and 2 (described below) were used to amplify part A, whereas primers 3 and 4 were used to amplify part B of EBNA-1. Primer 2 was designed to overlap part B, and primer 3 was designed to overlap part A. The partly overlapping PCR products from the two separate reactions were gel purified and mixed together with primers 1 and 4 in a second PCR in order to produce DNA encoding part A fused to part B (A-B) of EBNA-1. These primers were as follows: primer 1 (5′-GCCCGGAATTCTGCAGATACAGGAC-3′), primer 2 (5′-GGCCTCCACCTCCTGCTCCTGTTCC-3′), primer 3 (5′-AGGAGCAGGAGGTGGAGGCCG-3′), and primer 4 (5′-TCATCTGCGGGGCCCTGCTC-3′). Boldface letters indicate the part A sequence, and the part B sequence is underlined. Italics indicate restriction sites (EcoRI for primer 1 and ApaI for primer 2). The PCR product from the second reaction (A-B sequence) was digested with EcoRI and ApaI and cloned into pEGFP-C1-EBNA-1 plasmid in place of A-GlyAla-B sequence to generate the pEGFP-C1-EBNA-1 ΔGlyAla deletion construct. The region encoding EBNA-1 ΔGlyAla was digested with EcoRI and BamHI and cloned into pDsRed1-C1 plasmid to generate the pDsREd1-C1-EBNA-1 ΔGlyAla plasmid. pEGFP-N1-EBNA-1 ΔGlyAla and pDsRed1-N1-EBNA-1 ΔGlyAla were obtained as follows. The region encoding EBNA-1 ΔGlyAla was generated by PCR from pEGFP-C1-EBNA-1 ΔGlyAla with the EBNA-1 upstream primer 5′-AGCCGGAATTCCCACCATGACAGGACCT-3′ and the EBNA-1 downstream primer 5′-CGGGATCCTTCTCCTGCCCTTCCTC-3′. The EBNA-1 upstream primer has been designed for inserting an EcoRI restriction site, a Kozak sequence, and a translation initiation codon upstream of EBNA-1 ΔGlyAla. The EBNA-1 downstream primer has been designed for suppressing the stop codon at the 3′-terminal sequence of EBNA-1 ΔGlyAla and for inserting a BamHI restriction site. The resulting PCR product was digested by HindIII and BamHI, gel purified, and cloned into pEGFP-N1 and pDsRed1-N1.
The EBNA-1Δ90-376 deletion construct was obtained by using the pEGFP-C1-EBNA-1 from which the central GlyAla repeated region (aa 90 to 325) and the EBP2 binding region (aa 325 to 376) were removed by two-step PCR. Briefly, EB1 upstream primer (5′-GGAATTCCTCTGACGAGGGGCCAGGTAC-3′) and EB2 downstream primer (5′-TGTTCCACCGTGGGTCCCTTTGC-3′) were used to amplify the 8-89 EBNA-1 region, whereas EB3 upstream primer (5′-GGAGAAAAGAGGCCCAGGAGT-3′) and EB4 downstream primer (5′-CGGGATCCCGTCACTCCTGCCCTTCCTCACCCTC-3′) were used to amplify the 377-641 EBNA-1 region. Primers EB2 and EB3 were drawn in order to generate blunted end. Both regions were then fused by a ligation reaction; the resulting product was amplified by primers EB1 and EB4 in a second PCR to produce DNA encoding the EBNA-1 sequence deleted from the aa 90 to 376. The PCR product from the second reaction was digested with EcoRI and BamHI and cloned into pEGFP-C1 plasmid to generate the pEGFP-C1-EBNA-1Δ90-376 construct.
The cDNAs encoding human HMGB1and HMGB2 were cloned from HeLa cells. Both proteins were expressed as fusion proteins to the N and C termini of EGFP or the N and C termini of DsRed, as described previously (46).
Cell culture, DNA transfection, and cell cycle synchronization.
Human HeLa cells (ATCC CCL-2) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 2 mM l-glutamine in a 95% air–5% CO2 incubator at 37°C. Raji cells (ATCC CCL-86) were grown in advanced RPMI medium supplemented with 10% FCS and 2 mM l-glutamine in a 95% air–5% CO2 incubator at 37°C. For transfection experiments, HeLa cells were grown on glass coverslips in six-well plates until they reached ca. 80% confluence. Plasmids were transfected with the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's recommendations. For cell cycle synchronization at M phase, the cells were treated at 44 h posttransfection with nocodazole (40 ng/ml) for 4 h. After two washes with DMEM/F-12 to eliminate nocodazole, mitotic cells were observed by fluorescence microscopy.
Time-lapse microscopy.
Cells grown on glass coverslips were mounted in a cell observation chamber filled with complete medium without phenol red. The microscope chamber was kept at 37°C in a 95% air–5% CO2 atmosphere. Images were acquired with a Leica DMI6000 microscope (100× oil PlanApo, NA 1.4), a Piezo electric translator (Physik Instrument, Karlsruhe, Germany), a lambda DG-4 illuminator (Sutter Instrument, Novato, CA), and a CoolSnapHQ2 (10 MHz, binning of 2, mode Alt-Normal; Roper Scientific, Evry, France) driven by Metamorph7.3. The dichroic sets 470AF20/500DRLP/535AF35 and 525AF45/560DRLP/595AF60 were, respectively, used for collecting GFP and mRFP fluorescence. Time-lapse imaging was done by acquiring images every minute, at 10 different positions. A 0.3-μm z-step and a range of 20 μm were used in each position. Images were deconvoluted using Huygens Software Pr (SVI, Hilversum, Netherlands). Colocalization analyses were performed using ImageJ software (W. S. Rasband, ImageJ [U.S. National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij, 1997-2009]).
Confocal microscopy.
For confocal analysis, a Leica Sp5 microscope equipped with a 63× oil PlanApo (NA 1.4) was used. Cells were scanned at 400 Hz with a resolution of 1,024 × 512 pixels and a voxel size of 107 by 107 by 300 nm. The pinhole was adjusted so as to fit the airy disc. DAPI (4′,6′-diamidino-2-phenylindole), green fluorescent protein (GFP), and red fluorescent protein (RFP) were, respectively, excited at a 405-, 488-, and 561-nm wavelengths, and the emission fluorescence levels were detected by using a photo-multiplicator at, respectively, between 410 and 480 nm, 495 and 550 nm, and 590 and 670 nm. Three channels were recorded sequentially at each z-step.
Photobleaching.
Cells were kept at 37°C in a 5% CO2 atmosphere, and FRAP (fluorescence recovery after photobleaching) was performed on a confocal Zeiss LSM 710 microscope. A 488-nm laser and a C-Apochromat lens (×63, 1.2 NA) was used in the photobleaching experiments. A circular region of interest 3 μm in diameter was bleached with the 488-nm laser at full power, and subsequent scans were taken at 0.5% of full power. Five images were taken before bleaching, and then images were acquired every 1.15 s for 96 s. An average of eight data sets were analyzed for each result. Images of each data set were normalized and corrected for acquisitional photobleaching (47), plotted with time, and fitted individually with a diffusion model (59) to determine the characteristic diffusion time (τD).
FRET determination by tdFLIM measurement.
Two systems were used for FRET determination by time domain fluorescence lifetime imaging microscopy (tdFLIM) using the time- and space-correlated single-photon counting method (63). This method provides in picoseconds (ps) the time-resolved fluorescence decay for every pixel by counting and sampling single emitted photons according to (i) the time delay between photon arrival and laser pulse (ps time scale, 4,096 channels) and (ii) their xy coordinate (256 × 256 pixel image).
(i) A quadrant anode system was used for the FRET experiments performed in the absence of oriP region. It is based on a titanium sapphire laser (Millennia 5W/Tsunami 3960-M3BB-UPG kit; Spectra-Physics, Evry, France) that delivers ps pulses was tuned at 960 nm to obtain an excitation wavelength at 480 nm after frequency doubling. The repetition rate was 4 MHz after pulse-picker (Spectra-Physics, catalog no. 3980-35). The laser beam was expanded and inserted into an inverted epifluorescence microscope (DM IRBE; Leica, France) for wide-field illumination (a few mW/cm2). The microscope stage was equipped with an incubator system for temperature and CO2 regulation (37°C, 5% CO2). Green fluorescence decay images were obtained using a Leica Plan-Apochromat oil lens (×100, 1.3 NA), a dichroic beam splitter (505DRLP, Omega; Optophotonics, Eaubonne, France), an emission filter (535DF35, Omega; Optophotonics), and the quadrant anode TSCSPC detector (QA; Europhoton GmbH, Germany). A band-pass emission filter (518 nm < λem > 552 nm) was chosen to select the donor fluorescence (EGFP) and to reject the acceptor fluorescence (DsRed). The count rate was up to 30 kHz. Acquisition of fluorescence decay images was done after accumulation of sufficient single photon events, usually after 3 to 6 min. A temporal resolution of <100 ps and a spatial resolution of 500 nm were previously determined for this system (63).
