Abstract
Thermodynamic parameters for GU pairs are important for predicting the secondary structures of RNA and for finding genomic sequences that code for structured RNA. Optical melting curves were measured for 29 RNA duplexes with GU pairs to improve nearest neighbor parameters for predicting stabilities of helixes. The updated model eliminates a prior penalty assumed for terminal GU pairs. Six additional duplexes with the 5′GG/3′UU motif were added to the single representation in the previous database. This revises the ΔG°37 for the 5′GG/3′UU motif from an unfavorable 0.5 kcal/mol to a favorable −0.2 kcal/mol. Similarly, the ΔG°37 for the 5′UG/3′GU motif changes from 0.3 to −0.6 kcal/mol. The correlation coefficients between predicted and experimental ΔG°37, ΔH°, and ΔS° for the expanded database are 0.95, 0.89, and 0.87, respectively. The results should improve predictions of RNA secondary structure.
The explosion of biological data in the genomics era has filled databanks with large amounts of genetic information. Understanding of these data and making correlations are vital for maximally advancing the fields of biology and medicine. This necessitates accurate methods in bioinformatics and computational chemistry. One important area that bioinformatics and computational chemistry address is finding, predicting, and determining RNA structure from sequence.1
RNA participates in a variety of cellular functions involving gene expression and regulation. RNA typically folds in a hierarchical way.2,3 Base pairs form to generate motifs such as helixes and loops. Higher order interactions between these features result in three-dimensional structures. On that basis, knowledge of secondary structure is critical for the prediction of tertiary structure. Secondary structure prediction algorithms utilizing experimental thermodynamic data4−9 have relied on nearest neighbor models.10−13 Finding regions of genome sequences that code for structured RNA often also relies on nearest neighbor models.1,14−16 Because RNA molecules and their reverse complements can fold similarly, the thermodynamics of GU pairs provides information about the reading direction because their complement, CA, forms less stable base pairs.17
Prediction of GU pairs is also important because they are the most common non-Watson–Crick pair and have functions in a wide variety of RNAs. For example, GU pairs are found within two helical regions and at the junction of a helix and multibranch loop in eukaryotic 5S rRNA.18,19 A GU pair in the third position of the acceptor stem in tRNAAla20 distorts helix geometry21 and is important in Escherichia coli for recognition by alanine aminoacyl tRNA synthetase.22−24 Local helix geometry due to a conserved GU pair may also be important for binding of a yeast intron with hPrp8 or L32 protein.25,26 The U-rich tail of guide RNAs bind to a purine-rich region in unedited pre-mRNA to generate recurring 5′AGA/3′UUU motifs that may help RNA editing proteins bind to the major groove.27,28 The 5′ leader of HIV-1 can switch between helixes containing GU pairs to promote translation or packaging of its genome.29
GU pairs expose the exocyclic amino group of guanine in the minor groove, presenting a unique site for hydrogen bonding to facilitate function and molecular recognition. For example, a GU amino group at the splice site in the Tetrahymena thermophilia group I intron helps bind and align the splice site30−32 and stabilize the transition state of the splicing reaction.33 Šponer et al. reported a common tertiary interaction involving a GU pair, where the exocyclic NH2 of the G and the 2′OH of the U form hydrogen bonds, respectively, with the 2′OH and carbonyl oxygen of a cytidine in a GC pair of another helix.34
GU pairs can be metal ion binding sites.35−39 Colmenarejo and Tinoco observed that Co(NH3)63+ preferably binds to 5′GU/3′UG and 5′GG/3′UU over 5′UG/3′GU pairs, whereas Mg(H2O)62+ binds tightest to 5′UG/3′GU.40 This preference may explain why 5′UG/3′GU is the most prevalent tandem GU motif in rRNA.41 The propensity for binding metal ions allows design of sequences that bind heavy metals to facilitate solving of X-ray structures.37,38
Prediction of GU pairs often relies on a nearest neighbor model for folding stability. The database of RNA sequences from which GU nearest neighbor parameters were derived12 is relatively small, however, compared to that for Watson–Crick nearest neighbors.11 To expand the database, optical melting experiments were carried out on 29 oligoribonucleotide duplexes. Linear regression analysis on the expanded database provides a revised set of individual nearest neighbor (INN) parameters,42 which are reported herein. The parameters provide stability increments for internal and single terminal GU pairs. Stability increments for additional terminal GU pairs have been reported by Nguyen and Schroeder.43
Materials and Methods
Design of Oligonucleotides
Oligonucleotides were designed to expand the previous database12 to provide all possible combinations of base pair triplets containing GU pairs flanked by Watson–Crick pairs in different orientations (Table 1) and to have a substantial representation of each nearest neighbor containing a GU pair. An additional six sequences containing the 5′GG/3′UU doublet provided nine new representations for that motif, which had only one representation. Care was taken to select self-complementary sequences that do not favorably form alternative secondary structures, such as hairpins or loops.
Table 1. Occurrence of Each Base Pair Triplet 5′WGY/3′XUZ in the Database of RNA Sequences from Which INN Parameters for GU Pairs Were Derived.
| WX/YZ | AU | CG | GC | UA | GU | UG |
|---|---|---|---|---|---|---|
| AU | 2 | 3 | 2 | 4 | 1 | 6 |
| CG | 2 | 4 | 4 | 3 | 2 | 7 |
| GC | 8 | 2 | 3 | 2 | 4 | 1 |
| UA | 4 | 2 | 2 | 2 | 2 | 2 |
| GU | 2 | 5 | 1 | 1 | 0 | 0 |
| UG | 4 | 5 | 5 | 6 | 0 | 0 |
Synthesis and Purification of Oligoribonucleotides
Sequences for the following oligoribonucleotide duplexes were purchased from Integrated DNA Technologies (IDT): r(AGGCUU)2, r(AUGCGU)2, r(AGUCGAUU)2, r(CUGGCUAG)2, r(5′CAGAGGAGAC/3′GUCUUUUCUG), r(CAGUCGAUUG)2, r(CCGAAUUUGG)2, r(CGGAAUUUCG)2, r(CGGAUAUUCG)2, r(CGGGCGUUCG)2, r(CUGGAUUCAG)2, r(GAGAGCUUUC)2, r(GAGGAUCUUC)2, r(5′GAGUGGAGAG/3′CUCAUUUCUC), r(GGUUCGGGCC)2, and r(GUGAAUUUAC)2 (the / denotes a nonself-complementary duplex). Purity was checked by NMR except for those forming duplexes with adjacent GU pairs, which were checked by thin layer chromatography. All other sequences were synthesized and purified as previously described.44 All sequences were desalted with Sep-Pak C18 cartridges (Supporting Information).
UV Melting
RNA duplexes with concentrations from 10–6–10–3 M were melted in 0.5 mM Na2EDTA, 1 M NaCl, and 20 mM sodium cacodylate, pH 7, which maintains a stable pKa over a wide temperature range.45 Absorbance at 280 nm, typically from 15 to 80 °C, was measured on a Beckman Coulter DU 640 spectrophotometer.
NMR Experiments
Spectra were acquired on a Varian Inova 500 or 600 MHz spectrometer. The buffer for NMR was 80 mM NaCl, 18.8 mM NaH2PO4, 1.16 mM Na2HPO4, 0.02 mM Na2EDTA, pH 6.0, to which 15 μL of D2O was added to provide a lock signal. One-dimensional 1H spectra were acquired with the water 1H signal suppressed with a binomial 1:1 shaped pulse.46 Two-dimensional 1H–1H NOESY and 1H–1H TOCSY spectra were acquired with the water signal suppressed by a WATERGATE-type pulse sequence with flipback.47,48 Two-dimensional 1H–1H NOESY spectra for r(AUGCGU)2 were also measured in D2O.
Spectra were processed with NMRPipe49 and resonances were assigned with SPARKY.50 Proton chemical shifts were referenced to a temperature-dependent water chemical shift, δ,
| 1 |
where T is temperature in Kelvin.51 The internal reference standard for water was 2,2-dimethylsilapentate-5-sulfonic acid.
Melting Data Analysis
Melting curves for each duplex were fit to a two-state model with MeltWin 3.552 to derive values for ΔH° and ΔS°. The melting temperature, TM, was plotted against ln(CT/a) to provide another measure of ΔH° and ΔS°:
| 2 |
Here R is the gas constant (1.987 cal K–1 mol–1), CT is the total concentration of strands, and a is 1 for self-complementary duplexes and 4 for non-self-complementary duplexes. Sequences were added to the database if ΔH° values derived from averaging fits of melting curves agreed within 15% with these derived from eq (2), consistent with the two-state model.
Linear Regression to Fit Nearest Neighbor Parameters
Nearest neighbor thermodynamic parameters were obtained with a regression function reported by Xia et al.11 Matrix calculations were performed with R53 and independently verified with Mathematica 8.054 and Octave.55 All three software packages yielded nearly identical results.
Terms representing free energy contributions from non-GU nearest neighbors, that is, helix initiation (ΔG°init), symmetry (ΔG°sym), terminal AU pairs (ΔG°term AU), and Watson–Crick nearest neighbors (ΔG°j (WC NN)),11 were subtracted from the free energy found from the TM–1 vs ln(CT/a) plots (ΔG°i(duplex)) to provide an experimental free energy attributable to the GU components of each duplex:
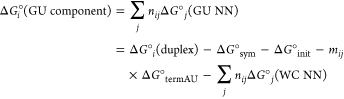 |
3 |
where i and j are labels for each different duplex and INN parameter, respectively, NN stands for nearest neighbor parameter, and mij is the number of terminal AU pairs. For example,
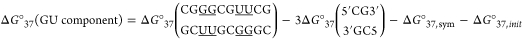 |
4 |
Here, ΔG°37(GU component) contains four 5′GG/3′CU nearest neighbors and two 5′GG/3′UU nearest neighbors. Values for Watson–Crick nearest neighbors from Xia et al.11 were used because experimental measurements on 22 duplexes not included in the fitting by Xia et al.11 are predicted within experimental error (Supporting Information). Making the new GU parameters consistent with the Xia et al.11 parameters provides compatibility with loop parameters derived with Xia et al.11 nearest neighbor parameters and allows easy adoption by programs using those parameters.
Each experimental duplex ΔG°37 was given an error limit of ±4% to account for systematic errors unless the percent difference between parameters found from TM–1 vs ln(CT/a) and averaged curve fits was greater. For the seven latter cases, this percent difference was doubled to provide an error limit. Error limits for ΔH° were assumed to be 12%.11 The symmetry contribution, 0.43 kcal/mol in ΔG°37, has no error56 and was therefore subtracted from ΔG°37 of self-complementary duplexes before calculating the error limit.
The GU component free energies were placed into M × 1 matrix G, where M is the number of duplexes.
| 5 |
S is an M × N matrix containing the counts of each nearest neighbor doublet in a duplex, where N is the number of GU nearest neighbor parameters being fit. GNN is an N × 1 matrix that contains the nearest neighbor parameters to be derived from G and S.
The general law of error propagation was used to calculate the variances for each duplex.57,58 Multiplication of both sides of eq (5) by an M × M matrix, σ–1, containing the variances in the diagonals yielded error-weighted matrices from which thermodynamic parameters were derived.
| 6 |
The values in GNN are thus Sσ–1·Gσ. The variances of each INN parameter are obtained with singular value decomposition (SVD) (ref (11), Supporting Information). Nearest neighbor parameters for ΔH° were found through the same process, and ΔS° parameters were calculated from ΔS° = (ΔH° – ΔG°)/TM.
