Abstract
The tandem PHD finger–bromodomain, found in many chromatin-associated proteins, has an important role in gene silencing by the human co-repressor KRAB-associated protein 1 (KAP1). Here we report the three-dimensional solution structure of the tandem PHD finger–bromodomain of KAP1. The structure reveals a distinct scaffold unifying the two protein modules, in which the first helix, αZ, of an atypical bromodomain forms the central hydrophobic core that anchors the other three helices of the bromodomain on one side and the zinc binding PHD finger on the other. A comprehensive mutation-based structure-function analysis correlating transcriptional repression, ubiquitin-conjugating enzyme 9 (UBC9) binding and SUMOylation shows that the PHD finger and the bromodomain of KAP1 cooperate as one functional unit to facilitate lysine SUMOylation, which is required for KAP1 co-repressor activity in gene silencing. These results demonstrate a previously unknown unified function for the tandem PHD finger–bromodomain as an intramolecular small ubiquitin-like modifier (SUMO) E3 ligase for transcriptional silencing.
Chemical modifications of chromatin on the DNA (for example, methylation of cytosine) and DNA-packing histones (for example, acetylation, methylation, phosphorylation, ubiquitination and SUMOylation) are important in the epigenetic control of gene transcription in response to physiological and environmental stimuli1–3. An emerging model suggests that there is an ‘epigenetic code’ embedded within chromatin to signify regions of distinct nuclear activities, such as heterochromatin formation or transcriptional activation4–6. It is thought that the epigenetic code is established by chromatin-modifying enzymes and interpreted by proteins that bind the chromatin in a modification-sensitive manner. The discovery of methyl-CpG binding domains7, bromodomains as acetyllysine binding domains8–10 and the ‘royal’ family of chromodomains, Tudor and MBT domains11–13, as well as PHD fingers as methyllysine binding domains14–16 provides support for this hypothesis.
To realize their full potential in epigenetic control, these conserved structural domains are expected to perform different molecular functions or work in a combinatorial or integrated fashion. Recent reports on the recognition of unmodified histone H3 by the BHC80 PHD finger in gene repression17 and the simultaneous methyllysine and methylarginine recognition of histone H3 by the RAG2 PHD finger in V(D)J recombination18,19 exemplify the functional versatility of a conserved structural fold. Integrated functions in histone interactions have also been shown for tandem modules of the same fold, such as the double bromodomain in human transcriptional proteins BRD2 and BRD4 (ref. 20,21) and Rsc4 of the yeast RSC remodeling complex22, the double chromodomain of Drosophila melanogaster CHD1 (chromo-ATPase/helicase-DNA binding)23 and the double Tudor domain of JMJD2A24. However, although tandem modules of different structural folds such as the PHD finger and the bromodomain are found in many transcriptional proteins25,26, molecular mechanisms for their possible interactive functions are much less understood.
The tandem bromodomain–PHD finger of the human transcriptional coactivator p300/CBP has been shown to be interdependent in interactions with nucleosomes27. A more common, reversely connected motif, the PHD finger–bromodomain, is found in many chromatin-associated proteins including histone lysine methyl-transferase MLL1 (ref. 28), Williams syndrome transcription factor (WSTF) in the chromatin-remodeling complex WINAC29, and the TIF1 family proteins (α, β, γ and δ; note that TIF1β is also known as KAP1 or TRIM28)30. This tandem PHD finger–bromodomain is also found in Sp140, a leukocyte-specific protein in the nuclear body that is involved in the pathogenesis of acute promyelocytic leukemia and viral infection31. Mutations of PHD fingers, particularly those that disrupt zinc coordination, have been linked to tumor formation and genetic disorders32. More recently, it has been reported that the PHD finger of the human co-repressor KAP1 functions as a unique SUMO E3 ligase that is required for KAP1’s gene transcription repression activity33.
To understand its molecular function in gene silencing, we solved the three-dimensional solution structure of the PHD finger–bromodomain of human KAP1 using NMR spectroscopy. The new structure reveals a uniquely unified tandem-domain architecture that is completely unlike that of the PHD finger–bromodomain of bromodomain–PHD finger transcription factor (BPTF)14. We show that the PHD finger and the bromodomain of KAP1 function interdependently in catalyzing SUMOylation of lysine residues within this tandem module. Our new structural and mechanistic insights into the molecular function of the tandem PHD finger–bromodomain provide a framework for the functional understanding of KAP1 as the co-repressor for the Kruppel-associated box (KRAB) C2H2 zinc-finger family of proteins, many of which are involved in the regulation of cell differentiation and development34–36.