(ii) A MicroTime 200 (PicoQuant, Berlin, Germany) was used to measure FRET between GFP–EBNA-1 and DsRed-EBP2 with or without oriP-containing plasmid. A pulsed diode laser operating at 470 nm (PicoQuant) is focused at the center of the optical axis in the sample through a water objective with a high-numerical aperture objective (×60, NA 1.3) mounted on an inverted microscope (Olympus, Tokyo, Japan). The fluorescence emission passes through a double dichroic mirror (DM 470/471 nm) and is focused on a pinhole. An avalanche photodiode (SPADs 14; Perkin-Elmer), in front of which is placed an emission filter (525 ± 25 nm), constitutes the GFP fluorescence detector. The subcellular position of the region of interest is chosen by first acquiring a prescan image of the field of view. A continuous DPSS laser operating at 470 nm is used to acquire the DsRed fluorescence into the red channel, in front of which is placed the emission filter (593 ± 20 nm). A TimeHarp 300PC board (PicoQuant) is used in a time-tagged time-resolved (TTTR) mode and coupled to a PC that pilots data acquisition and analysis using Symphotime software (PicoQuant). A single photo-counting TTTR trace was acquired over 20 s.
tdFLIM data analysis.
For qualitative determination of FRET, the fluorescence decays of EGFP within the regions of interest were extracted from the acquisition matrix, and the decays of EGFP-tagged proteins (donor) in the presence of DsRed-tagged proteins (acceptor) were compared to the control decays of the EGFP-tagged proteins measured in the absence of acceptors. Since the DsRed protein emits a weak green fluorescence before maturation in vitro as well as in vivo, the decays of the green species of each DsRed-tagged protein in the BP emission filter were analyzed in cells cotransfected with GFP. In these cells, no other component than GFP lifetime could be detected, indicating that immature DsRed from each DsRed-tagged protein cannot contribute to decay obtained with GFP.
To perform quantitative analyses, the experimental fluorescence decays were further deconvoluted with the instrument response function and fitted to a monoexponential and a biexponential function using a Marquardt nonlinear least-squares algorithm (Globals Unlimited Software/University of Illinois at Urbana-Champaign) or using the Symphotime software (PicoQuant). The fluorescence decay of EGFP-tagged proteins in monotransfected cells was monoexponential with a Τ1 lifetime. In cotransfected cells in which the fluorescence decay was faster than in monotransfected cells, the fluorescence decay of EGFP-tagged proteins was biexponential with a fixed T1 lifetime and a faster T2 lifetime. From the relative amplitudes of the lifetime components, we quantified the respective proportions of the two donor populations namely, α1 (Τ1 lifetime corresponding to the unbound donor) and α2 (T2 faster lifetime corresponding to the donor interacting with an acceptor).
Yeast two-hybrid screening.
A cDNA encoding EBNA-1 truncated form 8-410Δ95-314 was fused with the region encoding GAL4 DB domain into pAS2.1 (Clontech) between EcoRI and BamHI sites. A premade HeLa cell random cDNA library fused with GAL4 AD in pGAD-GH (Clontech) was used. A yeast two-hybrid assay was performed according to the manufacturer's protocol. Briefly, Saccharomyces cerevisiae CG-1945 strain was cotransformed with cDNA HeLa/pGAD-GH and 1-90/327-410 EBNA-1/pAS2.1 or pAS2.1 control. The transformed yeasts were selected on agar plates with a double (−Leu/−Trp) or a triple (−Leu/−Trp/−His) dropout of the essential amino acids. Prey plasmids from dropout-resistant yeast clones were isolated, and cDNA inserts were sequenced for identification. Plasmids encoding for putative EBNA-1 partners were further tested in Y2H assay with plasmids encoding three different baits: 1-90/327-410 EBNA-1, EBNA-1 ΔGA, and EBNA-1 full length. Transformed CG1945 cells were screened with high stringency by plating on −Leu/−Trp/−His selective dropout minimal medium containing 25 mM 3-aminotriazole.
Cell transduction by lentiviral vectors for delivery of shRNA to HMGB2.
Two different pLKO.1 lentiviral vectors targeting distinct regions of HMGB2 mRNA were used to transduce Raji and HeLa cells. Both constructs have been previously used to inactivate HMGB2 expression in UT-7 cells (31). After 3 weeks of continuous selection with puromycin, HMGB2 expression was monitored by Western blot as described previously (31). EBV episomes were quantified by using Kappa SYBR Fast qPCR Master Mix (Kapa Biosystems, Boston, MA) with the primers 5′-GGAGATACTGTTAGCCCTG-3′ and 5′-GTGTGTTATAAATCTGTTCCAAG-3′ designed within the region encoding BHRF1. Cellular DNA was quantified by using the primers 5′-TAGCAACCTCAAACAGACACCA-3′ and 5′-CAGCCTAAGGGTGGGAAAAT-3′ designed within the region encoding the β-globin.
RESULTS
EBNA-1 and EBP2 partly colocalize in living cells throughout the cell cycle.
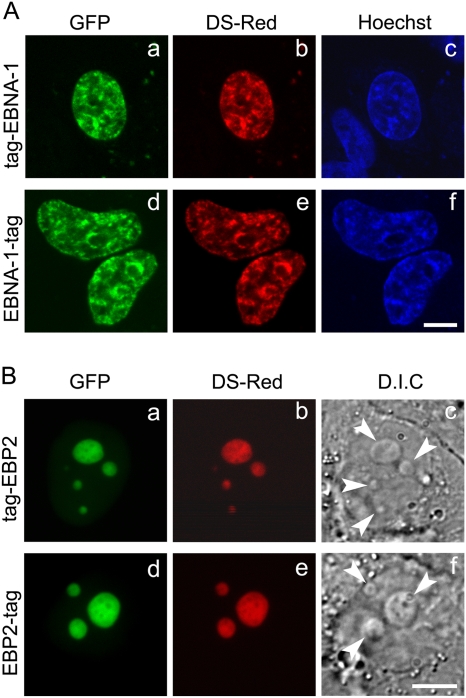
EGFP- and DsRed-tagged forms of EBNA-1 and EBP2 were expressed in human HeLa cells and observed by live cell imaging during interphase and mitosis. This approach was chosen because it prevents artifactual mislocalization that may result from the use of chemical fixative or drugs that irreversibly block cell cycle progression, such as demecholcine, as previously observed (46). During interphase, EBNA-1 colocalized mainly with condensed and dispersed forms of the chromatin, which are characterized by intense and weak Hoechst staining, respectively (Fig. 1A). In addition, a weak nucleolar staining was observed in most cells. EBP2 concentrated mainly, albeit not exclusively, in the nucleoli during interphase, as confirmed by differential interference contrast microscopy (Fig. 1B). Importantly, the subcellular localization of the resulting tagged proteins was not influenced by either the nature of the fluorophores or their relative positions in the fusion proteins.
Fig 1.
Subcellular localization of tagged proteins by confocal microscopy in living cells. (A) HeLa cells were cotransfected with either GFP–EBNA-1 (a) and DsRed–EBNA-1 (b) or with EBNA-1–GFP (d) and EBNA-1–DsRed (e). The DNA was stained with Hoechst. Images a to f show single confocal z-section. Whatever the nature and the position of the fluorophore, EBNA-1 colocalizes with chromatin (c and f). Scale bar, 10 μm. (B) HeLa cells were cotransfected with either GFP-EBP2 (a) and DsRed-EBP2 (b) or with EBP2-GFP (d) and EBP2-DsRed (e). Images a to f show single confocal z-section. Whatever the nature and the position of the fluorophore, EBP2 colocalizes perfectly with nucleoli (arrowheads) observed by differential interference contrast (D.I.C.) (c and f). Scale bar, 10 μm.
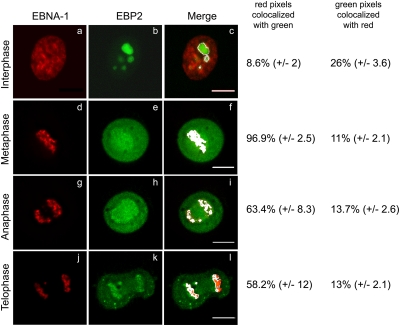
A time course analysis was performed by quantitative video microscopy on cells coexpressing DsRed–EBNA-1 and GFP-EBP2. During interphase, EBNA-1 and EBP2 partly colocalized both within and outside the nucleolus (Fig. 2a to c). From metaphase to cytokinesis EBNA-1 diffusely coated the mitotic chromosomes, as reported previously (26, 36, 42, 67, 68). Importantly, only 13 to 15% of the EBP2 colocalized with EBNA-1 on chromosomes. Indeed, EBP2 dispersed within the nucleoplasm after nucleolus breakdown and exhibited two localizations during mitosis (Fig. 2e, h, and k). In addition to a weak diffuse staining in the nucleoplasm, we also detected EBP2 around the chromosomes and close to a structure that was highly reminiscent of the mitotic spindle. At the end of mitosis, the fluorescence of GFP-EBP2 decreased in the vicinity of the mitotic spindle, an observation consistent with the progressive depolymerization of spindle microtubules between daughter cells (Fig. 2k and l). Based on these observations, we evaluated that an average of 96% of the EBNA-1 colocalized with EBP2 during metaphase, 61% during anaphase, and 58% at the end of mitosis.
Fig 2.
Dynamic localization of EBNA-1 and EBP2 throughout the cell cycle. Living HeLa cells coexpressing GFP-EBP2 and DsRed–EBNA-1 were observed from interphase (a to c) to mitosis (d to l) by video microscopy. All images are deconvoluted z-optical sections. Colocalized pixels (in white) from the merge images were quantified with ImageJ software. The standard deviations are indicated (n = 4). Scale bars, 10 μm.