Nearest neighbor parameters for Watson–Crick pairs were obtained from fitting published data for 112 duplexes, which included the 90 duplexes that Xia et al. previously fit, and 22 additional duplexes (Supporting Information). The symmetry contribution, if present, was subtracted from each thermodynamic parameter derived from the TM–1 vs ln(CT/a) plot. Matrix calculations were carried out as described above to generate ΔG°37 and ΔH° for each nearest neighbor parameter, with all three software packages yielding similar results.
The F-test was used to test the hypothesis that a least-squares model can fit the dependence of Gσ on Sσ and GNN.59,60 If the F-value is larger than the critical F-value for N and N − v degrees of freedom at the 5% significance level, where N is the number of duplexes and v the number of nearest neighbor parameters, or if the p-value is less than 0.05, then the hypothesis that there is a dependence of Gσ on GNN may be accepted.60
The paired t-test was used to evaluate the significance of the differences between predictions of thermodynamic properties with the updated parameters and those reported by Mathews et al.12 and the difference between experimental values and predictions by each set of nearest neighbor parameters. The difference between each pair of a set with b values of a variable, X, before and after treatment is defined as μ(XD) = μ(X1) – μ(X2), where X2 represents the response of X1 to treatment.61 The null hypothesis states that μ(XD) = 0. To test this and the alternative hypothesis that μ(XD) ≠ 0, the mean and standard deviation of the difference between each block of values is found.
| 7 |
| 8 |
A t-ratio is defined as
| 9 |
If the t-ratio is greater than t-value for (b – 1) degrees of freedom or less than its negative, then the null hypothesis is rejected at the 0.05 significance level.
For example, in using the paired t-test to evaluate how well experimental ΔG°’s are predicted by nearest neighbor parameters, b is the number of duplexes whose ΔG°’s are being tested and XD is the difference between the predicted and experimental ΔG° for each sequence.
The probability density function (PDF), f(t), of the Student’s t-distribution was used as a measure of how significantly a given INN parameter contributes to the model,11,59 with smaller values of f(t) indicating greater contribution,
| 10 |
where Γ is the gamma function, r = N – v degrees of freedom and t = ΔG°j(NN)/σj(NN), that is, the quotient of the free energy of the INN parameter over the estimate of its error. Calculations were carried out with R53 using the anova and t-test functions, and the critical t-value was determined with the qt function in R.
Results
Table 2 lists results for duplexes in the database used for determination of nearest neighbor parameters for GU pairs. Most of the duplexes are six to eight base pairs in length and have melting temperatures in the 30–70 °C range. For the 29 new duplexes reported here, the average difference between ΔG°37, ΔH°, and ΔS° derived from TM–1 vs ln(CT/a) plots and averaged curve fits are 2%, 7%, and 8%, respectively. Three duplexes, r(AGGCUU)2, r(AUGCGU)2, and r(GUCGUAC/), with TM’s less than 25 °C that were included in the database of Mathews et al.12 were omitted from the new database. Determination of thermodynamics from optical melting curves is difficult when the TM is less than 25 °C.
Table 2. Thermodynamic Parameters for Duplex Formation in 1 M NaCla.
| TM–1 vs log
CT |
average of curve fits |
predicted |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sequenceb | –ΔG°37(kcal/mol) | –ΔH° (kcal/mol) | –ΔS° (eu) | TMc(°C) | –ΔG°37(kcal/mol) | –ΔH° (kcal/mol) | –ΔS° (eu) | TMc(°C) | –ΔG°37(kcal/mol) | –ΔH° (kcal/mol) | –ΔS° (eu) | TMc(°C) | ref |
| Two State Sequences Used in Regression Analysis | |||||||||||||
| CGGCUG | 5.55 | 43.20 | 121.4 | 35.7 | 5.51 | 45.40 | 128.6 | 35.9 | 4.94 | 41.27 | 117.1 | 31.7 | (101) |
| CUGCGG | 4.31 | 41.40 | 119.6 | 26.8 | 4.55 | 36.00 | 101.4 | 27.4 | 4.94 | 41.27 | 117.1 | 31.7 | (101) |
| GCCGGUp | 9.17 | 58.20 | 158.1 | 57.0 | 9.44 | 60.40 | 164.3 | 57.0 | 8.66 | 55.99 | 152.7 | 54.4 | (102) |
| GCGUGC | 5.11 | 46.18 | 132.4 | 33.2 | 5.15 | 49.69 | 143.6 | 33.7 | 4.10 | 51.10 | 151.5 | 27.9 | 86 |
| GCUGGC | 6.47 | 59.10 | 169.7 | 41.5 | 6.59 | 59.10 | 169.3 | 41.9 | 6.43 | 57.67 | 165.0 | 41.4 | (101) |
| GGCGCU | 8.42 | 56.40 | 154.7 | 52.9 | 8.47 | 55.40 | 151.3 | 53.1 | 8.22 | 55.67 | 152.8 | 52.1 | (102) |
| GGCGUC | 4.67 | 38.10 | 107.8 | 29.0 | 4.92 | 37.30 | 104.4 | 30.2 | 5.74 | 43.27 | 120.9 | 37.6 | (101) |
| GUGCAU | 5.10 | 47.50 | 136.9 | 33.1 | 5.10 | 47.00 | 135.0 | 33.4 | 4.92 | 46.93 | 135.5 | 32.0 | (43) |
| UCCGCC/ | 6.71 | 57.00 | 162.2 | 38.0 | 6.69 | 54.30 | 153.4 | 37.9 | 7.73 | 48.54 | 131.6 | 44.7 | (84) |
| UCCGGGp | 7.44 | 47.70 | 129.8 | 48.5 | 7.34 | 47.10 | 128.2 | 48.7 | 7.96 | 47.87 | 128.7 | 52.5 | (102) |
| UGGCCGp | 8.56 | 53.00 | 143.3 | 54.7 | 8.11 | 46.60 | 124.1 | 55.1 | 7.92 | 49.17 | 133.0 | 51.8 | (102) |
| UUGCAG | 4.20 | 37.20 | 106.5 | 25.3 | 4.30 | 35.50 | 100.4 | 25.7 | 3.82 | 40.07 | 116.9 | 23.3 | (43) |
| CUCGCUC/ | 7.78 | 64.20 | 181.8 | 43.1 | 8.00 | 70.30 | 200.8 | 43.6 | 8.17 | 58.88 | 163.4 | 46.0 | (103) |
| GCGGGAC/ | 9.00 | 45.20 | 116.8 | 55.0 | 9.30 | 50.60 | 133.1 | 54.7 | 9.85 | 62.22 | 168.7 | 54.8 | this work |
| AGUCGAUU | 6.00 | 53.30 | 152.6 | 38.9 | 6.03 | 58.20 | 168.3 | 38.9 | 4.14 | 47.17 | 138.8 | 27.1 | (104) |
| AUGCGCGUp | 9.31 | 54.90 | 147.0 | 58.6 | 9.05 | 53.90 | 144.6 | 58.8 | 8.08 | 55.25 | 152.1 | 51.1 | (101) |
| AUGCGUAUp | 5.27 | 46.80 | 133.9 | 34.4 | 5.29 | 42.60 | 120.3 | 34.8 | 4.22 | 42.45 | 123.3 | 26.6 | (101) |
| AUGUGCAUp | 6.17 | 57.10 | 164.2 | 39.5 | 6.08 | 51.30 | 145.8 | 39.9 | 5.87 | 63.41 | 185.7 | 37.7 | (101) |
| CAGGGCUC/ | 11.10 | 62.80 | 166.6 | 61.4 | 11.50 | 65.60 | 174.2 | 61.0 | 11.52 | 71.58 | 193.6 | 60.4 | this work |
| CCAGUUGG | 5.70 | 61.10 | 178.6 | 37.1 | 5.80 | 60.40 | 176.3 | 37.4 | 6.20 | 65.80 | 192.3 | 39.3 | (95) |
| CCAUGUGG | 7.80 | 70.50 | 202.1 | 46.5 | 7.80 | 71.20 | 204.3 | 46.9 | 8.59 | 71.47 | 202.9 | 50.0 | (95) |
| CCUGUAGG | 6.81 | 71.10 | 207.3 | 42.0 | 6.81 | 66.10 | 191.1 | 42.4 | 6.22 | 59.88 | 173.1 | 39.7 | (104) |
| CGGAUUCG | 6.56 | 72.60 | 213.0 | 40.8 | 6.59 | 70.30 | 205.4 | 41.1 | 5.92 | 61.87 | 180.3 | 38.3 | (104) |
| CGUUGACG | 6.93 | 73.50 | 214.6 | 42.4 | 6.93 | 68.40 | 198.0 | 42.8 | 6.27 | 73.87 | 217.9 | 39.7 | (104) |
| CUCGGCUC/ | 8.22 | 73.90 | 211.8 | 44.2 | 8.30 | 76.80 | 220.9 | 44.3 | 8.42 | 76.70 | 220.1 | 44.9 | (104) |
| CUGGCUAG | 7.10 | 60.38 | 171.8 | 44.4 | 7.06 | 62.56 | 179.0 | 44.0 | 7.38 | 53.03 | 147.1 | 47.5 | this work |
| GACGCCAG/ | 10.50 | 63.80 | 171.8 | 57.5 | 11.00 | 74.60 | 205.1 | 56.9 | 11.35 | 71.19 | 192.9 | 59.5 | this work |
| GACGCGUU | 9.50 | 62.20 | 169.9 | 57.4 | 9.60 | 62.70 | 171.4 | 57.6 | 9.12 | 76.11 | 215.9 | 51.9 | this work |
| GAGGUGAG/ | 7.63 | 78.40 | 228.2 | 41.4 | 7.63 | 76.10 | 220.8 | 41.6 | 7.10 | 68.65 | 198.4 | 39.7 | (105) |
| GAGUGCUC | 9.40 | 83.00 | 237.4 | 51.6 | 9.20 | 77.40 | 220.0 | 51.8 | 9.21 | 77.05 | 218.7 | 52.0 | (52) |
| GAGUGGAG/ | 9.66 | 82.30 | 234.1 | 49.3 | 9.59 | 80.40 | 228.2 | 49.3 | 9.26 | 75.40 | 213.2 | 48.8 | (105) |
| GAUGCAUUp | 6.82 | 62.90 | 180.8 | 42.6 | 6.84 | 58.70 | 167.2 | 43.2 | 6.34 | 71.67 | 210.7 | 39.9 | (106) |
| GCAGCUGU | 10.30 | 72.30 | 199.8 | 58.3 | 10.40 | 72.00 | 198.4 | 58.9 | 11.58 | 75.29 | 205.5 | 63.3 | this work |
| GCAGUUGC | 5.90 | 64.80 | 190.0 | 38.1 | 6.00 | 69.30 | 203.9 | 38.6 | 6.52 | 68.78 | 200.7 | 40.9 | (85) |
| GCAUGUGC | 8.40 | 72.40 | 206.1 | 49.2 | 8.50 | 73.00 | 208.0 | 49.4 | 8.91 | 74.45 | 211.3 | 51.1 | (85) |
| GCUGGUGC/ | 7.60 | 69.40 | 199.1 | 42.0 | 7.70 | 71.80 | 206.6 | 42.1 | 8.48 | 73.24 | 208.8 | 45.5 | (85) |
| GGAGCUCU | 10.50 | 66.57 | 180.9 | 61.1 | 10.60 | 67.60 | 184.0 | 61.3 | 11.30 | 75.99 | 208.4 | 62.0 | this work |
| GGAGUUCC | 6.43 | 73.10 | 214.9 | 40.2 | 6.44 | 68.40 | 199.6 | 40.5 | 6.68 | 69.80 | 203.5 | 41.5 | (104) |
| GGAUGUCC | 8.39 | 73.00 | 208.4 | 49.0 | 8.59 | 78.00 | 223.4 | 49.0 | 9.07 | 75.47 | 214.1 | 51.6 | (104) |
| GGCGGGGC/ | 13.80 | 76.50 | 202.2 | 69.5 | 14.00 | 77.40 | 204.4 | 70.1 | 12.54 | 85.75 | 236.0 | 60.4 | (44) |
| GGCGUGCC | 9.72 | 73.40 | 206.9 | 55.0 | 9.75 | 74.20 | 208.0 | 55.0 | 10.62 | 77.88 | 216.9 | 58.0 | (104) |
| GGCUGGCC | 13.10 | 87.20 | 238.8 | 65.9 | 13.30 | 90.30 | 248.4 | 65.4 | 12.95 | 84.45 | 230.4 | 66.4 | (52) |
| GGUUGACC | 8.30 | 78.30 | 225.9 | 47.6 | 8.30 | 77.40 | 222.8 | 47.7 | 8.07 | 79.37 | 229.9 | 46.7 | (52) |
| GUAGCUAU | 7.30 | 50.30 | 138.7 | 47.4 | 7.40 | 57.90 | 162.8 | 46.7 | 7.52 | 62.39 | 176.9 | 46.5 | this work |
| GUCGGGCC/ | 15.00 | 96.00 | 261.3 | 66.9 | 14.20 | 83.30 | 222.7 | 68.6 | 13.11 | 75.61 | 201.4 | 66.8 | this work |
| GUCGUGAC | 6.05 | 69.10 | 203.3 | 38.7 | 6.08 | 64.70 | 188.9 | 39.0 | 6.44 | 69.02 | 201.7 | 40.7 | (104) |
| GUCUAGAU | 7.70 | 70.00 | 201.0 | 46.3 | 7.70 | 68.70 | 196.8 | 46.3 | 7.47 | 64.70 | 184.5 | 45.9 | (107) |
| UACCGGUG | 9.70 | 51.70 | 135.4 | 63.1 | 10.00 | 54.70 | 144.1 | 63.6 | 9.62 | 58.53 | 157.7 | 59.5 | this work |
| UAUGCAUGp | 6.44 | 62.30 | 180.1 | 41.0 | 6.55 | 53.10 | 150.1 | 41.8 | 6.10 | 52.83 | 150.7 | 39.5 | (106) |
| UCACGUGG | 8.40 | 46.90 | 124.2 | 56.0 | 8.60 | 45.00 | 117.1 | 58.7 | 10.14 | 64.77 | 176.1 | 60.1 | this work |
| UGACGUCG | 10.40 | 63.80 | 172.3 | 61.6 | 10.60 | 67.30 | 182.7 | 61.7 | 9.52 | 65.83 | 181.4 | 56.5 | this work |
| UUACGUAG | 6.20 | 44.60 | 124.0 | 40.5 | 6.20 | 49.20 | 138.9 | 40.1 | 5.68 | 53.13 | 152.9 | 37.2 | this work |
| CAGAGGAGAC/ | 9.43 | 98.95 | 288.6 | 46.4 | 9.39 | 95.19 | 276.7 | 46.6 | 10.23 | 96.23 | 277.3 | 49.4 | this work |
| CAGCGCGUUG | 12.31 | 77.02 | 208.6 | 66.2 | 11.59 | 66.84 | 178.2 | 67.1 | 12.84 | 83.53 | 227.9 | 66.1 | this work |
| CAGUCGAUUG | 8.70 | 92.33 | 269.7 | 47.5 | 8.54 | 84.54 | 245.0 | 47.9 | 9.26 | 75.49 | 213.6 | 52.4 | this work |
| CCAGCGUCCU/ | 11.60 | 87.90 | 246.0 | 55.9 | 11.70 | 90.10 | 252.7 | 55.9 | 13.17 | 80.81 | 218.2 | 64.6 | 108 |
| CCGAAUUUGG | 6.99 | 76.76 | 225.0 | 42.6 | 7.00 | 76.35 | 223.5 | 42.6 | 8.48 | 78.07 | 224.5 | 48.4 | this work |
| CGGAAUUUCG | 7.88 | 90.52 | 266.5 | 44.7 | 7.77 | 84.47 | 247.3 | 44.9 | 7.78 | 75.51 | 218.3 | 45.9 | this work |
| CGGAUAUUCG | 8.78 | 88.20 | 256.2 | 48.3 | 8.49 | 80.30 | 231.4 | 48.5 | 8.35 | 78.94 | 227.5 | 48.0 | this work |
| CGGGCGUUCG | 11.55 | 101.66 | 290.5 | 56.0 | 11.65 | 101.34 | 289.2 | 56.4 | 10.96 | 100.19 | 287.7 | 54.3 | this work |
| CGGUGCAUCG | 14.76 | 102.42 | 282.6 | 64.1 | 14.18 | 94.72 | 259.7 | 64.2 | 13.24 | 82.27 | 222.6 | 68.4 | this work |
| CUGGAUUCAG | 10.15 | 97.81 | 282.7 | 51.8 | 9.97 | 92.86 | 267.3 | 52.0 | 9.58 | 82.43 | 234.9 | 52.4 | this work |
| GAGAGCUUUC | 8.82 | 86.57 | 250.6 | 48.9 | 8.72 | 82.81 | 238.9 | 48.9 | 9.48 | 85.79 | 245.9 | 51.5 | this work |
| GAGGAUCUUC | 9.83 | 93.86 | 270.9 | 51.3 | 9.40 | 83.92 | 240.3 | 51.4 | 10.22 | 82.33 | 232.3 | 55.4 | this work |
| GAGUGGAGAG/ | 9.87 | 96.93 | 280.7 | 48.1 | 9.79 | 93.52 | 270.0 | 48.2 | 10.24 | 93.27 | 267.6 | 49.9 | this work |
| GGUUCGGGCC | 13.59 | 115.71 | 329.3 | 59.8 | 13.56 | 113.36 | 321.8 | 60.2 | 12.76 | 105.69 | 299.7 | 59.2 | this work |
| GUGAAUUUAC | 4.78 | 62.63 | 186.4 | 32.8 | 4.60 | 72.40 | 218.6 | 32.5 | 4.72 | 64.89 | 193.9 | 32.6 | this work |
| GUGUGCAUAC | 8.90 | 58.60 | 160.1 | 55.0 | 9.20 | 66.70 | 185.6 | 54.2 | 10.18 | 71.65 | 198.2 | 57.9 | this work |
| GUUAGCUGAC | 8.60 | 69.60 | 196.7 | 50.4 | 8.50 | 66.90 | 188.5 | 50.5 | 9.34 | 77.71 | 220.3 | 52.5 | this work |
| UCGCCAGAGG/ | 15.32 | 93.46 | 252.0 | 69.2 | 15.42 | 94.49 | 254.9 | 69.2 | 16.35 | 92.38 | 245.2 | 73.8 | (109) |
Duplexes with a 5′GGUC/3′CUGG motif are not listed because they were not used in fitting nearest neighbor parameters and no new sequences were measured.
Listed in order of length of the oligoribonucleotide and then alphabetically for sequences of the same length. All non-self-complementary sequences have a slash and only one sequence shown. GU base pairs are underlined.
Calculated at a total strand concentration of 1 × 10–4 M.
With the Exclusion of the 5′GGUC/3′CUGG Motif, the Experimental Results Can Be Fit to a Nearest Neighbor Model
The results in Table 2 were fit to a nearest neighbor model for GU pairs after subtracting contributions from Watson–Crick nearest neighbors (eq 3). This method avoids conflating thermodynamic parameters for Watson–Crick pairs with the idiosyncrasies of GU pairs. Published thermodynamics for duplexes with all Watson–Crick pairs (Supporting Information) were used to test published parameters for Watson–Crick pairs.11 Fitting the expanded database of 112 duplexes gave INN parameters within error of the values reported by Xia et al.11 (Table 3). Most free energy parameters did not change by more than 0.05 kcal/mol at 37 °C. Consequently, the GU component values were calculated from eq (3) by subtracting the previously published Watson–Crick thermodynamic values11 so that the GU parameters are consistent with the widely used Watson–Crick parameters.
Table 3. INN Parameters for Canonical Base Pairs in 1 M NaCl without a Separate Parameter for Terminal GU Pairs.
| INN | INN counts | ΔG°37(kcal/mol) | ΔG°37 error (kcal/mol) | ΔH° (kcal/mol) | ΔH° error (kcal/mol) | ΔS°a(eu) | ΔS° error (eu) |
|---|---|---|---|---|---|---|---|
| Watson–Crick Nearest Neighbor Doublets | |||||||
| 5′AA3′ | –0.93 (−0.96) | 0.03 (0.03) | –6.82 (−7.09) | 0.79 (0.77) | –19.0 (−19.8) | 2.5 (2.4) | |
| 3′UU5′ | |||||||
| 5′AU3′ | –1.10 (−1.09) | 0.08 (0.07) | –9.38 (−9.11) | 1.68 (1.56) | –26.7 (−25.8) | 5.2 (4.8) | |
| 3′UA5′ | |||||||
| 5′UA3′ | –1.33 (−1.39) | 0.09 (0.08) | –7.69 (−8.50) | 2.02 (1.86) | –20.5 (−22.9) | 6.3 (5.7) | |
| 3′AU5′ | |||||||
| 5′CU3′ | –2.08 (−2.07) | 0.06 (0.06) | –10.48 (−10.90) | 1.24 (1.15) | –27.1 (−28.5) | 3.8 (3.5) | |
| 3′GA5′ | |||||||
| 5′CA3′ | –2.11 (−2.11) | 0.07 (0.06) | –10.44 (−11.03) | 1.28 (1.18) | –26.9 (−28.8) | 3.9 (3.6) | |
| 3′GU5′ | |||||||
| 5′GU3′ | –2.24 (−2.27) | 0.06 (0.05) | –11.40 (−11.98) | 1.23 (1.12) | –29.5 (−31.3) | 3.9 (3.5) | |
| 3′CA5′ | |||||||
| 5′GA3′ | –2.35 (−2.39) | 0.06 (0.05) | –12.44 (−13.21) | 1.20 (1.05) | –32.5 (−34.9) | 3.7 (3.2) | |
| 3′CU5′ | |||||||
| 5′CG3′ | –2.36 (−2.38) | 0.09 (0.09) | –10.64 (−10.88) | 1.65 (1.54) | –26.7 (−27.4) | 5.0 (4.7) | |
| 3′GC5′ | |||||||
| 5′GG3′ | –3.26 (−3.31) | 0.07 (0.06) | –13.39 (−14.18) | 1.24 (1.07) | –32.7 (−35.0) | 3.8 (3.3) | |
| 3′CC5′ | |||||||
| 5′GC3′ | –3.42 (−3.46) | 0.08 (0.07) | –14.88 (−16.04) | 1.58 (1.33) | –36.9 (−40.6) | 4.9 (4.0) | |
| 3′CG5′ | |||||||
| GU Nearest Neighbor Doublets | |||||||
| 5′GU3′ | 8 | 0.72 | 0.19 | –13.83 | 4.21 | –46.9 | 13.0 |
| 3′UG5′ | |||||||
| 5′GG3′ | 9 | –0.25 | 0.16 | –17.82 | 3.75 | –56.7 | 11.6 |
| 3′UU5′ | |||||||
| 5′AG3′ | 22 | –0.35 | 0.08 | –3.96 | 1.73 | –11.6 | 5.3 |
| 3′UU5′ | |||||||
| 5′UG3′ | 18 | –0.39 | 0.09 | –0.96 | 1.80 | –1.8 | 5.5 |
| 3′AU5′ | |||||||
| 5′UU3′ | 26 | –0.51 | 0.08 | –10.38 | 1.79 | –31.8 | 5.5 |
| 3′AG5′ | |||||||
| 5′UG3′ | 10 | –0.57 | 0.19 | –12.64 | 4.01 | –38.9 | 12.3 |
| 3′GU5′ | |||||||
| 5′AU3′ | 24 | –0.90 | 0.08 | –7.39 | 1.65 | –21.0 | 5.1 |
| 3′UG5′ | |||||||
| 5′CG3′ | 26 | –1.25 | 0.09 | –5.56 | 1.68 | –13.9 | 5.1 |
| 3′GU5′ | |||||||
| 5′CU3′ | 21 | –1.77 | 0.09 | –9.44 | 1.76 | –24.7 | 5.4 |
| 3′GG5′ | |||||||
| 5′GG3′ | 24 | –1.80 | 0.09 | –7.03 | 1.75 | –16.8 | 5.4 |
| 3′CU5′ | |||||||
| 5′GU3′ | 25 | –2.15 | 0.10 | –11.09 | 1.78 | –28.8 | 5.4 |
| 3′CG5′ | |||||||
| 5′GGUC3′b | 3 | –4.12 | 0.54 | –30.80 | 8.87 | –86.0 | 23.7 |
| 3′CUGG5′ | |||||||
| Other Nearest Neighbor Parameters | ||||||
|---|---|---|---|---|---|---|
| initiationc | 4.09 (4.23) | 0.22 (0.20) | 3.61 (6.40) | 4.12 (3.56) | –1.5 (6.99) | 12.7 (10.9) |
| terminal AU penaltyc | 0.45 (0.43) | 0.04 (0.04) | 3.72 (3.85) | 0.83 (0.77) | 10.5 (11.04) | 2.6 (2.4) |
| symmetryc | 0.43 | 0 | 0 | 0 | –1.4 | 0 |
Values for ΔS° were derived from ΔS° = (ΔH° – ΔG°37)/310.15.