EBNA-1 and EBP2 interact in nucleoli and nucleoplasm during interphase.
We investigated a possible interaction between EBNA-1 and EBP2 by measuring Förster resonance energy transfer (FRET). FRET is a nonradiative energy transfer that can occur when a donor and a compatible acceptor fluorophore are located at a distance lower than 10 nm from each other (15). FRET can be detected via tdFLIM and used to monitor protein-protein interactions in living cells (11). Indeed, if the interacting proteins are conjugated to suitable donor and acceptor fluorophores in such a way that the fluorescence emission spectrum of the donor overlaps the absorption spectrum of the acceptor and their dipoles align, FRET occurs and reduces the lifetime of the donor fluorescence (11). In the present case, a tdFLIM setup was used to measure FRET with a spatial accuracy of a few hundred nanometers, which is therefore well adapted for studies at a subcellular scale. Since tdFLIM measurement is not sensitive to fluorophore concentration or unintended photobleaching, it is especially appropriate for detecting FRET in transiently transfected cells. Finally, tdFLIM analysis can give access to the proportion of GFP-tagged molecules involved in a FRET at the single-cell level (63).
FRET efficiency relies on the relative positions and distance of the donor and acceptor fluorophores even when fusion proteins are in close interaction. Therefore, FRET was performed with the eight possible pairs obtained when combining the different donors (GFP-EBP2, EBP2-GFP, GFP–EBNA-1, and EBNA-1–GFP) with the different acceptors (DsRed–EBNA-1, EBNA-1–DsRed, DsRed-EBP2, and EBP2-DsRed) to minimize false-negative results. Since identical results were obtained with the eight possible donor/acceptor pairs, we only present here data collected from cells coexpressing GFP-EBP2 and DsRed–EBNA-1.
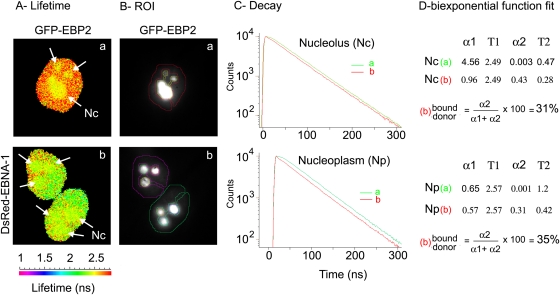
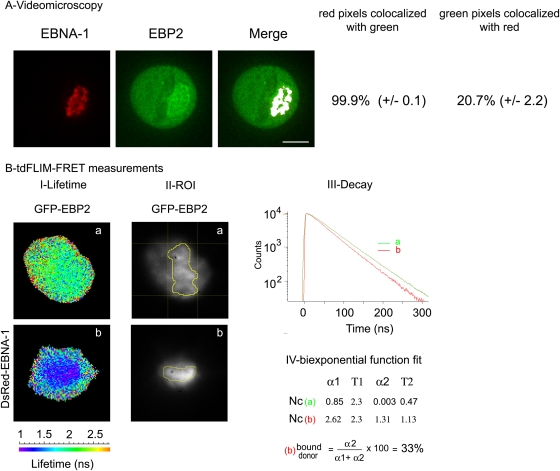
As illustrated in Fig. 3, we first performed a pixel-by-pixel acquisition of GFP fluorescence decay in cells expressing GFP-EBP2 alone (Fig. 3A, condition a) or coexpressing DsRed–EBNA-1 (Fig. 3A, condition b). Two mean lifetime values were measured for GFP-EBP2 alone, depending on its subnuclear localization: 2.57 ± 0.02 ns (n = 8 cells) in the nucleoplasm and 2.49 ± 0.015 ns (n = 8 cells) in the nucleolus. This result was consistent with the well-known effect of the local environment of the fluorophore on its fluorescence lifetime (30). Subsequently, regions of interest (ROIs) corresponding to nucleolar and nucleoplasmic areas were manually drawn from tdFLIM images (Fig. 3B) and GFP-EBP2 fluorescence decay was analyzed. In the nucleoli, GFP-EBP2 fluorescence decay was significantly faster in the presence (see the red curves in Fig. 3C) than in the absence (see the green curves in Fig. 3C) of DsRed–EBNA-1, demonstrating that EBNA-1 and EBP2 interact with each other in this compartment. Subsequent analysis allowed us to estimate that 31% of the GFP-EBP2 interacted with DsRed–EBNA-1 in the nucleoli (Fig. 3). Surprisingly, whereas only low amounts of GFP-EBP2 were detected in the nucleoplasm (Fig. 2), a significant FRET was also be measured in this compartment (Fig. 3C, Np), and we calculated that 35% of the GFP-EBP2 interacted with DsRed–EBNA-1 out of the nucleoli.
Fig 3.
EBP2 and EBNA-1 interact in the nucleolus and the nucleoplasm during interphase. tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images for GFP fluorescence (515 nm < λ > 560 nm), in HeLa cells transfected with GFP-EBP2 alone (images Aa and Ba) or in the presence of DsRed–EBNA-1 (images Ab and Bb). (A) tdFLIM images were obtained by analyzing the fluorescence decays with a single lifetime pixel by pixel and are displayed as fluorescence lifetime pseudocolor maps. A lifetime colored scale is presented. Each arrow indicates a nucleolus. (B) Regions of interest (ROIs) corresponding to the nucleolus and nucleoplasm have been selected from tdFLIM images. (C) The fluorescence decay of GFP-EBP2 was measured in each ROI in the absence (condition a, green curves) or in the presence of DsRed–EBNA-1 (condition b, red curves). (D) The experimental curves were further fitted with a biexponential function using Globals Unlimited software for quantifying the relative proportions of the two donor populations: α1 with a slow Τ1 lifetime corresponding to the unbound donor and α2 with a fast T2 lifetime corresponding to the donor interacting with the acceptor (n = 12).
Altogether, these results demonstrated for the first time that EBNA-1 and EBP2 interact specifically in living cells during interphase, a finding which is in agreement with previous data obtained by yeast double-hybrid screening and biochemical approaches (42, 58). In addition, we provided the first evidence that EBNA-1 and EBP2 interacted in human living cells both in the nucleolus and in the nucleoplasm.
EBNA-1 and EBP2 do not interact during mitosis.
Since we previously demonstrated by FRET that EBP2 interacted with EBNA-1 during interphase, we wondered whether this interaction was conserved upon mitosis. This question is of the utmost importance since EBP2 is currently considered as a docking protein for EBNA-1 onto chromosomes during mitosis.
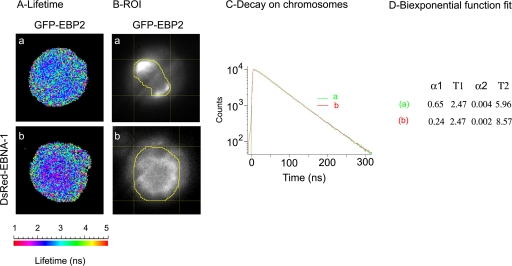
Figure 4 shows that the fluorescence decay of GFP-EBP2 in an ROI defined around mitotic chromosomes (Fig. 4A) was identical in the absence (green curve drawn from tdFLIM image a) and in the presence of DsRed–EBNA-1 (red curves drawn from tdFLIM image b). In both cases, we measured undistinguishable monoexponential decays with a unique fluorescence lifetime T1 of 2.47 ns, indicating that FRET did not occur between GFP-EBP2 and DsRed–EBNA-1. This result was confirmed on 12 mitotic cells. Therefore, EBNA-1 and EBP2 did not interact during mitosis in living cells, whereas they efficiently interacted during interphase and colocalized at least in part on mitotic chromosomes in living cells.
Fig 4.
FRET measurement between EBP2 and EBNA-1 during mitosis in living cells. tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images of the GFP donor (515 nm < λ > 560 nm) in mitotic HeLa cells expressing GFP-EBP2 alone (images a, and green curves) or in the presence of DsRed–EBNA-1 (images b, and red curves). (A) The tdFLIM images were obtained by analyzing the fluorescence decays with a single lifetime pixel by pixel and are displayed as fluorescence lifetime pseudocolor maps. A lifetime colored scale is presented. (B) The ROI corresponding to mitotic chromosomes has been manually drawn from tdFLIM images. (C) The decays of GFP-EBP2 in the presence of DsRed–EBNA-1 (condition b or red curves) were compared to the control decays of GFP-EBP2 alone (condition a or green curves) in each ROI. (D) The experimental curves were further fitted with a biexponential function for quantifying the proportion of two donor populations: α1 with a slow Τ1 lifetime corresponding to the unbound donor and α2 with a fast T2 lifetime corresponding to the donor interacting with an acceptor (n = 12).