Ref (12).
Values for initiation, terminal AU, and nearest neighbors with only Watson–Crick pairs are from ref (11) when not in parentheses and derived from an expanded database when in parentheses. Values for nearest neighbors with GU pairs were derived using the Xia et al. parameters11 for Watson–Crick nearest neighbors.
For fitting GU parameters, duplexes containing the motif, 5′GGUC/3′CUGG, were excluded from the regression due to its poor fit in the nearest neighbor model.12 For the other 70 duplexes, 12 GU INN parameters were initially derived by linear regression, which included a penalty term for terminal GU pairs to correct for the fact that two duplexes with the same nearest neighbors can have different numbers of GU pairs and therefore different number of hydrogen bonds. A similar term was required for terminal AU pairs.11 Fitting of additional parameters would not give a unique fit.42 This 12 parameter fit gave values of −0.02 ± 0.06 kcal/mol and 2.34 ± 1.17 kcal/mol for the terminal GU penalty ΔG°37 and ΔH°, respectively (Supporting Information). The PDF for the terminal GU penalty ΔG°37 and ΔH° were 0.38 and 5.6 × 10–2, respectively, indicating that the term is not statistically significant. Therefore, the data were fit without a terminal GU term. The resulting nearest neighbor parameters are listed in Table 3.
The free energy parameters at 37 °C for 5′UG/3′AU, 5′UU/3′AG, and 5′AU/3′UG are less favorable than previously reported12 by at least 0.43 kcal/mol. This corresponds to at least a factor of 2 for an equilibrium constant at 37 °C. The ΔG°37 values for each of the tandem GU motifs are more favorable than previously reported. The 5′UG/3′GU nearest neighbor contributes favorably to helix stability by −0.57 kcal/mol, whereas previous data provided an unfavorable increment of 0.30 kcal/mol.12 Similarly increased stability from 0.47 to −0.25 kcal/mol at 37 °C was found for 5′GG/3′UU, which was previously represented by a single duplex (Table 3).
Estimated errors of the free energy parameters for most nearest neighbor motifs are less than 0.10 kcal/mol (Table 3). The p-value for the F-test is less than 2.2 × 10–16, indicating that there is a linear dependence of the free energy of a duplex on the occurrence of each nearest neighbor parameter in it at the 5% significance level.60 The PDF values from the Student t-distribution (Table 4) are small for ΔG°37 except for the 5′GG/3′UU motif. The relatively large PDF for the 5′GG/3′UU motif may be attributed to the small magnitude of its free energy and large error compared to those of most of the other INN parameters.
Table 4. Probability Density Function (PDF) of Student’s t-Distribution for ΔG°37 and ΔH° for Each GU INN Motif without a Separate Parameter for Terminal GU Pairsa.
| motif | PDF, ΔG°37 | PDF, ΔH° | motif | PDF, ΔG°37 | PDF, ΔH° |
|---|---|---|---|---|---|
| 5′GU3′ | 5.8 × 10–4 | 2.6 × 10–3 | 5′AU3′ | 4.7 × 10–16 | 6.1 × 10–5 |
| 3′UG5′ | 3′UG5′ | ||||
| 5′GG3′ | 1.2 × 10–1 | 2.4 × 10–5 | 5′CG3′ | 4.9 × 10–20 | 2.4 × 10–3 |
| 3′UU5′ | 3′GU5′ | ||||
| 5′AG3′ | 8.7 × 10–5 | 3.1 × 10–2 | 5′CU3′ | 1.8 × 10–27 | 2.7 × 10–6 |
| 3′UU5′ | 3′GG5′ | ||||
| 5′UG3′ | 1.0 × 10–4 | 3.4 × 10–1 | 5′GG3′ | 7.4 × 10–28 | 2.8 × 10–4 |
| 3′AU5′ | 3′CU5′ | ||||
| 5′UU3′ | 5.9 × 10–8 | 5.3 × 10–7 | 5′GU3′ | 1.6 × 10–29 | 1.0 × 10–7 |
| 3′AG5′ | 3′CG5′ | ||||
| 5′UG3′ | 5.6 × 10–3 | 3.7 × 10–3 | |||
| 3′GU5′ |
Table 5 lists results for apparently two-state duplexes that were omitted from the database fitted because their TM’s are less than 25 °C. The predicted thermodynamic parameters for r(AGGCUU)2 and r(AUGCGU)2 do not agree well with those measured. The NMR spectra of r(AGGCUU)2 and r(AUGCGU)2 have strong H2′-H6/8 cross peaks and a sequential H2′, H1′-H6/8 proton walk in 2D 1H–1H NMR (Supporting Information) that indicate the duplexes adopt a largely A-form conformation.62 For r(AUGCGU)2, however, the presence of broad on-diagonal peaks and exchange cross peaks in the aromatic region of the 2D spectra and of more imino resonances in a 1D spectrum than the number of imino protons in the sequence indicates the presence of alternate conformations. The presence of broad on-diagonal peaks, particularly for A1H8 and H2, in the aromatic region of the 2D spectra for r(AGGCUU)2 also suggests multiple conformations at 0 °C.
Table 5. Thermodynamic Parameters for Formation of Duplexes Not Included in Fitting Nearest Neighbor Parametersa.
|
TM–1 vs log CT |
average of
curve fits |
predicted |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sequenceb | –ΔG°37(kcal/mol) | –ΔH° (kcal/mol) | –ΔS° (eu) | TMc (°C) | –ΔG°37(kcal/mol) | –ΔH° (kcal/mol) | –ΔS° (eu) | TMc (°C) | –ΔG°37(kcal/mol) | –ΔH°(kcal/mol) | –ΔS° (eu) | TMc (°C) | ref |
| Sequences with TM < 25 °C | |||||||||||||
| AGGCUU | 4.07 | 37.10 | 106.6 | 24.2 | 4.24 | 34.60 | 97.9 | 24.7 | 2.24 | 30.63 | 91.5 | 5.8 | (104) |
| AUGCGUp | 4.22 | 24.50 | 65.4 | 19.5 | 4.37 | 24.10 | 63.6 | 21.1 | 2.30 | 29.73 | 88.5 | 5.2 | (101) |
| GUCGUAC/ | 4.20 ± 0.65 | 47.30 ± 9.01 | 139.1 ± 30.1 | 22.2 | 3.80 ± 0.48 | 54.20 ± 11.09 | 162.3 ± 36.1 | 22.2 | 3.66 | 51.98 | 155.7 | 21.0 | this work |
| Non-Two-State Sequences | |||||||||||||
| UGGCUA | 5.36 | 49.70 | 143.0 | 35.0 | 5.54 | 36.10 | 98.5 | 35.8 | 2.32 | 24.63 | 71.9 | –0.1 | (104) |
| CAUGUGC/ | 7.70 ± 0.14 | 49.10 ± 4.46 | 133.5 ± 14.2 | 44.3 | 8.10 ± 0.14 | 60.00 ± 11.18 | 167.4 ± 35.6 | 45.1 | 5.92 | 59.57 | 173.0 | 33.8 | this work |
| GUGGUCG/ | 7.93 ± 0.18 | 56.21 ± 4.47 | 155.7 ± 14.4 | 44.9 | 8.05 ± 0.18 | 69.80 ± 6.64 | 199.1 ± 18.1 | 43.9 | 7.67 | 55.73 | 154.9 | 43.6 | this work |
| GGUGUACC | 5.94 | 65.80 | 193.1 | 38.3 | 5.99 | 49.60 | 140.6 | 39.0 | 6.54 | 61.72 | 177.9 | 41.4 | (104) |
| GUAGCUGC | 6.57 | 73.10 | 214.4 | 40.9 | 6.41 | 50.70 | 142.8 | 41.6 | 8.14 | 56.33 | 155.3 | 51.3 | (104) |
| GAGGCGCGGAG/ | 9.52 ± 0.24 | 136.60 ± 11.90 | 409.7 ± 37.7 | 43.9 | 8.28 ± 0.56 | 59.27 ± 6.93 | 164.4 ± 21.0 | 46.4 | 10.88 | 111.46 | 324.3 | 49.6 | this work |
| GCUUUGCGGAGC | 13.22 ± 0.35 | 141.60 ± 7.01 | 414.0 ± 21.5 | 54.4 | 10.39 ± 0.34 | 82.58 ± 14.35 | 232.7 ± 45.3 | 55.8 | 13.92 | 129.51 | 372.6 | 58.1 | this work |
Experimental errors are listed for sequences melted in this study.
Listed in order of length of the oligoribonucleotide and then alphabetically for sequences of the same length. All non-self-complementary sequences have a slash and only one sequence shown. GU base pairs are underlined.
Calculated at a total strand concentration of 1 × 10–4 M.
Table 5 also lists duplexes that do not melt in a two-state manner. There are many possible reasons for this.56,63−65 The average difference between experimental and predicted TM for these sequences is 10.0 °C, while the predicted free energy is, on average, within 1.33 kcal/mol of the experimental free energy (Table 5). Evidently, the INN model may provide useful predictions for non-two-state sequences even though ΔH° from the van’t Hoff equation is erroneous.64
The Expanded Database Improves Predictions of Duplex Stability
Using the previous parameters,12 the correlation coefficients between experimental values for ΔG°37, ΔH°, and ΔS° and those predicted for the 70 duplexes in Table 2 are 0.89, 0.86, and 0.85, respectively. Comparisons of the values of ΔG°37, ΔH°, and ΔS° of the 70 duplexes as predicted with the previous parameters,12 and those in Table 3yielded, respectively, means of the differences of −0.36 kcal/mol, −1.75 kcal/mol, and −4.5 eu. The paired t-test gives t-values of −3.386, −2.528, and −2.257, respectively, which have absolute magnitudes greater than 1.995, indicating that the two sets of parameters differ with a significance level of 0.05 for 69 degrees of freedom.61 Furthermore, the respective p-values of 1.2 × 10–3, 1.4 × 10–2, and 2.7 × 10–2 are less than 0.05. This again indicates that the new parameters predict the thermodynamics of RNA duplexes significantly differently from those published previously.12
Using the set of GU parameters in Table 3, the correlation coefficients between experimental values for ΔG°37, ΔH°, and ΔS° and those predicted for the 70 duplexes in Table 2 are 0.95, 0.89, and 0.87, respectively. Comparison of experimental values of ΔG°37, ΔH°, and ΔS° of the 70 duplexes and those predicted with the set of GU parameters in Table 3 yielded means of differences of −0.05 kcal/mol, 0.58 kcal/mol, and 2.1 eu and t-values of −0.544, 0.590, and 0.686, respectively, which have absolute magnitudes less than 1.995. The corresponding p-values of 0.59, 0.56, and 0.50 are greater than 0.05. These results show that the thermodynamic properties predicted with the new INN parameters are not significantly different from experiment.61 The same analysis yielded means of differences of −0.41 kcal/mol, −1.17 kcal/mol, and −2.4 eu and t-values of −2.752, −1.096, and −0.736, with corresponding p-values of 7.6 × 10–3, 0.28, and 0.46 for ΔG°37, ΔH°, and ΔS°, respectively, when experimental properties were compared with predicted properties using previous parameters.12 Evidently, the expanded database provides improved modeling of the thermodynamics of GU pairs.
The Nearest Neighbor Model Is Not Perfect
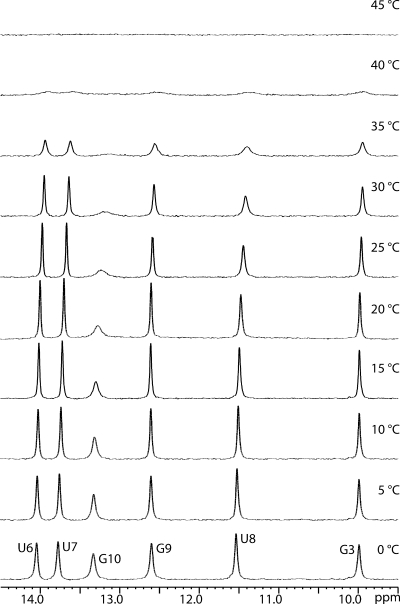
While the nearest neighbor model predicts well the ΔG°37 for most of the duplexes in Table 2, there are likely to be other terms that partially control stability. For example, there are four duplexes, r(GGCGUC)2, r(AGUCGAUU)2, r(UCACGUGG)2, and r(CCGAAUUUGG)2 with predicted ΔG°37 values not within 1.0 kcal/moland 20% of the 1/TM vs ln(CT/a) experimental values. No pattern is evident for these duplexes. A series of 1D spectra were acquired for r(CCGAAUUUGG)2 at different temperatures (Figure 1) because its predicted free energy is 1.5 kcal/mol more favorable at 37 °C than measured. These spectra show that the imino protons of all but U8, which is in the GU pair, and G10, which is in the terminal base pair disappear with each other, consistent with the duplex melting in a two-state manner. The results suggest that the nearest neighbor model does not include all factors that determine stabilities of duplexes with GU pairs.
Figure 1.
1D 1H imino spectra of r(CCGAAUUUGG)2 from 0 to 45 °C.
The expanded database allows preliminary testing of models beyond the nearest neighbor model. For example, terminal GU pairs could be considered separately43 and a base pair triplet model used for internal GU pairs. Comparison of measured values of ΔG°37 for terminal GU pairs with those predicted from the parameters in Table 3 give a standard deviation within 0.30 kcal/mol at 37 °C (Supporting Information). For the 16 triplets, 5′WGY/3′XUZ, with WX and YZ as Watson–Crick pairs, 12 measured ΔG°37(GU component) values are within 0.5 kcal/mol of the predicted values and the others within 1.0 kcal/mol (Supporting Information). The nearest neighbor model is apparently a reasonable approximation, and considerably more data would be required to develop a triplet model.
One clear exception to the nearest neighbor model is multiple terminal GU pairs.43 Thus, the parameters in Table 3 cannot be used beyond the first terminal GU pair at a helix end. Parameters for additional terminal GU pairs have been published by Nguyen and Schroeder.43
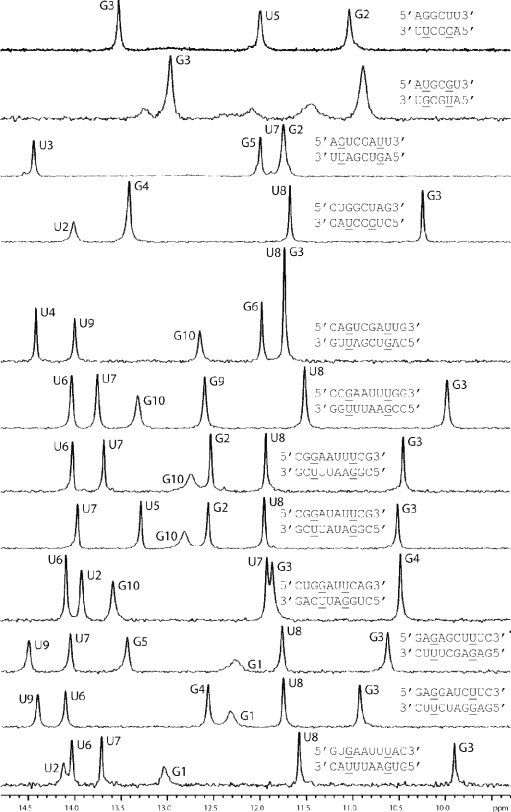
Imino Proton NMR Spectra of Several Duplexes Are Consistent with the Expected Base Pairing
To check for expected base pairing, NMR imino proton spectra were measured for 12 duplexes. All had chemical shifts from 10 to 15 ppm (Figure 2). Chemical shifts for GH1 and UH3 of GU pairs were relatively upfield (10–12 ppm), consistent with expectations.66 Chemical shifts for UH3 in AU pairs resonated between 13 and 15 ppm and GH1 in GC pairs resonated from 12 to 13.5 ppm, as expected.67,68 The absence of an imino peak for a terminal base pair in r(CUGGCUAG)2 indicates exchange with water. The G3-H1 and U7–H3 resonances of r(CUGGAUUCAG)2 appear to overlap, as evident by the presence of a single large peak. These chemical shift signatures show that the RNA sequences form the expected duplexes.
Figure 2.
1D imino NMR spectra for some RNA duplexes with GU pairs. Spectra were acquired at 0 °C for r(AGGCUU)2 and r(AUGCGU)2; 1 °C for r(AGUCGAUU)2; 5 °C for others.
Discussion
GU pairs are the most common non-Watson–Crick base pairs in RNA structures. Thus, the thermodynamics of GU pairs are important for finding regions of RNA that are structured,1,14−16,69 predicting the secondary structure12 or determining structure on the basis of chemical modification8,70 and/or NMR data.71
GU pairs can serve as binding sites for proteins or metal ions and participate in tertiary interactions.72,73 Thus, a better characterization of the thermodynamic properties of GU pairs can improve prediction of secondary and tertiary structure and help predict binding sites for metal ions and target sites for therapeutics. For example, GU pairs in group I introns can bind cations, including Mg2+, Co3+, and Os3+.38,40,74,75 Divalent metal ion binding by GU pairs, which have greater negative potential in the major groove than other base pairs, was postulated as important for activating RNA catalysis.76 Divalent ions that interact with a GU pair help catalyze splicing by group I and group II introns77−82 and cleavage by HDV ribozyme.35 Metal ion binding with RNA neutralizes negative potential, which may promote higher order RNA folding.75 The 5′GG/3′UU and 5′GU/3′UG motifs particularly contain greater negative potential in the major groove than their Watson–Crick counterparts.83
The Database of Sequences for Determining GU Thermodynamic Parameters Was Expanded
Not including sequences containing the 5′GGUC/3′CUGG motif, the database in Table 2 expands from 35 to 70 the duplexes used to fit nearest neighbor parameters for GU pairs. This expansion includes published data not included in the original database43,84−86 along with 29 new measurements (Table 2). Two of the original 35 duplexes were removed from the database because their melting temperatures were below 25 °C, which makes it difficult to analyze the melting curves. A third duplex, r(AUCUAGGU)2, was omitted because two-state melting could not be confirmed. The expanded database contains GU pairs flanked by Watson–Crick pairs in all possible orientations (Table 1). The new set of GU INN parameters were obtained with consideration for propagated errors from experiment and from Watson–Crick nearest neighbor parameters. Errors for the free energies of individual nearest neighbors were less than 0.2 kcal/mol for tandem GU pairs and 0.1 kcal/mol for other GU motifs. The 5′GG/3′UU motif, which was previously represented by a single sequence, was added to the fitting. The favorable free energy of −0.25 ± 0.16 kcal/mol for 5′GG/3′UU is in better agreement with the value of −0.5 kcal/mol used by Mathews et al.12 to optimize secondary structure prediction than with the previous single experimental measurement of 0.47 kcal/mol.
GU Pairs Are Generally Less Stable than GC and AU Pairs
The free energies of formation for many of the duplexes with GU pairs (Table 2) can be compared with the free energies when the U or G of the GU pairs is replaced with a C or A, respectively, to form GC or AU pairs (Table 6). Because many of the latter duplexes terminated with a 3′ phosphate, the comparisons assume that the 3′ phosphate has negligible effect on ΔG°37 at 1 M NaCl.87,88 Duplexes containing GC pairs in place of GU pairs are more stable at 37 °C by 1.8 ± 0.8 kcal/mol per GU pair (Table 6). This is presumably due to the presence of an additional hydrogen bond in GC pairs and unfavorable backbone distortion due to GU pairs. Terminal substitutions all have a less than average effect while internal substitutions have a larger than average effect, as expected if backbone distortion is less important for a terminal GU.