These results strongly contrasted with previous findings of Wu et al. and others (26, 42, 67, 68). Such discrepancies might result at least in part from technical differences. In contrast to previous studies, colocalization and subsequent interactions between EBNA-1 and EBP2 were analyzed in the context of living mitotic cells in the absence of any drug promoting cell cycle arrest. We hypothesized that such drugs, e.g., demecholcine, could modify EBP2 or EBNA-1 localization and therefore promote artifactual interactions between these proteins. Indeed, demecholcine induces a metaphase block by preventing the assembly of spindle fibers during mitosis. To test this assumption, mono- and double-transfected HeLa cells were treated with demecholcine and analyzed by video microscopy and tdFLIM as described previously. As illustrated in Fig. 5, EBNA-1 concentrated in the close vicinity of the equatorial chromosome plate in demecholcine-treated cells. Strikingly, EBP2 colocalization with EBNA-1 markedly increased on chromosomes in these cells. Whereas 11% ± 2.1% of the EBP2 colocalized with EBNA-1 in untreated cells (Fig. 2), this proportion rose to 20.7% ± 2.2% in demecholcine-arrested cells (Fig. 5A). Importantly, a significant FRET was detected in demecholcine-treated cells, and we estimated that 33% of the GFP-EBP2 interacted with DsRed–EBNA-1 in this condition (Fig. 5B). Therefore, we concluded from these experiments that demecholcine promoted EBP2 mislocalization, which in turn favored the subsequent interaction of EBP2 with EBNA-1.
Fig 5.
Demecholcine induces artifactual interaction between EBNA-1 and EBP2. (A) HeLa cells coexpressing GFP-EBP2 and DsRed–EBNA-1 were blocked in metaphase by demecholcine (10 μg/ml) and observed by video microscopy as indicated. The images represent deconvoluted optical z-sections. Colocalized pixels (in white) from the merge images were quantified by using ImageJ software. The error corresponds to the standard deviation (n = 4). Scale bars, 10 μm. (B) tdFLIM measurements were carried out as described in the legend of Fig. 3. (I) Lifetime images were obtained by analyzing the fluorescence decays with a single lifetime pixel by pixel and are displayed as fluorescence lifetime pseudocolor maps. A colored lifetime scale is presented. (II) ROI corresponding to chromosomes have been manually defined from tdFLIM images. (III) In each ROI, the decays of GFP-EBP2 were measured in the absence (condition a or green curves) and in the presence of DsRed–EBNA-1 (condition b or red curves). (IV) The experimental curves were further fitted with a biexponential function using Globals Unlimited software in order to quantify the proportion of two donor populations: α1 with a slow Τ1 lifetime corresponding to the unbound donor and α2 with a fast T2 lifetime corresponding to the donor interacting with an acceptor (n = 4).
EBNA-1 and EBP2 interaction cannot be observed even in the presence of oriP.
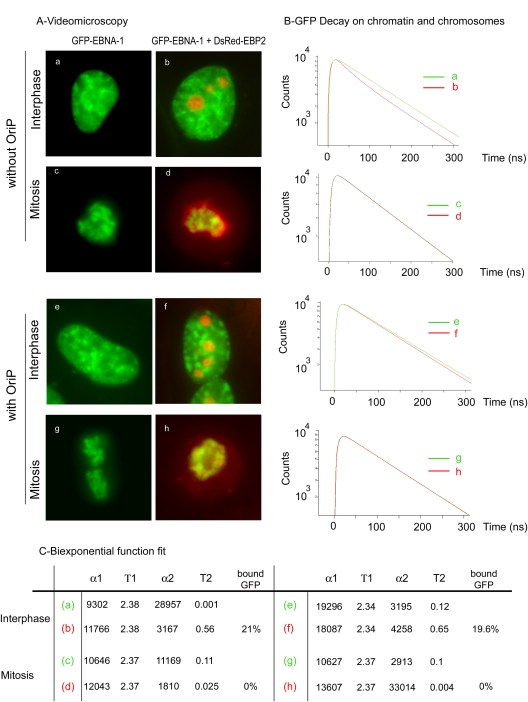
Another major difference between our experiments and previous studies in which EBP2 was claimed to mediate the tethering of EBNA-1 to chromosomes (26, 42, 67, 68) was the absence of a plasmid containing oriP. Indeed, EBNA-1 binds not only to cellular chromatin and chromosomes but also to oriP during the cell cycle (12). Even though oriP and cellular chromatin binding involve two different regions of EBNA-1, its C-terminal and N-terminal parts, respectively, one may suggest that the binding of EBNA-1 to oriP promotes EBNA-1 interaction with EBP2 during mitosis. To address this question, we constructed an oriP plasmid that could encode for DsRed-EBP2. In this way it was possible to evaluate FRET in cells containing different ratios of EBNA-1 over oriP by modulating the relative ratios of GFP–EBNA-1/DsRed-EBP2-oriP plasmids from 2 to 0.5. Since identical results were obtained in all tested conditions, we only present here data obtained for a 1:1 plasmid ratio, which corresponded to the experimental conditions used in previous works that investigated EBNA-1 binding to EBP2 in the presence of oriP (26, 42, 67, 68). We first examined the localization of EBNA-1 and EBP2 in interphase and mitosis by video microscopy. As seen in Fig. 6A, GFP–EBNA-1 and DsRed-EBP2 staining patterns were indistinguishable with (e to h) or without (a to d) the presence of oriP. Interactions between GFP–EBNA-1 and DsRed-EBP2 were analyzed by FRET in the absence or presence of oriP. During interphase, GFP–EBNA-1 fluorescence decay was significantly faster in the presence of DsRed—EBP2 on the chromatin (see red curves corresponding to panels b and f) independently of the presence of oriP. The same results were obtained in nucleoli (data not shown). During mitosis, the presence of oriP did not significantly increase EBNA-1 interaction with EBP2 since no FRET could be detected in the chromosome region. Therefore, we concluded from these experiments that the presence of oriP plasmids did not significantly increase the association of EBNA-1 with EBP2, as detected by FLIM-FRET, either during interphase or during mitosis.
Fig 6.
oriP does not promote interaction between EBNA-1 and EBP2. (A) HeLa cells expressing GFP–EBNA-1 or coexpressing GFP–EBNA-1 and DsRed-EBP2 with or without oriP were observed by video microscopy as indicated. (B) tdFLIM measurements were carried out using scanning confocal time-resolved microscope by acquiring fluorescence decay images for GFP fluorescence, on six different points of HeLa cells transfected with GFP–EBNA-1 alone (green curves a, c, e, and g) or in the presence of DsRed-EBP2 without oriP (red curves b and d) or with oriP (red curves f and h). (C) The experimental curves were further fitted with a biexponential function using Symphotime software in order to quantify the proportion of two donor populations: α1 with a slow Τ1 lifetime corresponding to the unbound donor and α2 with a fast T2 lifetime corresponding to the donor interacting with an acceptor (n = 15).
EBNA-1 interacts with HMGB2 on chromatin during interphase and mitosis.
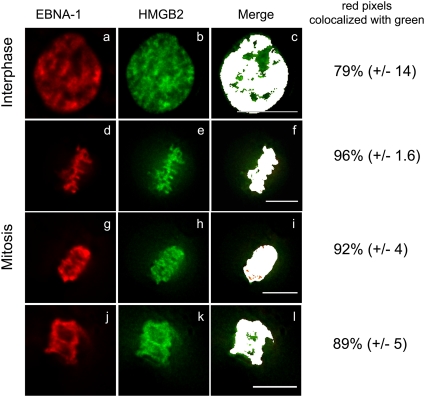
The exact nature and time course of EBNA-1 interaction with cellular chromatin is still highly debated. Since EBP2 did not form detectable complexes with EBNA-1 during mitosis, we wondered whether other cellular proteins might interact with EBNA-1 on mitotic chromosomes. To address this question, we performed a yeast two-hybrid assay using a truncation mutant covering aa 8 to 410 (Δ95-314) as a bait. This truncated form of EBNA-1 lacked the Gly-Arg central repeats, as well as the C-terminal DNA-binding domain, but it encompassed the three known chromosome-binding sites of EBNA-1. Importantly, this truncated form has previously been shown to colocalize with chromatin during interphase and mitosis similarly to wild-type EBNA-1 (36). A total of 75 independent clones encoding 35 different proteins were identified. Ten of these encoded known partners of EBNA-1, including SF2/P32/TAP (66) and EBP-2. We focused on new putative interactors that were known to interact with cellular chromatin. One of them was HMGB2, an abundant and conserved component of the chromatin that is endowed with architectural activities (60). Importantly, HMGB2 has been previously shown to associate with chromosomes during interphase and mitosis (46). As illustrated in Fig. 7, GFP-HMGB2 and DsRed–EBNA-1 colocalized almost perfectly in living cells during interphase and throughout mitosis. Similar observations were made in cells coexpressing DsRed-HMGB2 and GFP–EBNA-1 (data not shown).
Fig 7.
Dynamic localization of EBNA-1 and HMGB2 throughout the cell cycle. Living HeLa cells coexpressing GFP-HMGB2 and DsRed–EBNA-1 were observed during interphase (a to c) and throughout mitosis (d to l) by video microscopy. All images are deconvoluted z-optical sections. Colocalizing pixels (in white) from the merge images were quantified using ImageJ software. The error corresponds to the standard deviation (n = 4). Scale bars, 10 μm.