Table 6. Free Energy Differences When GU Pairs Are Replaced with AU or GC Pairs.
| GC duplex | ref | ΔG°37 GC duplex | GU duplex. | ref | ΔG°37 GU duplex | ΔΔG°37 per GU pair (kcal/mol) |
|---|---|---|---|---|---|---|
| CCGCGG | (11) | 9.84 | CUGCGG | (101) | 4.31 | 2.77 |
| CGGCCGp | (110) | 9.90 | CGGCUG | (101) | 5.55 | 2.18 |
| CGGCCGp | (110) | 9.90 | UGGCCGp | (102) | 8.56 | 0.67 |
| CUGCAGp | (111) | 7.11 | UUGCAG | (43) | 4.20 | 1.46 |
| GCCGGCp | (110) | 11.20 | GCCGGUp | (102) | 9.17 | 1.02 |
| GGCGCCp | (112) | 11.33 | GGCGCU | (102) | 8.42 | 1.46 |
| GGCGCCp | (112) | 11.33 | GGCGUC | (101) | 4.67 | 3.33 |
| GUGCAC | (111) | 7.65 | GUGCAU | (43) | 5.10 | 1.28 |
| AUGCGCAUp | (101) | 10.17 | AUGCGUAUp | (101) | 5.27 | 2.45 |
| CAUGCAUGp | (113) | 9.67 | UAUGCAUGp | (106) | 6.44 | 1.62 |
| GAUGCAUCp | (113) | 10.12 | GAUGCAUUp | (106) | 6.82 | 1.65 |
| GCAGCUGC | (114) | 13.87 | GCAGCUGU | this work | 10.30 | 1.79 |
| average | 1.80 ± 0.76 | |||||
| AU duplex | ref | ΔG°37 AU duplex | GU duplex. | ref | ΔG°37 GU duplex | ΔΔG°37 per GU pair (kcal/mol). |
|---|---|---|---|---|---|---|
| ACCGGUp | (115) | 8.51 | GCCGGUp | (102) | 9.17 | –0.33 |
| AGCGCU | (112) | 7.99 | GGCGCU | (102) | 8.42 | –0.22 |
| CAGCUGp | (111) | 6.68 | CGGCUG | (101) | 5.55 | 0.57 |
| CUGCAGp | (111) | 7.11 | CUGCGG | (101) | 4.31 | 1.40 |
| GACGUC | (116) | 7.35 | GGCGUC | (101) | 4.67 | 1.34 |
| UCCGGAp | (88) | 7.99 | UCCGGGp | (102) | 7.44 | 0.28 |
| CUCACUC/ | (11) | 9.71 | CUCGCUC/ | (117) | 7.78 | 1.93 |
| AAUGCAUUp | (113) | 7.18 | GAUGCAUUp | (106) | 6.82 | 0.18 |
| AUACGUAU | (101) | 6.53 | AUGCGUAUp | (101) | 5.27 | 0.63 |
| AUGCGCAUp | (101) | 10.17 | AUGCGCGUp | (101) | 9.31 | 0.43 |
| UAUGCAUAp | (113) | 7.27 | UAUGCAUGp | (106) | 6.44 | 0.42 |
| Average | 0.60 ± 0.70 | |||||
The effect of replacing GU with GC pairs can be compared to replacing AU pairs with GC pairs (Supporting Information). On average, replacing an AU pair with a GC pair stabilized a duplex by 1.5 ± 0.4 kcal/mol per AU pair. In this case, there was no apparent difference between terminal and internal substitutions.
Duplexes containing AU pairs in place of GU pairs are more stable at 37 °C by 0.6 ± 0.7 kcal/mol per GU pair (Table 6). While the difference is zero within the standard deviation, in only 2 of 11 cases is the GU duplex more stable than the AU duplex and in both cases the difference is within the experimental error of 4%.
Unlike terminal AU pairs, no penalty for terminal GU pairs is required to account for base pair composition. The terminal AU penalty of 0.45 kcal/mol at 37 °C was considered to account for numbers of base pairing hydrogen bonds.11 Thus, the penalty for terminal GU pairs was assumed to be equal to that of AU pairs,12 consistent with wobble GU pairs at the end of a helix having two hydrogen bonds.89 When the terminal GU parameter was included in the reparameterization of GU nearest neighbor thermodynamic parameters, the free energy of each nearest neighbor parameter differed by no more than 0.01 kcal/mol from that calculated without it (Table 2 and Supporting Information). The lack of a terminal GU penalty may arise from the flexibility of a terminal GU pair which allows optimization of hydrogen bonding and stacking interactions without incurring the energetic penalty associated with an interior GU distorting the backbone.43 For example, even for an internal GU pair, optimal stability may be found with only one hydrogen bond due to stacking energies.90,91 Thus, flexibility of terminal GU’s may compensate for the difference between the free energy of formation of two and three hydrogen bonds in GU and GC pairs, respectively.
Tandem GU Pairs Have Structural Features That Correlate with Their Thermodynamic Properties
With the exception of 5′GGUC/3′CUGG, the 5′UG/3′GU motif is more stable than 5′GU/3′UG (Table 3). Available structures show that 5′UG/3′GU contains interstrand stacking between the guanines,90−93 whereas 5′GU/3′UG does not.91,94,95 The favorability of the 5′UG/3′GU motif relative to the 5′GU/3′UG motif is consistent with molecular dynamics (MD) simulations91 that predict a one hydrogen bond GU pair90 predominates in duplexes containing the 5′GU/3′UG motif while a two hydrogen bond model predominates in duplexes containing the 5′UG/3′GU motif. There is also less overlap of negative potentials in 5′UG/3′GU than in 5′GU/3′UG.95 In two different sequences containing the 5′UG/3′GU motif, there is also intrastrand stacking between each GU pair and its Watson–Crick neighbors.92,93 By comparison, the 5′GU/3′UG motif contains less overlap between the GU pairs and Watson–Crick purine neighbors, but has intrastrand stacking between the tandem GU pairs.94 Furthermore, the 5′UG/3′GU motif preserves the A-form of RNA more than 5′GU/3′UG.96
The 5′GGUC/3′CUGG motif is an exception to the above generalizations. NMR spectra and modeling indicate that the GU pairs of r(GAGGUCUC)2 contain two hydrogen bonds,52 whereas the GU pairs in r(GGCGUGCC)2 contain only one hydrogen bond.90 This difference would contribute to the favorable free energy of 5′GGUC/3′CUGG compared to that of 5′GU/3′UG in other contexts, such as 5′CGUG/3′GUGC. Pan et al. saw similar hydrogen-bonding scenarios in MD simulations.91 Additional stability for the 5′GGUC/3′CUGG motif may also be conferred from less overlap of its negative electrostatic potentials between a GC and GU pair than for its related motif, 5′CGUG/3′GUGC.52 These patterns may explain the poor fit of nearest neighbor parameters for the 5′GU/3′UG motif when duplexes containing the 5′GGUC/3′CUGG motif are included in the fit. Alternatively, the extra stability of the 5′GGUC/3′CUGG motif over 5′CGUG/3′GUGC may arise from poor cross-strand overlap between the U in a GU pair and the C in its neighboring GC pair in 5′CGUG/3′GUGC.97 Stacking interactions alone do not contribute to the stability of nearest neighbor motifs comprised of the same base pairs, however, as evident from the comparable stability of 5′UG/3′GU and 5′GG/3′UU. This contrasts with the expectation that the free energy of 5′GG/3′UU is between the other tandem GU motifs because its base stacking is intermediate among them.98 Understanding the interactions responsible for the observed sequence dependence of thermodynamics presents a challenge to computational chemists.
GU Pairs of RNA Are More Stable than GT Pairs of DNA
Comparison of ΔG°37 values for GT nearest neighbors in DNA74 with those measured for GU nearest neighbors show that GT nearest neighbors are on average 0.84 ± 0.36 kcal/mol less stable than their GU counterparts. The extra stability of GU relative to GT is also evident from comparisons of ΔG°37 (GU or GT component) for duplexes containing comparable triplet motifs (Table 7). This may reflect a possible hydrogen bond between the amino group of guanine and the O2′ of uracil,99 which is not possible with DNA. MD simulations utilizing residual dipolar coupling (RDC) restraints suggest that the 5′TG/3′GT motif contains a bifurcated hydrogen bond100 similar to that in the 5′GU/3′UG motif.90,91 Another difference between GT and GU is that the 5′GGTC/3′CTGG motif fits the nearest neighbor parameters for the 5′GT/3′TG motif better than their respective uracil-substituted RNA motifs.74 Consistent with the relative stabilities of GT and GU nearest neighbors, component free energies of GT pairs in duplexes are consistently less favorable than those of GU pairs flanked by the same Watson–Crick pairs (Table 7).
Table 7. Component Free Energies of GU and GT Pairsa.
| sequencesb | ΔG°37(1/TM vs ln(CT/a)) (kcal/mol) | experimental ΔG°37(component) (kcal/mol) | predicted ΔG°37(component)c(kcal/mol)d |
|---|---|---|---|
| GCGUGC | –5.11 | –2.79 | –1.78 |
| GACCGTGCAC/ | –7.17 | –0.40 | 0.20 |
| AUGCGUAUc | –5.27 | –3.07 | –2.54 |
| CCATGCGTAACG/ | –8.94 | –0.90 | –0.30 |
| CTTGCATGTAAGc,e | –6.10 | –0.55 | –0.15 |
| CUCGGCUC/ | –8.22 | –3.45 | –3.65 |
| GACGTTGGAC/ | –7.91 | –1.40 | –0.30 |
| CUGGCUAG | –7.10 | –4.04 | –4.32 |
| CTTGGATCTAAG | –5.89 | –1.20 | 0.20 |
| GAGUGCUC | –9.40 | –5.06 | –4.87 |
| GGAGTGCTCC | –7.66 | –2.20 | –0.70 |
| GCAGUUGC | –5.90 | 0.64 | 0.02 |
| GGAGUUCC | –6.43 | 0.27 | 0.02 |
| GGCAGTTCGC/ | –6.87 | 1.40 | 2.60 |
| GCAUGUGC | –8.40 | –1.86 | –2.37 |
| GGAUGUCC | –8.39 | –1.69 | –2.37 |
| GCGATGTCGCe | –7.98 | 0.10 | 0.70 |
| CAGUCGAUUGc | –8.70 | –0.97 | –1.25 |
| GTACAGTGATC/ | –7.78 | –1.20 | 0.80 |
| CGAGTCGATTCGc,e | –7.71 | 0.35 | 0.80 |
| GAGAGCUUUC | –8.82 | –1.06 | –1.72 |
| CGAGACGTTTCG | –6.96 | 1.70 | 2.10 |
| GAGGAUCUUCc | –9.83 | –1.93 | –2.12 |
| CATGAGGCTAC/ | –8.57 | –0.90 | 0.40 |
| GAGUGGAGAG/ | –9.87 | –0.78 | –1.15 |
| GACTGGAGAG/e | –4.61 | 0.30 | 1.50 |
| GUGAAUUUAC | –4.78 | –1.86 | –1.80 |
| GUUAGCUGAC | –8.60 | –1.06 | –1.80 |
| CGTGACGTTACG | –8.19 | 0.70 | 1.50 |
| CGTTACGTGACG | –7.86 | 1.10 | 1.50 |
| GUGUGCAUAC | –8.90 | –1.30 | –2.58 |
| CGTGTCGATACG | –8.42 | 0.20 | 1.00 |
| CGTGTCTAGATACGe | –9.40 | 0.20 | 1.00 |
Data for DNA duplexes were referenced from ref (74).
Listed in order of length of the oligoribonucleotide and then alphabetically for sequences of the same length. All nonself-complementary sequences have a slash and only one sequence shown. GU and GT base pairs are underlined.
Component free energies were divided by 2.
Calculated with free energies in Table 3.
Marginally non-two-state.74
Acknowledgments
The authors thank Zhenjiang Xu for suggesting the paired t-test and Dr. Susan Schroeder for comments on the manuscript.
Glossary
Abbreviations
- 1D
one-dimensional
- 2D
two-dimensional
- HIV-1
human immunodeficiency virus-1
- INN
individual nearest neighbor
- MD
molecular dynamics
- NN
nearest neighbor
- NOESY
nuclear Overhauser effect spectroscopy
probability density function
- RDC
residual dipolar coupling
- SVD
singular value decomposition
- TOCSY
total correlation spectroscopy
- WC
Watson–Crick
Supporting Information Available
(I) Thermodynamic parameters for duplex formation of Watson–Crick sequences. (II) Experimental thermodynamic parameters and error limits for newly measured sequences. (III) Component free energies and enthalpies of GU pairs. (IV) Free energies of doublets and triplets containing GU pairs calculated as component ΔG°37 of their sequences. (V) Free energy differences between sequences where GC pair(s) were replaced by AU pair(s). (VI) INN parameters for GU pairs calculated with a separate term for terminal GU pairs. (VII) Probability density function of the Student’s t-distribution for each INN motif with a separate parameter for terminal GU pairs. (VIII) 2D NOESY spectra for r(AGGCUU)2 showing H2′, H1′, and H6/H8 regions. (IX) 2D NOESY spectra for r(AUGCGU)2 showing H2′, H1′, and H6/H8 regions. (X) Desalting procedure for oligoribonucleotides. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by NIH Grant GM22939 (D.H.T.)