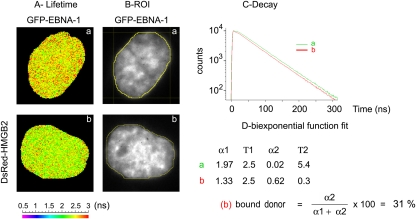
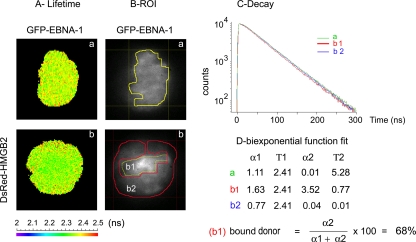
In order to confirm that EBNA-1 effectively interacts with HMGB2 in living human cells, we performed a FRET analysis as described previously. FRET was assessed with the eight possible pairs obtained when combining the four different donors (GFP-HMGB2, HMGB2-GFP, GFP–EBNA-1, EBNA-1–GFP) with the four acceptors (DsRed–EBNA-1, EBNA-1–DsRed, DsRed-HMGB2, HMGB2-DsRed). Whatever the relative positions of the fluorophore in the fusions proteins, a significant FRET was observed when HMGB2 was fused to DsRed and EBNA-1 to GFP (Fig. 8). We could evaluate that an average of 31% of the GFP–EBNA-1 interacted with DsRed-HMGB2 during interphase. This proportion increased up to 68% on mitotic chromosomes (Fig. 9). In contrast, no FRET could be detected when tdFLIM analysis was performed in the nucleoplasm of mitotic cells (Fig. 9).
Fig 8.
HMGB2 and EBNA-1 interact during interphase in living cells. tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images of the GFP donor (515 nm < λ > 560 nm) in mitotic HeLa cells expressing GFP–EBNA-1 alone (images a and green curves) or in the presence of DsRed-HMGB2 (images b and red curves). (A) The tdFLIM images were obtained by analyzing the fluorescence decays with a single lifetime pixel-by-pixel and are displayed as fluorescence lifetime pseudocolor maps. A lifetime colored scale is presented. (B) ROIs corresponding to the nucleus have been manually drawn from tdFLIM images. (C) In each ROI, the decays of GFP–EBNA-1 in the presence of DsRed-HMGB2 (condition b or red curves) were compared to the control decays of GFP–EBNA-1 alone (condition a or green curves). (D) The experimental curves were further fitted with a biexponential function using Globals Unlimited software for quantifying the proportion of two donor populations: α1 with a slow Τ1 lifetime corresponding to the unbound donor and α2 with a fast T2 lifetime corresponding to the donor interacting with an acceptor (n = 10).
Fig 9.
HMGB2 and EBNA-1 interact throughout mitosis in living cells. tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images of the GFP donor (515 nm < λ > 560 nm) in mitotic HeLa cells transfected with GFP–EBNA-1 alone (images a and green curves) or in the presence of DsRed acceptor after a cotransfection with DsRed-HMGB2 (images b and red curves). (A) The tdFLIM images were obtained by analyzing the fluorescence decays with a single lifetime pixel by pixel and are displayed as fluorescence lifetime pseudocolor maps. A lifetime colored scale is presented. (B) ROI corresponding to the chromosomes or to the cytoplasm has been manually drawn from tdFLIM images. (C) In each ROI, the decays of GFP–EBNA-1 in the presence of DsRed-HMGB2 (condition b or red and blue curves) were compared to the control decays of GFP–EBNA-1 alone (condition a or green curves). (D) The experimental curves were further fitted with a biexponential function using Globals Unlimited software for quantifying the proportion of two donor populations: α1 with a slow Τ1 lifetime corresponding to the unbound donor and α2 with a fast T2 lifetime corresponding to the donor interacting with an acceptor (n = 10).
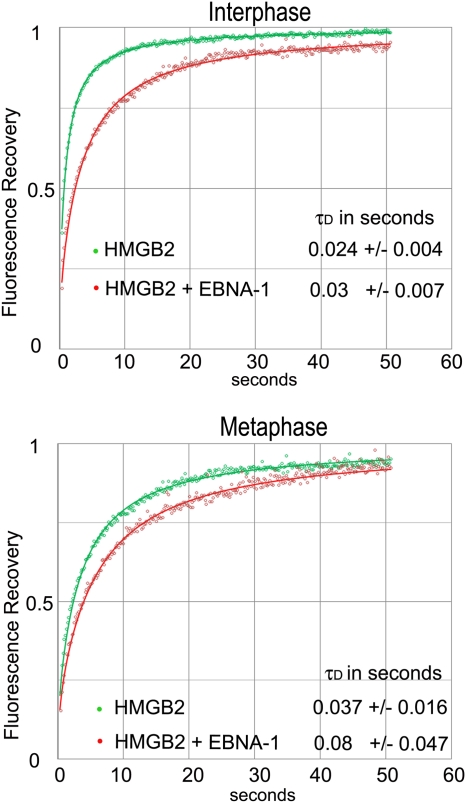
The absence of FRET when HMGB2 was fused to GFP and EBNA-1 was fused to DsRed, whatever their position, may be explained by parameters that reduce FRET efficiency, such as an excessive distance between the fluorophores and/or fluorophore dipoles that do not align even when fusion proteins are in close interaction. To substantiate this hypothesis, we used fluorescence recovery after photobleaching (FRAP) to measure the mobility of HMGB2-GFP in the presence or in the absence of DsRed–EBNA-1. Preliminary experiments indicated that HMGB2-GFP was extremely mobile in living cells, whereas EBNA-1 was stably bound onto chromatin (data not shown). Therefore, HMGB2 mobility was expected to decrease in cells coexpressing EBNA-1 only if both proteins formed a complex. The results of FRAP experiments are depicted in Fig. 10. As expected, EBNA-1–DsRed markedly decreased HMGB2-GFP mobility in mitotic and interphase cells, confirming that HMGB2-GFP and EBNA-1–DsRed interaction occurred in the absence of FRET. These experiments demonstrated that EBNA-1 increased HMGB2 residence time on mitotic chromatin, where they are in close interaction.
Fig 10.
FRAP analysis indicates that HMGB2 mobility is reduced in the presence of EBNA-1. For the upper graph, calculated from 14 cells of a representative experiment, the average fluorescence intensities in the bleached region for GFP-HMGB2 alone (green circles) or GFP-HMGB2 plus DsRed–EBNA-1 (red circles) were measured over time during interphase. The τD values are averages ± the standard deviation. Recovery is faster for GFP-HMGB2 alone: τD of 0.024 s versus 0.03 s in the presence of DsRed–EBNA-1 (Student t test; n = 14, P < 0.01). For the lower graph, calculated from 14 cells of a representative experiment, the average fluorescence intensities in the bleached region for GFP-HMGB2 alone (green circles) and GFP-HMGB2 plus DsRed–EBNA-1 (red circles) were measured during mitosis. The τD values are averages ± the standard deviation. Recovery is faster for GFP-HMGB2 alone: τD of 0.037 s versus 0.08 s in the presence of DsRed–EBNA-1 (Student t test; n = 14, P < 0.01).
To determine the level of specificity of EBNA-1 interaction with HMGB2, we investigated the possible interaction of EBNA-1 with HMGB1, a highly abundant variant of HMGB that is nearly 80% identical to HMGB2 (60). As illustrated in Fig. 11, no FRET could be detected in living HeLa cells coexpressing DsRed-HMGB1 and GFP–EBNA-1 in interphase or in mitosis. Similar results were obtained in cells coexpressing EBNA-1 and HMGB1 fused to GFP or DsRed at their N- or C-terminal extremities (data not shown). Taken together, the results of these experiments established that HMGB2 and EBNA-1 interacted in a highly specific manner both during interphase and mitosis, notably on mitotic chromosomes.
Fig 11.
No interaction can be detected by FRET between HMGB1 and EBNA-1 during interphase and during mitosis in living cells. tdFLIM-FRET measurements were carried out by acquiring fluorescence decay images of the GFP donor (515 nm < λ > 560 nm) in mitotic HeLa cells transfected with GFP–EBNA-1 alone (images a and c and green curves) or in the presence of DsRed-HMGB2 (images b and d and red curves). (A) The tdFLIM images were obtained by analyzing the fluorescence decays with a single lifetime pixel by pixel and are displayed as fluorescence lifetime pseudocolor maps. A lifetime colored scale is presented. (B) ROIs corresponding to the nucleus (images a and b) or to chromosomes (images c and d) have been manually drawn from tdFLIM images. (C) In each ROI, the decays of GFP–EBNA-1 in the presence of DsRed-HMGB1 (condition b during interphase and condition d during mitosis or red curves) were compared to the control decays of GFP–EBNA-1 alone (condition a during interphase and condition c during mitosis or green curves). (D) The experimental curves were further fitted with a biexponential function using Globals Unlimited software for quantifying the proportion of two donor populations: α1 with a slow Τ1 lifetime corresponding to the unbound donor and α2 with a fast T2 lifetime corresponding to the donor interacting with an acceptor (n = 10).
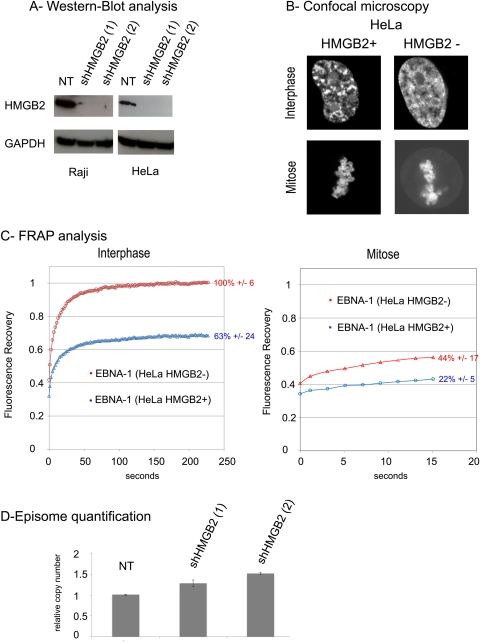
Inhibition of HMGB2 expression affects EBNA-1 stability on chromatin and chromosomes but has no significant impact on episome maintenance.