Author Present Address
⊥ Department of Chemistry, Northwestern University, Evanston, Illinois, 60208, USA.
Author Present Address
∥ Roswell Park Cancer Institute, Buffalo, New York 14263.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Mathews D. H., Moss W. N., and Turner D. H. (2010) Folding and finding RNA secondary structure, in RNA Worlds: From Life’s Origins to Diversity in Gene Regulation (Atkins J. F., Gesteland R. F., Cech T. R., Eds.) 4th ed., pp 293–308, Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H.; Sugimoto N.; Freier S. M. (1988) RNA structure prediction. Annu. Rev. Biophys. Biophys. Biochem. 17, 167–192. [DOI] [PubMed] [Google Scholar]
- Tinoco I.; Bustamante C. (1999) How RNA folds. J. Mol. Biol. 293, 271–281. [DOI] [PubMed] [Google Scholar]
- Andronescu M.; Aguirre-Hernández R.; Condon A.; Hoos H. H. (2003) RNAsoft: a suite of RNA secondary structure prediction and design software tools. Nucleic Acids Res. 31, 3416–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I. L.; Fontana W.; Stadler P. F.; Bonhoeffer L. S.; Tacker M.; Schuster P. (1994) Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125, 167–188. [Google Scholar]
- Lück R.; Gräf S.; Steger G. (1999) ConStruct: a tool for thermodynamic controlled prediction of conserved secondary structure. Nucleic Acids Res. 27, 4208–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D. H.; Turner D. H. (2002) Dynalign: An algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 317, 191–203. [DOI] [PubMed] [Google Scholar]
- Mathews D. H.; Disney M. D.; Childs J. L.; Schroeder S. J.; Zuker M.; Turner D. H. (2004) Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U.S.A. 101, 7287–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D. H. (2004) Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 10, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer P. N.; Dengler B.; Tinoco I.; Uhlenbeck O. C. (1974) Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 86, 843–853. [DOI] [PubMed] [Google Scholar]
- Xia T. B.; SantaLucia J.; Burkard M. E.; Kierzek R.; Schroeder S. J.; Jiao X. Q.; Cox C.; Turner D. H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- Mathews D. H.; Sabina J.; Zuker M.; Turner D. H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288, 911–940. [DOI] [PubMed] [Google Scholar]
- Turner D. H. (2000) Conformational changes, in Nucleic Acids: Structures, Properties, and Functions (Bloomfield V. A., Crothers D. M., Tinoco J., I., Eds.) pp 259–334, University Science Books, Herndon, VA. [Google Scholar]
- Washietl S.; Hofacker I. L.; Stadler P. F. (2005) Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 102, 2454–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzilov A.; Keegan J.; Mathews D. H. (2006) Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics 7, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R.; Neuböck R.; Hofacker I. L.; Washietl S. (2007) The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Res. 35, W335–W338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiche K.; Stadler P. F. (2007) RNAstrand: reading direction of structured RNAs in multiple sequence alignments. Algorithm. Mol. Biol. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. A.; Nilges M.; Huang A.; Brunger A. T.; Moore P. B. (1992) NMR analysis of helix-I from the 5S RNA of Escherichia coli. Biochemistry 31, 1610–1621. [DOI] [PubMed] [Google Scholar]
- Szymański M.; Barciszewska M. Z.; Erdmann V. A.; Barciszewski J. (2000) An analysis of G-U base pair occurrence in eukaryotic 5S rRNAs. Mol. Biol. Evol. 17, 1194–1198. [DOI] [PubMed] [Google Scholar]
- Sprinzl M.; Vassilenko K. S. (2005) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33, D139–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmer S.; Reif B.; Ott G.; Arnold L.; Sprinzl M. (1996) NMR evidence for helix geometry modifications by a G-U wobble base pair in the acceptor arm of E-coli tRNA(Ala). FEBS Lett. 385, 15–20. [DOI] [PubMed] [Google Scholar]
- Hou Y. M.; Schimmel P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333, 140–145. [DOI] [PubMed] [Google Scholar]
- McClain W. H.; Foss K. (1988) Changing the identity of a transfer RNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240, 793–796. [DOI] [PubMed] [Google Scholar]
- Mueller U.; Schubel H.; Sprinzl M.; Heinemann U. (1999) Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G·U wobble base pair at 1.16 angstrom resolution. RNA 5, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. A.; Li H. (1996) Yeast ribosomal protein L32 recognizes an RNA G:U juxtaposition. RNA 2, 226–234. [PMC free article] [PubMed] [Google Scholar]
- Reyes J. L.; Gustafson E. H.; Luo H. R.; Moore M. J.; Konarska M. M. (1999) The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA 5, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung S. S.; Koslowsky D. J. (2001) Interactions of mRNAs and gRNAs involved in trypanosome mitochondrial RNA editing: structure probing of an mRNA bound to its cognate gRNA. RNA 7, 1803–1816. [PMC free article] [PubMed] [Google Scholar]
- Mooers B. H. M.; Singh A. (2011) The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate. RNA 17, 1870–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.; Heng X.; Garyu L.; Monti S.; Garcia E. L.; Kharytonchyk S.; Dorjsuren B.; Kulandaivel G.; Jones S.; Hiremath A.; Divakaruni S. S.; LaCotti C.; Barton S.; Tummillo D.; Hosic A.; Edme K.; Albrecht S.; Telesnitsky A.; Summers M. F. (2011) NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knitt D. S.; Narlikar G. J.; Herschlag D. (1994) Dissection of the role of the conserved G·U pair in group I RNA self-splicing. Biochemistry 33, 13864–13879. [DOI] [PubMed] [Google Scholar]
- Pyle A. M.; Moran S.; Strobel S. A.; Chapman T.; Turner D. H.; Cech T. R. (1994) Replacement of the conserved G·U with a G-C pair at the cleavage site of the tetrahymena ribozyme decreases binding, reactivity, and fidelity. Biochemistry 33, 13856–13863. [DOI] [PubMed] [Google Scholar]
- Strobel S. A.; Cech T. R. (1995) Minor groove recognition of the conserved G·U pair at the tetrahymena ribozyme reaction site. Science 267, 675–679. [DOI] [PubMed] [Google Scholar]
- Strobel S. A.; Cech T. R. (1996) Exocyclic amine of the conserved G·U pair at the cleavage site of the Tetrahymena ribozyme contributes to 5′-splice site selection and transition state stabilization. Biochemistry 35, 1201–1211. [DOI] [PubMed] [Google Scholar]
- Šponer J.; Šponer J. E.; Petrov A. I.; Leontis N. B. (2010) Quantum chemical studies of nucleic acids: Can we construct a bridge to the RNA structural biology and bioinformatics communities?. J. Phys. Chem. B 114, 15723–15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-H.; Gong B.; Bevilacqua P. C.; Carey P. R.; Golden B. L. (2009) A catalytic metal ion interacts with the cleavage site G·U wobble in the HDV ribozyme. Biochemistry 48, 1498–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-H.; Yajima R.; Chadalavada D. M.; Chase E.; Bevilacqua P. C.; Golden B. L. (2010) A 1.9 Å crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry 49, 6508–6518. [DOI] [PubMed] [Google Scholar]
- Keel A. Y.; Rambo R. P.; Batey R. T.; Kieft J. S. (2007) A general strategy to solve the phase problem in RNA crystallography. Structure 15, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft J. S.; Tinoco I. (1997) Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexammine. Structure 5, 713–721. [DOI] [PubMed] [Google Scholar]
- Wang W. M.; Zhao J. W.; Han Q. W.; Wang G.; Yang G. C.; Shallop A. J.; Liu J.; Gaffney B. L.; Jones R. A. (2009) Modulation of RNA metal binding by flanking bases: N-15 NMR evaluation of GC, tandem GU, and tandem GA sites. Nucleosides Nucleotides Nucleic Acids 28, 424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenarejo G.; Tinoco I. Jr. (1999) Structure and thermodynamics of metal binding in the P5 helix of a group I intron ribozyme. J. Mol. Biol. 290, 119–135. [DOI] [PubMed] [Google Scholar]
- Gautheret D.; Konings D.; Gutell R. R. (1995) G·U base pairing motifs in ribosomal RNA. RNA 1, 807–814. [PMC free article] [PubMed] [Google Scholar]
- Gray D. M. (1997) Derivation of nearest-neighbor properties from data on nucleic acid oligomers. 1. Simple sets of independent sequences and the influence of absent nearest neighbors. Biopolymers 42, 783–793. [DOI] [PubMed] [Google Scholar]
- Nguyen M.-T.; Schroeder S. J. (2010) Consecutive terminal GU pairs stabilize RNA helices. Biochemistry 49, 10574–10581. [DOI] [PubMed] [Google Scholar]
- Serra M. J.; Smolter P. E.; Westhof E. (2004) Pronouced instability of tandem IU base pairs in RNA. Nucleic Acids Res. 32, 1824–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada H.; Takahashi K. (1998) Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins 33, 159–166. [PubMed] [Google Scholar]
- Smallcombe S. H. (1993) Solvent suppression with symmetrically-shifted pulses. J. Am. Chem. Soc. 115, 4776–4785. [Google Scholar]
- Grzesiek S.; Bax A. (1993) The importance of not saturating H2O in protein NMR - application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115, 12593–12594. [Google Scholar]
- Piotto M.; Saudek V.; Sklenář V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665. [DOI] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Goddard T. D., and Kneller D. G. (2004) SPARKY, NMR Assignment and Integration Software, 3rd ed., University of California, San Francisco. [Google Scholar]
- Cavanagh J., Fairbrother W. J., Palmer A. G. I., and Skelton N. J. (1996) Protein NMR Spectroscopy: Principles and Practice, Academic Press, San Diego. [Google Scholar]
- McDowell J. A.; Turner D. H. (1996) Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry 35, 14077–14089. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2010) R: A Language and Environment for Statistical Computing, x64 2.11.1 ed., R Foundation for Statistical Computing, Vienna, Austria.
- Wolfram Research (2010) Mathematica Edition: Version 8.0, Champaign, Illinois.
- Eaton J. W. (2002) GNU Octave Manual.