Considering that HMGB2 and EBNA-1 specifically interacted during interphase and mitosis, we wondered whether HMGB2 contributed to EBNA-1 interaction with cellular chromatin and/or to the maintenance of viral episomes. To address this point, we stably silenced HMGB2 expression in HeLa and Raji cells, a lymphoblastoid cell line that contains an average of 40 episomes per cell. As shown in Fig. 12A, HMGB2 expression was totally abrogated in both cell lines. In HeLa cells that did not express HMGB2, EBNA-1–GFP still colocalized with cellular chromatin both during interphase and mitosis; however, an additional weak diffuse staining was also observed (Fig. 12B), suggesting that the absence of HMGB2 might increase EBNA-1 mobility. To address this question, FRAP experiments were conducted on EBNA-1–GFP in HMGB2+ and HMGB2− HeLa cells (Fig. 12C). We first noticed in HMGB2+ cells during interphase that the FRAP recovery curve did not reach the initial level of fluorescence; only 63% of the bleached EBNA-1 presented a high turnover on chromatin and contributed to the fluorescence recovery (mobile fraction). Consequently, we deduced that 37% was immobile. In the absence of HMGB2, the EBNA-1 mobile fraction increased up to 100%, clearly indicating that HMGB2–EBNA-1 interaction is the determinant stabilizing EBNA-1 on chromatin during interphase. During mitosis, the fraction of immobile EBNA-1 was more important (78.3%), indicating that EBNA-1 interaction with chromosomes was more stable during mitosis. As before, we observed an increased mobility of EBNA-1 in mitotic HeLa cells that did not express HMGB2. Altogether, these results indicated that HMGB2 plays a determining role in stabilizing EBNA-1 binding to chromatin and chromosomes. However, since the immobile fraction did not totally disappear during mitosis, additional mechanisms probably contribute to stabilizing EBNA-1 chromosome binding. We next sought to determine whether HMGB2 inactivation impacts the maintenance of EBV episomes. To answer this question, we measured the relative amounts of viral and cellular genomes in Raji cells in which HMGB2 was inactivated for 4 weeks. The relative viral load was analyzed in two independent cell populations that did not express HMGB2, namely, shHMGB2 (1) and shHMGB2 (2). As shown in Fig. 12D, inactivating HMGB2 did not abrogate episome maintenance, at least over a short period of time. Instead, we observed a slight increase in the viral load in Raji cells that did not express HMGB2; this increase was not statistically significant in this assay.
Fig 12.
HMGB2 stabilizes EBNA-1 interactions with cellular chromatin and chromosomes but does not significantly alter episomes maintenance in Raji cells. (A) HMGB2 and GAPDH expression was performed on Raji and HeLa cells 3 weeks after transduction with a nontargeting shRNA (NT) or two different shHMGB2 expressing lentiviral vectors. (B) EBNA-1–GFP was transiently expressed in HeLa cells expressing HMGB2 (HMGB2+) or not (HMGB2−). The cells were observed by confocal fluorescence microscopy during interphase and mitosis. (C) FRAP analyzes of EBNA-1 were also performed on HeLa cells HMGB2+ (blue curves) and HMGB2− (red curves). Calculated from nine cells, the average fluorescence intensities in the bleached region were measured for 250 s in interphase and for 15 s at different time points along mitosis to avoid that the movement of chromosomes induces an artifactual fluorescence recovery measure. The percentages of the mobile fraction in front of each curve are averages ± the standard deviation. The differences in mobile fraction values between EBNA-1 in HMGB2+ and EBNA-1 in HMGB2− are statistically significant (Mann-Whitney nonparametric U test; n = 9, P < 0.05). (D) Raji cells expressing HMGB2 (NT) or not—“shHMGB2 (1)” and “shHMGB2 (2)”—were grown for 3 weeks. Viral and cellular DNA were quantified by quantitative PCR as described in the text. The results are expressed as the relative amount of viral over cellular DNA. The ratio was set to 1 for the original Raji cells (NT).The standard deviation is shown. No statically significant differences could be detected between the three cell lines (n = 3 repeats per cell line).
DISCUSSION
EBP2 is the only EBNA-1-interacting protein identified to date that has been proposed to mediate EBNA-1 docking onto mitotic chromosomes (58, 67). This model was initially based on the apparent colocalization of both proteins in demecholcine-arrested cells (26, 42, 67, 68). In addition, a set of convincing observations argued for a central role for EBP2 in episome maintenance (26, 58). Although different mechanisms have been proposed to explain EBNA-1 interaction with mitotic chromosomes, direct evidences for EBP2 being responsible for EBNA-1 docking onto mitotic chromosomes were still lacking. Moreover, a possible interaction of EBNA-1 with interphase chromatin has been much debated but was still lacking definitive proofs. A recent study established that EBP2 redistribution from the nucleolus to the chromosomal peripheral layer occurred at late prophase (42). We showed here that EBP2 localization during mitosis was not restricted to the periphery of chromosomes. Indeed, EBP2 was also detected in the interzone of the mitotic spindle and homogeneously dispersed within the nucleoplasm. This localization was highly reminiscent of previous observations made on B23 by Ochs et al. (45). B23 is one of the most abundant proteins of the nucleolus. Similarly to EBP2, B23 has been involved in rRNA processing and localized in the granular component of the nucleolus during interphase (7). Interestingly, B23 and some other nucleolar proteins such as the fibrillarin, nucleolin, and Ki-67 are components of the perichromosomal layer that covers each chromosome from one telomere to the other and seems to act as a peripheral skeleton around mitotic chromosomes (17, 56; for a review, see references 20 and 65). Similarities between EBP2 and B23 localization during mitosis suggest that EBP2 might also be a component of the perichromosomal layer. This structure might constitute a docking site, allowing proteins necessary for early nuclear assembly to segregate toward daughter cells equally (65). These observations were still compatible with EBP2 acting as a molecular bridge between EBNA-1 and mitotic chromosomes.
In the present study, we demonstrated by FRET that EBNA-1 and EBP2 interacted during interphase in living cells both in the nucleoplasm and in the nucleoli. However, despite a partial colocalization of both proteins during mitosis, no interaction between EBNA-1 and EBP2 was detected whatever the stage of mitosis. Importantly, reproducing experimental conditions depicted in previous experiments that studied EBNA-1–EBP2 binding with the presence of oriP plasmid, we obtained the same results, indicating that oriP is unlikely to promote EBNA-1 interaction with EBP2 in living cells. One might argue that FRET could provide false-negative results. Indeed, the efficiency of energy transfer depends not only on the distance between the donor and acceptor molecules but also on the relative orientation of their respective transition dipole moment (15). These factors are therefore influenced by the spatial arrangement of the fusion proteins within the complex they form. We do not favor this hypothesis for two reasons. First, FRET was observed in interphase but not in mitosis in cells expressing the same pair of fusion proteins. Second, FRET between EBP2 and EBNA-1 could be artificially restored by treating the cells with demecholcine, a drug that induces microtubule disassembly. Accordingly, demecholcine promoted the subsequent relocalization of EBP2 on chromosomes, which confirmed that EBNA-1–EBP2 interaction was hampered due to the spatial distribution of the cellular components during mitosis.
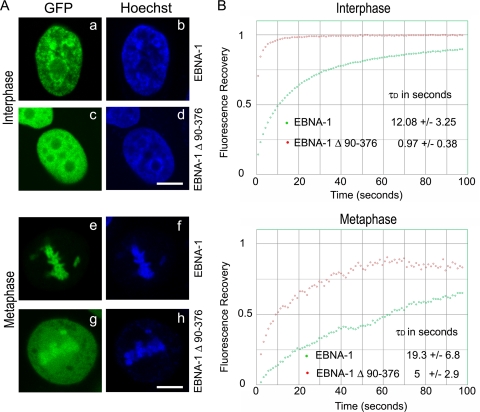
This observation strongly contradicted a current model, which stipulated that EBP2 was an obligate partner of EBNA-1 on mitotic chromosomes (26, 42, 67, 68). Other studies have already underlined some limits of this model. Indeed, a truncated form of EBNA-1 that lacked the EBP2 binding site (EBNA-1 Δ325-376) still localized on mitotic chromosomes (54). Sears et al. suggested that EBNA-1 could bind mitotic chromosomes independently of EBP2, a process that would involve an AT hook domain contained in the N-terminal CBS (54). Importantly, other experiments from our laboratory demonstrated that aa 325 to 376 were somewhat important for stabilizing EBNA-1 on chromosomes during mitosis. FRAP experiments were conducted on EBNA-1 and on a truncated form of EBNA-1 that did not contain EBP2-binding domain (Fig. 13). During interphase, the τD for the fluorescence recovery was 12.08 s for EBNA-1 and 12-fold shorter (τD = 0.97 s) for the truncated variant of EBNA-1, a finding in agreement with EBNA-1 interacting with EBP2 during interphase. In mitotic cells, the τD for EBNA-1–GFP was slightly longer (19.3 s), indicating that EBNA-1 interaction with chromosomes was more stable than during interphase, which was further confirmed by additional experiments (see Fig. 12). Although EBNA-1 did not interact with EBP2 during mitosis, EBP2-binding domain seems to be also important at that time, since the τD recovery for the truncated form of EBNA-1 was 5-fold shorter (5 s). Therefore, two non-mutually-exclusive hypotheses could be proposed. First, EBP2 might be required to load EBNA-1 onto chromatin during or at the end of interphase but would not be required later on. Second, aa 325 to 376 could be involved in another, as-yet-undiscovered interaction that would be necessary to stably maintain EBNA-1 on chromosomes during mitosis. The absence of an interaction between EBP2 and EBNA-1 during mitosis may not be inconsistent with an essential role for EBP2 in episome maintenance. Indeed, the localization of EBP2 around mitotic chromosomes and in the interzone of the mitotic spindle suggest a role for EBP2 in chromosome stabilization and/or in chromatid segregation. This may explain why oriP-containing plasmids are improperly transmitted in cells where EBP2 expression has been silenced or when EBP2 was dissociated from chromosomes by Aurora kinase (25). This would also be compatible with a possible role for EBP2 in chromosome stability, as suggested by Lee et al. (33).
Fig 13.
Contribution of the EBP2 binding site (or CBS2) to the localization and stability of EBNA-1 on chromatin and chromosomes. (A) HeLa cells transfected with GFP–EBNA-1 or GFP–EBNA-1Δ90-376 were observed by confocal microscopy during interphase and mitosis as described in Materials and Methods. (B) FRAP analysis of GFP–EBNA-1 and GFP–EBNA-1Δ90-376. For the upper graph, calculated from 12 cells of a representative experiment, the average fluorescence intensities in the bleached region for GFP–EBNA-1 (green circles) and GFP–EBNA-1Δ90-376 (red circles) were measured over time during interphase. The τD values are averages ± the standard deviation. Recovery is faster for GFP–EBNA-1Δ90-376 than for GFP–EBNA-1: τD of 0.97 s and 12.08 s, respectively (Student t test; n = 12, P < 0.005). For the lower graph, calculated from six cells of a representative experiment, the average fluorescence intensities in the bleached region for GFP–EBNA-1 (green circles) and GFP–EBNA-1Δ90-376 (red circles) were measured during mitosis. The τD values are averages ± the standard deviation. Recovery is faster for GFP–EBNA-1Δ90-376 than for GFP–EBNA-1: τD of 5 s and 19.3 s, respectively (Student t test; n = 6, P < 0.005).
We report here for the first time that EBP2 and EBNA-1 unexpectedly interact during interphase mainly within the nucleoli. EBP2 is a nucleolar protein that is highly conserved in eukaryotes. Its functions have been associated with the maturation of 25S rRNA and assembly of 60S ribosomal subunits in S. cerevisiae (64). EBP2 is likely to contribute to ribosome biogenesis in human as well, since it was identified in nucleoli from HeLa cells (53). A direct contribution of EBNA-1 in ribosome biogenesis is therefore possible, although this has not been formally demonstrated to our knowledge. The high proportion of EBP2 that is engaged in an interaction with EBNA-1 in the nucleoli suggests that EBP2–EBNA-1 interaction is functionally different from what has been proposed thus far and may be related to some unreported activity of EBNA-1 in the nucleolus. In the last few years, a growing list of studies has shown that viruses, expressed nucleolar proteins that are able to interfere with nucleolus functions, with major consequences on viral replication cycle and/or virus-induced pathology (for reviews, see references 21 and 22). EBP2 activities have also been linked to cell growth control and tumorigenesis. Indeed, EBP2 expression is required for human cell proliferation, but its overexpression has been associated with an increased expression of cyclin E1 and subsequent chromosome instability (26, 33). Whether EBNA-1 modulates such aspect of EBP2 functions during interphase merits a careful examination.
Similarly to EBV, other persistent viruses have developed a mechanism to anchor their episomal genomes to the cellular chromosomes during mitosis. This was notably described for Kaposi's sarcoma-associated herpesvirus and papillomaviruses, in which two proteins—LANA and E2, respectively—act similarly to EBNA-1 for episome maintenance. However, the nature of EBNA-1, LANA, and E2 interaction with chromatin is different. Bovine papillomavirus type 1 (BPV-1) E2 protein is randomly distributed over all chromosomes as small dots that costain with the cellular protein Brd4 (6, 37), a cellular protein that is essential for the maintenance of BPV-1 genomes (72). In contrast, the human papillomavirus type 8 (HPV8) E2 protein forms large speckles on the short arm of acrocentric chromosomes, a region corresponding to nucleolar organizing region (NOR). Whereas almost all components of nucleoli are dispersed throughout the cell during mitosis, components of fibrillar centers (mainly the rRNA gene and RNA polymerase I complex) remains associated to chromosomes at NORs. The colocalization of HPV8 E2 and NORs is consistent with its capacity to bind to the rRNA gene and colocalize with the ribosomal transcription factor UBF1 (49). LANA fused with GFP diffusely paints all chromosomes and directly binds core histone H2A-H2B (5, 48). As observed in our work and by others, EBNA-1–GFP associated diffusely with all chromosomes independently of the presence of oriP region.
To identify other proteins that could modulate EBNA-1 interaction with the cellular chromatin, we performed a double-hybrid screen that identified HMGB2 as a new partner of EBNA-1. A combination of FRET and FRAP approaches allowed us to demonstrate that HMGB2 and EBNA-1 established a highly specific interaction both during interphase and mitosis. To our knowledge, this is the first evidence of EBNA-1 interacting directly with a known chromatin component. HMGB2, as well as its highly homologous variant HMGB1, binds within the minor groove of AT-rich B-form DNA with no sequence specificity and increases the flexibility of the DNA to which they are bound (60). HMGB may also modulate nucleosome positioning and stability and therefore influence DNA accessibility (40). Altogether, these activities are thought to modulate the local architecture of the chromatin and to facilitate the access of many proteins to their target sequence, including Hox proteins (74), steroid hormone receptors (8), P53 (23, 61), P73 (61), Oct1, Oct2, and Oct3/4 (9, 75), Rel family transcription factors (2), and others. Importantly, HMGB interaction with DNA and/or proteins is usually extremely weak both in vitro and in vivo. In vitro binary or ternary complexes with DNA are often difficult to detect by conventional double-hydride or biochemical approaches (2). In living cells, HMGB1 and HMGB2 interact with chromatin during interphase and mitosis but this interaction is labile and both proteins are highly mobile in the nucleus (46). Recent work by Agresti et al. demonstrated that HMGB1 interaction with glucocorticoid receptor (GR) occurs only on chromatin and decreased both HMGB1 and GR mobility (2). This model would be in agreement with our data as well. Indeed, EBNA-1–DsRed markedly decreased HMGB2-GFP mobility in mitotic and interphase cells, suggesting that HMGB2 residence time was increased on chromatin when it associated with EBNA-1. Our results clearly indicated that HMGB2–EBNA-1 interaction took place on condensed chromosomes during mitosis, but we cannot exclude that this interaction occurs also out of the chromatin during interphase, although EBNA-1 mostly colocalized with DNA Hoechst staining. Considering the known HMGB2 properties, we believe that HMGB2 may provide favorable conditions for promoting and/or stabilizing EBNA-1 binding to cellular DNA. This cooperating mechanism would be compatible with other models proposed to mediate EBNA-1 association with chromatin and mitotic chromosomes. Indeed, EBNA-1 contain AT-hook structures that could bind directly to AT-rich DNA (54) and also RGG motifs that allow DNA loading or DNA stability by the way of G-quadruplex RNA (44). As a matter of fact, whereas EBNA-1 mobility was markedly increased in the absence of HMGB2, EBNA-1 ability to stably interact with mitotic chromosomes was not entirely abrogated. This result confirmed the notion that EBNA-1 anchorage on chromatin is likely to involve additional partners. Whereas similar experimental conditions were used to investigate EBNA-1–HMGB1 and EBNA-1–HMGB2 interactions, we did not observe any interaction between HMGB1 and EBNA-1. Therefore, we consider that HMGB1 cannot substitute for HMGB2 for EBNA-1 interaction on chromatin, although HMGB1 is abundantly expressed and functionally redundant to HMGB2 (Fig. 11).
Importantly, inactivating HMGB2 in Raji cells had no dramatic effect on the average viral load over a 3- to 4-week period. This result indicates that HMGB2 would be dispensable for the functions of EBNA-1 implicated in episome replication, partitioning and stability. Alternatively, the maintenance of viral episomes over a few weeks may still be compatible with abnormalities in the episome segregation, a process that merits a careful examination in the context of a HMGB2-negative background. Moreover, a longer period of growth may be required to detect a significant defect in episome maintenance.
ACKNOWLEDGMENTS
We thank the core facility Imagerie des Processus Dynamiques en Biologie Cellulaire et Biologie du Développement of the Institut Jacques Monod and the cell imagery facility of the UMRS 872. We also thank Thérèse Pothier for carefully reading and correcting the manuscript.