- Cantor C. R., Schimmel P. R. (1980) Biophysical Chemistry, Part III: The Behavior of Biological Macromolecules, pp. 1197–1198, W. H. Freeman and Company, San Francisco. [Google Scholar]
- Bevington P. R., Robinson D. K. (2002) Data Reduction and Error Analysis for the Physical Sciences, 3rd ed., McGraw-Hill, New York. [Google Scholar]
- Drosg M. (2007) Dealing with Uncertainties: A Guide to Error Analysis, Springer-Verlag, Heidelberg. [Google Scholar]
- Crawley M. J. (2007) The R Book, 1st ed., John Wiley & Sons, West Sussex. [Google Scholar]
- Kinney J. J. (2002) Statistics for Science and Engineering, 1st ed., Addison-Wesley, Boston. [Google Scholar]
- Devore J., and Peck R. (2005) Statistics: the Exploration and Analysis of Data, 5th ed., Brooks/Cole - Thomson Learning, Belmont, CA. [Google Scholar]
- Varani G.; Aboulela F.; Allain F. H. T. (1996) NMR investigation of RNA structure. Prog. Nucl. Mag. Res. Spectrosc. 29, 51–127. [Google Scholar]
- Chaires J. B. (1997) Possible origin of differences between van’t Hoff and calorimetric enthalpy estimates. Biophys. Chem. 64, 15–23. [DOI] [PubMed] [Google Scholar]
- Mergny J.-L.; Lacroix L. (2003) Analysis of thermal melting curves. Oligonucleotides 13, 515–537. [DOI] [PubMed] [Google Scholar]
- SantaLucia J.; Turner D. H. (1997) Measuring the thermodynamics of RNA secondary structure formation. Biopolymers 44, 309–319. [DOI] [PubMed] [Google Scholar]
- Fürtig B.; Richter C.; Wohnert J.; Schwalbe H. (2003) NMR spectroscopy of RNA. ChemBioChem 4, 936–962. [DOI] [PubMed] [Google Scholar]
- Reid B. R.; McCollumn L.; Ribeiro N. S.; Abbate J.; Hurd R. E. (1979) Identification of tertiary base pair resonances in the nuclear magnetic resonance spectra of transfer ribonucleic acid. Biochemistry 18, 3996–4005. [DOI] [PubMed] [Google Scholar]
- Johnston P. D.; Redfield A. G. (1981) Nuclear magnetic resonance and nuclear Overhauser effect study of yeast phenylalanine transfer ribonucleic acid imino protons. Biochemistry 20, 1147–1156. [DOI] [PubMed] [Google Scholar]
- Cockerill M. (1993) Not much to malign - Multalin 4.0. Trends Biochem. Sci. 18, 106–107. [Google Scholar]
- Deigan K. E.; Li T. W.; Mathews D. H.; Weeks K. M. (2009) Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U.S.A. 106, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart J. M.; Kennedy S. D.; Mathews D. H.; Turner D. H. (2008) NMR-assisted prediction of RNA secondary structure: Identification of a probable pseudoknot in the coding region of an R2 Retrotransposon. J. Am. Chem. Soc. 130, 10233–10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R. T.; Rambo R. P.; Doudna J. A. (1999) Tertiary motifs in RNA structure and folding. Angew. Chem., Int. Ed. 38, 2327–2343. [DOI] [PubMed] [Google Scholar]
- Varani G.; McClain W. H. (2000) The G·U wobble base pair: a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allawi H. T.; SantaLucia J. (1997) Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- Cate J. H.; Doudna J. A. (1996) Metal-binding sites in the major groove of a large ribozyme domain. Structure 4, 1221–1229. [DOI] [PubMed] [Google Scholar]
- Konforti B. B.; Abramovitz D. L.; Duarte C. M.; Karpeisky A.; Beigelman L.; Pyle A. M. (1998) Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell 1, 433–441. [DOI] [PubMed] [Google Scholar]
- Adams P. L.; Stahley M. R.; Kosek A. B.; Wang J.; Strobel S. A. (2004) Crystal structure of a self-splicing group I intron with both exons. Nature 430, 45–50. [DOI] [PubMed] [Google Scholar]
- Forconi M.; Sengupta R. N.; Piccirilli J. A.; Herschlag D. (2010) A rearrangement of the guanosine-binding site establishes an extended network of functional interactions in the tetrahymena group I ribozyme active site. Biochemistry 49, 2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipchock S. V.; Strobel S. A. (2008) A relaxed active site after exon ligation by the group I intron. Proc. Natl. Acad. Sci. U.S.A 105, 5699–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley M. R.; Adams P. L.; Wang J.; Strobel S. A. (2007) Structural metals in the group I intron: A ribozyme with a multiple metal ion core. J. Mol. Biol. 372, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S. A.; Ortoleva-Donnelly L. (1999) A hydrogen-bonding triad stabilizes the chemical transition state of a group I ribozyme. Chem. Biol. 6, 153–165. [DOI] [PubMed] [Google Scholar]
- Toor N.; Keating K. S.; Taylor S. D.; Pyle A. M. (2008) Crystal structure of a self-spliced group II intron. Science 320, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D.; Landon T.; Greenbaum N. L.; Fenley M. O. (2007) The electrostatic characteristics of G·U wobble base pairs. Nucleic Acids Res. 35, 3836–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Znosko B. M.; Jiao X. Q.; Turner D. H. (2004) Factors affecting thermodynamic stabilities of RNA 3 × 3 internal loops. Biochemistry 43, 12865–12876. [DOI] [PubMed] [Google Scholar]
- Serra M. J.; Baird J. D.; Dale T.; Fey B. L.; Retatagos K.; Westhof E. (2002) Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA 8, 307–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A. E.; Wu M.; Turner D. H. (1994) The stability and structure of tandem GA mismatches in RNA depend on closing base-pairs. Biochemistry 33, 11349–11354. [DOI] [PubMed] [Google Scholar]
- Freier S. M.; Burger B. J.; Alkema D.; Neilson T.; Turner D. H. (1983) Effects of 3′ dangling end stacking on the stability of GGCC and CCGG double helixes. Biochemistry 22, 6198–6206. [Google Scholar]
- Freier S. M.; Alkema D.; Sinclair A.; Neilson T.; Turner D. H. (1985) Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry 24, 4533–4539. [DOI] [PubMed] [Google Scholar]
- Crick F. H. C. (1966) Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555. [DOI] [PubMed] [Google Scholar]
- Chen X. Y.; McDowell J. A.; Kierzek R.; Krugh T. R.; Turner D. H. (2000) Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G·U pairs: One hydrogen bond for each G·U pair in r(GGCGUGCC)2 and two for each G·U pair in r(GAGUGCUC)2. Biochemistry 39, 8970–8982. [PubMed] [Google Scholar]
- Pan Y. P.; Priyakumar U. D.; MacKerell A. D. (2005) Conformational determinants of tandem GU mismatches in RNA: Insights from molecular dynamics simulations and quantum mechanical calculations. Biochemistry 44, 1433–1443. [DOI] [PubMed] [Google Scholar]
- Biswas R.; Wahl M. C.; Ban C.; Sundaralingam M. (1997) Crystal structure of an alternating octamer r(GUAUGUA)dC with adjacent G·U wobble pairs. J. Mol. Biol. 267, 1149–1156. [DOI] [PubMed] [Google Scholar]
- Utsunomiya R.; Suto K.; Balasundaresan D.; Fukamizu A.; Kumar P. K. R.; Mizuno H. (2006) Structure of an RNA duplex r(GGCG(Br)UGCGCU)2 with terminal and internal tandem G·U base pairs. Acta Crystallogr. D 62, 331–338. [DOI] [PubMed] [Google Scholar]
- Biswas R.; Sundaralingam M. (1997) Crystal structure of r(GUGUGUA)dC with tandem G·U/U·G wobble pairs with strand slippage. J. Mol. Biol. 270, 511–519. [DOI] [PubMed] [Google Scholar]
- McDowell J. A.; He L. Y.; Chen X. Y.; Turner D. H. (1997) Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2. Biochemistry 36, 8030–8038. [DOI] [PubMed] [Google Scholar]
- Masquida B.; Westhof E. (2000) On the wobble G·U and related pairs. RNA 6, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. B.; Hung L. W.; Jeong M. S.; Holbrook E. L.; Chen X. Y.; Turner D. H.; Holbrook S. R. (2006) The crystal structure at 1.5 Å resolution of an RNA octamer duplex containing tandem G·U basepairs. Biophys. J. 90, 4530–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J. P.; Sundaralingam M. (2000) Synthesis and crystal structure of an octamer RNA r(guguuuac)/r(guaggcac) with G·G/U·U tandem wobble base pairs: comparison with other tandem G·U pairs. Nucleic Acids Res. 28, 4376–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K.; Wahl M. C.; Sundaralingam M. (1999) Crystal structure of an RNA duplex r(GGGCGCUCC)2 with non-adjacent G·U base pairs. Nucleic Acids Res. 27, 2196–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Salgado F.; Desvaux H.; Boulard Y. (2006) NMR assessment of the global shape of a non-labelled DNA dodecamer containing a tandem of G·T mismatches. Magn. Reson. Chem. 44, 1081–1089. [DOI] [PubMed] [Google Scholar]
- Sugimoto N.; Kierzek R.; Freier S. M.; Turner D. H. (1986) Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry 25, 5755–5759. [DOI] [PubMed] [Google Scholar]
- Freier S. M.; Kierzek R.; Caruthers M. H.; Neilson T.; Turner D. H. (1986) Free energy contributions of G·U and other terminal mismatches to helix stability. Biochemistry 25, 3209–3213. [DOI] [PubMed] [Google Scholar]
- Testa S. M.; Disney M. D.; Turner D. H.; Kierzek R. (1999) Thermodynamics of RNA-RNA duplexes with 2-or 4-thiouridines: Implications for antisense design and targeting a group I intron. Biochemistry 38, 16655–16662. [DOI] [PubMed] [Google Scholar]
- He L.; Kierzek R.; SantaLucia J.; Walter A. E.; Turner D. H. (1991) Nearest-neighbor parameters for G·U mismatches - 5′GU3′/3′UG5′ is destabilizing in the contexts CGUG/GUGC, UGUA/AUGU, and AGUU/UUGU but stabilizing in GGUC/CUGG. Biochemistry 30, 11124–11132. [DOI] [PubMed] [Google Scholar]
- Xia T. B.; McDowell J. A.; Turner D. H. (1997) Thermodynamics of nonsymmetric tandem mismatches adjacent to G·C base pairs in RNA. Biochemistry 36, 12486–12497. [DOI] [PubMed] [Google Scholar]
- Sugimoto N.; Kierzek R.; Turner D. H. (1987) Sequence dependence for the energetics of terminal mismatches in ribonucleic acid. Biochemistry 26, 4559–4562. [DOI] [PubMed] [Google Scholar]
- Ziomek K.; Kierzek E.; Biala E.; Kierzek R. (2002) The thermal stability of RNA duplexes containing modified base pairs placed at internal and terminal positions of the oligoribonucleotides. Biophys. Chem. 97, 233–241. [DOI] [PubMed] [Google Scholar]
- Schroeder S. J.; Turner D. H. (2001) Thermodynamic stabilities of internal loops with GU closing pairs in RNA. Biochemistry 40, 11509–11517. [DOI] [PubMed] [Google Scholar]
- Schroeder S. J.; Turner D. H. (2000) Factors affecting the thermodynamic stability of small asymmetric internal loops in RNA. Biochemistry 39, 9257–9274. [DOI] [PubMed] [Google Scholar]
- Freier S. M.; Sinclair A.; Neilson T.; Turner D. H. (1985) Improved free energies for G·C base-pairs. J. Mol. Biol. 185, 645–647. [DOI] [PubMed] [Google Scholar]
- Freier S. M.; Kierzek R.; Jaeger J. A.; Sugimoto N.; Caruthers M. H.; Neilson T.; Turner D. H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. U.S.A. 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M.; Sugimoto N.; Sinclair A.; Alkema D.; Neilson T.; Kierzek R.; Caruthers M. H.; Turner D. H. (1986) Stability of XGCGCp, GCGCYp, and XGCGCYp helixes: an empirical estimate of the energetics of hydrogen bonds in nucleic acids. Biochemistry 25, 3214–3219. [DOI] [PubMed] [Google Scholar]
- Sugimoto N.; Kierzek R.; Turner D. H. (1987) Sequence dependence for the energetics of dangling ends and terminal base pairs in ribooligonucleotides. Biochemistry 26, 4554–4558. [DOI] [PubMed] [Google Scholar]
- Burkard M. E.; Turner D. H. (2000) NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry 39, 11748–11762. [DOI] [PubMed] [Google Scholar]
- Petersheim M.; Turner D. H. (1983) Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry 22, 256–263. [DOI] [PubMed] [Google Scholar]
- Kierzek R.; Caruthers M. H.; Longfellow C. E.; Swinton D.; Turner D. H.; Freier S. M. (1986) Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry 25, 7840–7846. [DOI] [PubMed] [Google Scholar]
- Kierzek R.; Burkard M. E.; Turner D. H. (1999) Thermodynamics of single mismatches in RNA duplexes. Biochemistry 38, 14214–14223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.