This study was supported by funding from UPMC and INSERM to the laboratory of V.M.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Adams A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agresti A, Lupo R, Bianchi ME, Muller S. 2003. HMGB1 interacts differentially with members of the Rel family of transcription factors. Biochem. Biophys. Res. Commun. 302:421–426 [DOI] [PubMed] [Google Scholar]
- 3. Altmann M, et al. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. U. S. A. 103:14188–14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ambinder RF, Shah WA, Rawlins DR, Hayward GS, Hayward SD. 1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 64:2369–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barbera AJ, et al. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861 [DOI] [PubMed] [Google Scholar]
- 6. Baxter MK, McPhillips MG, Ozato K, McBride AA. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 79:4806–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biggiogera M, et al. 1989. Simultaneous immunoelectron microscopic visualization of protein B23 and C23 distribution in the HeLa cell nucleolus. J. Histochem. Cytochem. 37:1371–1374 [DOI] [PubMed] [Google Scholar]
- 8. Boonyaratanakornkit V, et al. 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18:4471–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Butteroni C, De Felici M, Scholer HR, Pesce M. 2000. Phage display screening reveals an association between germline-specific transcription factor Oct-4 and multiple cellular proteins. J. Mol. Biol. 304:529–540 [DOI] [PubMed] [Google Scholar]
- 10. Canaan A, et al. 2009. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc. Natl. Acad. Sci. U. S. A. 106:22421–22426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clegg RM, Holub O, Gohlke C. 2003. Fluorescence lifetime-resolved imaging: measuring lifetimes in an image. Methods Enzymol. 360:509–542 [DOI] [PubMed] [Google Scholar]
- 12. Daikoku T, et al. 2004. In vivo dynamics of EBNA1-oriP interaction during latent and lytic replication of Epstein-Barr virus. J. Biol. Chem. 279:54817–54825 [DOI] [PubMed] [Google Scholar]
- 13. Dhar SK, et al. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287–296 [DOI] [PubMed] [Google Scholar]
- 14. Dresang LR, Vereide DT, Sugden B. 2009. Identifying sites bound by Epstein-Barr nuclear antigen 1 (EBNA1) in the human genome: defining a position-weighted matrix to predict sites bound by EBNA1 in viral genomes. J. Virol. 83:2930–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Förster T. 1946. Energiewanderung und fluorezenz. Naturwissenschaften 6:166–175 [Google Scholar]
- 16. Gardner JS, Chiu AL, Maki NE, Harris JF. 1985. A quantitative stability analysis of human monoclonal antibody production by heteromyeloma hybridomas, using an immunofluorescent technique. J. Immunol. Methods 85:335–346 [DOI] [PubMed] [Google Scholar]
- 17. Gautier T, et al. 1994. Fate of specific nucleolar perichromosomal proteins during mitosis: cellular distribution and association with U3 snoRNA. Biol. Cell 82:81–93 [DOI] [PubMed] [Google Scholar]
- 18. Grogan EA, et al. 1983. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc. Natl. Acad. Sci. U. S. A. 80:7650–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrison S, Fisenne K, Hearing J. 1994. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 68:1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hernandez-Verdun D, Gautier T. 1994. The chromosome periphery during mitosis. Bioessays 16:179–185 [DOI] [PubMed] [Google Scholar]
- 21. Hiscox JA. 2002. The nucleolus: a gateway to viral infection? Arch. Virol. 147:1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hiscox JA. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jayaraman L, et al. 1998. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12:462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanda T, Otter M, Wahl GM. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kapoor P, Frappier L. 2005. Methods for measuring the replication and segregation of Epstein-Barr virus-based plasmids. Methods Mol. Biol. 292:247–266 [DOI] [PubMed] [Google Scholar]
- 26. Kapoor P, Lavoie BD, Frappier L. 2005. EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol. Cell. Biol. 25:4934–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kapoor P, Shire K, Frappier L. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaschka-Dierich C, Falk L, Bjursell G, Adams A, Lindahl T. 1977. Human lymphoblastoid cell lines derived from individuals without lymphoproliferative disease contain the same latent forms of Epstein-Barr virus DNA as those found in tumor cells. Int. J. Cancer 20:173–180 [DOI] [PubMed] [Google Scholar]
- 29. Krysan PJ, Haase SB, Calos MP. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lakowicz JR. 1980. Fluorescence spectroscopic investigations of the dynamic properties of proteins, membranes and nucleic acids. J. Biochem. Biophys. Methods 2:91–119 [DOI] [PubMed] [Google Scholar]
- 31. Laurent B, et al. 2010. High-mobility group protein HMGB2 regulates human erythroid differentiation through trans-activation of GFI1B transcription. Blood 115:687–695 [DOI] [PubMed] [Google Scholar]
- 32. Lee MA, Diamond ME, Yates JL. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 73:2974–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee MC, et al. 2008. Ectopic EBP2 expression enhances cyclin E1 expression and induces chromosome instability in HEK293 stable clones. BMB Rep. 41:716–721 [DOI] [PubMed] [Google Scholar]
- 34. Lindahl T, et al. 1976. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J. Mol. Biol. 102:511–530 [DOI] [PubMed] [Google Scholar]
- 35. Lu F, et al. 2010. Genome-wide analysis of host-chromosome binding sites for Epstein-Barr virus nuclear antigen 1 (EBNA1). Virol. J. 7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marechal V, et al. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McPhillips MG, Ozato K, McBride AA. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 79:8920–8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Middleton T, Sugden B. 1994. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J. Virol. 68:4067–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 71:4882–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muller S, et al. 2001. New EMBO members' review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 20:4337–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nanbo A, Sugden A, Sugden B. 2007. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 26:4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nayyar VK, Shire K, Frappier L. 2009. Mitotic chromosome interactions of Epstein-Barr nuclear antigen 1 (EBNA1) and human EBNA1-binding protein 2 (EBP2). J. Cell Sci. 122:4341–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Niederman JC, McCollum RW, Henle G, Henle W. 1968. Infectious mononucleosis: clinical manifestations in relation to EB virus antibodies. JAMA 203:205–209 [DOI] [PubMed] [Google Scholar]
- 44. Norseen J, Johnson FB, Lieberman PM. 2009. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 83:10336–10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ochs R, Lischwe M, O'Leary P, Busch H. 1983. Localization of nucleolar phosphoproteins B23 and C23 during mitosis. Exp. Cell Res. 146:139–149 [DOI] [PubMed] [Google Scholar]
- 46. Pallier C, et al. 2003. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol. Biol. Cell 14:3414–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Phair RD, Gorski SA, Misteli T. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375:393–414 [DOI] [PubMed] [Google Scholar]
- 48. Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poddar A, Reed SC, McPhillips MG, Spindler JE, McBride AA. 2009. The human papillomavirus type 8 E2 tethering protein targets the ribosomal DNA loci of host mitotic chromosomes. J. Virol. 83:640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rawlins DR, Milman G, Hayward SD, Hayward GS. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859–868 [DOI] [PubMed] [Google Scholar]
- 51. Ritzi M, et al. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971–3984 [DOI] [PubMed] [Google Scholar]
- 52. Sample J, Henson EB, Sample C. 1992. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J. Virol. 66:4654–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scherl A, et al. 2002. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13:4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sears J, et al. 2004. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78:11487–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shah KM, Young LS. 2009. Epstein-Barr virus and carcinogenesis: beyond Burkitt's lymphoma. Clin. Microbiol. Infect. 15:982–988 [DOI] [PubMed] [Google Scholar]
- 56. Sheval EV, Polyakov VY. 2008. The peripheral chromosome scaffold, a novel structural component of mitotic chromosomes. Cell Biol. Int. 32:708–712 [DOI] [PubMed] [Google Scholar]
- 57. Shirakata M, Hirai K. 1998. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J. Biochem. 123:175–181 [DOI] [PubMed] [Google Scholar]
- 58. Shire K, Ceccarelli DF, Avolio-Hunter TM, Frappier L. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soumpasis DM. 1983. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41:95–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stros M. 2010. HMGB proteins: interactions with DNA and chromatin. Biochim. Biophys. Acta 1799:101–113 [DOI] [PubMed] [Google Scholar]
- 61. Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. 2002. HMGB1 and HMGB2 cell-specifically downregulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 277:7157–7164 [DOI] [PubMed] [Google Scholar]
- 62. Thompson MP, Kurzrock R. 2004. Epstein-Barr virus and cancer. Clin. Cancer Res. 10:803–821 [DOI] [PubMed] [Google Scholar]
- 63. Tramier M, et al. 2002. Picosecond-hetero-FRET microscopy to probe protein-protein interactions in live cells. Biophys. J. 83:3570–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsujii R, et al. 2000. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells 5:543–553 [DOI] [PubMed] [Google Scholar]
- 65. Van Hooser AA, Yuh P, Heald R. 2005. The perichromosomal layer. Chromosoma 114:377–388 [DOI] [PubMed] [Google Scholar]
- 66. Wang Y, Finan JE, Middeldorp JM, Hayward SD. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18–29 [DOI] [PubMed] [Google Scholar]
- 67. Wu H, Ceccarelli DF, Frappier L. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu H, Kapoor P, Frappier L. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yates J, Warren N, Reisman D, Sugden B. 1984. A cis-Acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 81:3806–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yates JL, Guan N. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yates JL, Warren N, Sugden B. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812–815 [DOI] [PubMed] [Google Scholar]
- 72. You J, Croyle JL, Nishimura A, Ozato K, Howley PM. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349–360 [DOI] [PubMed] [Google Scholar]
- 73. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]
- 74. Zappavigna V, Falciola L, Helmer-Citterich M, Mavilio F, Bianchi ME. 1996. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 15:4981–4991 [PMC free article] [PubMed] [Google Scholar]
- 75. Zwilling S, Konig H, Wirth T. 1995. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14:1198–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]