RESULTS
Structure of the PHD finger–bromodomain of KAP1
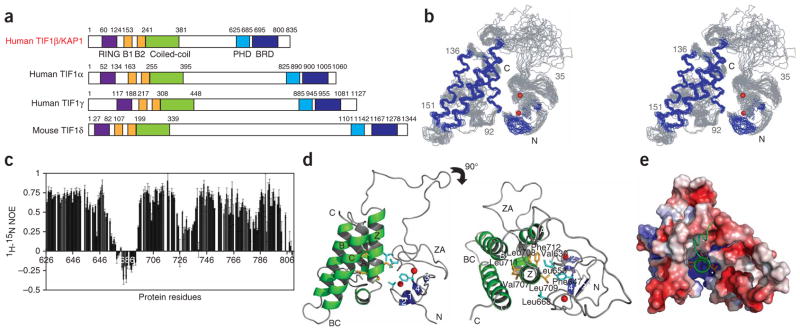
We determined the three-dimensional solution structure of the tandem PHD finger–bromodomain, encompassing residues 624–812 of human KAP1 (Fig. 1a) with a total of 3,726 NOE-derived distance and torsion-angle restraints obtained from heteronuclear multidimensional NMR37. An ensemble of the 20 lowest-energy NMR structures is depicted in Figure 1b. Excluding the linker residues 674–695, the structures of the PHD finger (residues 623–673) and the bromodomain (residues 696–801) are well defined. The linker showed high mobility in solution as evidenced by reduced or negative backbone 1H-15N heteronuclear NOEs compared to the structural regions of the protein (Fig. 1c).
Figure 1.
Three-dimensional structure of the tandem PHD finger–bromodomain of human KAP1. (a) Protein organization of the TIF1 family. The conserved protein domains are color coded: RING (purple), B1 and B2 (gold), coiled coil (green), PHD finger (blue) and bromodomain (BRD, dark blue). (b) Stereoview of the backbone atoms (N, Cα and C′) of 20 superimposed NMR structures of the PHD finger–bromodomain of KAP1 (showing residues 627–800). Secondary-structural elements and loops are respectively colored in blue and gray, and two zinc atoms are shown as red spheres. (c) The backbone 1H-15N heteronuclear NOEs of the KAP1 PHD finger–bromodomain (residues 623–801). Error bars represent the s.d. of NOE values measured in three data sets. (d) Ribbon representation of the average minimized NMR structure of the PHD finger (blue) and the bromodomain (green) of KAP1 in front and top views. The ribbon structure was prepared using Pymol (http://www.pymol.org). (e) Surface electrostatic potential representation of the PHD finger–bromodomain, highlighting the central helix αZ in a unified tandem-domain structure in the top view.
The PHD finger–bromodomain forms an integrated structural unit, despite the flexible linker. The two individual modules have conserved structural folds, as seen in the corresponding domain family8,32. The bromodomain adopts a left-handed four-helical bundle (helices αZ, αA, αB and αC), whereas the Cys4-His-Cys3 PHD finger contains an antiparallel β-sheet of two β-turn-β units that coordinate two zinc atoms in a cross-brace topology (Fig. 1d and Supplementary Fig. 1 online). In the tandem motif, the two domains are brought together to anchor around the first helix, αZ, of the bromodomain (Fig. 1d). Unlike an amphipathic αZ in most bromodomains, αZ in the atypical KAP1 bromodomain consists of a contiguous stretch of amino acids with large hydrophobic and aromatic side chains in its C-terminal half (that is, Val707–Phe712; Supplementary Fig. 1). The hydrophobic C-terminal segment of αZ in KAP1 is buried in the center of the tandem-domain structure. Packed on one side of αZ are residues from the other three helices of the bromodomain bundle, and on the other side are Val630, Phe647, Leu653 and Leu668 of the PHD finger on a twisted antiparallel β-sheet (Fig. 1d,e). Thus, although both hydrophobic and electrostatic interactions mediate domain-domain association, αZ serves as the hydrophobic core of the unified tandem-domain structure.
Structural integration of the PHD finger–bromodomain
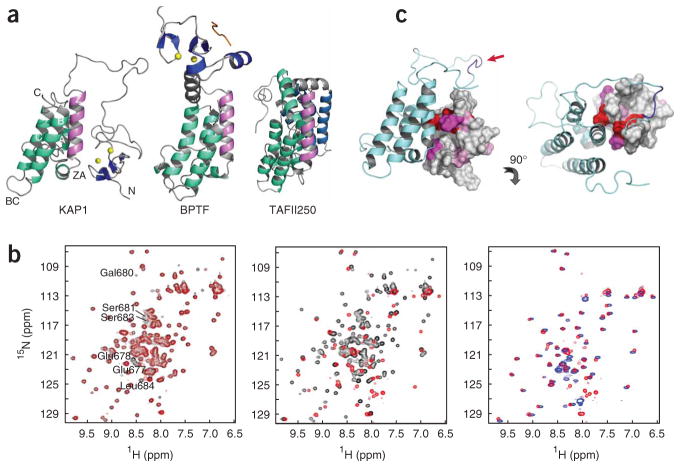
The integrated structure of the KAP1 PHD finger–bromodomain is unique and different from the structure of the PHD finger–bromodomain of human BPTF14, or those of the tandem bromodomains of human transcriptional protein TAFII250 (ref. 9) and yeast remodeling protein Rsc4 (ref. 22). As seen by structural comparisons, the PHD finger and bromodomain in BPTF do not contact each other, but they are connected by a rigid three-turn α-helix that spans about 18 Å (Fig. 2a, middle). In TAFII250, the corresponding αZ in the second bromodomain is a typical amphipathic helix packed to the hydrophobic core of the helical bundle on one side and exposed to the solvent on the other (Fig. 2a, right). The two bromodomains are connected by a short loop followed by a two-turn α-helix, and they interact between αB of the first bromodomain and αA of the second. The domain-domain association is reinforced by interactions between the ZA loops of the two domains. In BPTF or TAFII250, the acetyllysine binding pocket located between the ZA and BC loops is accessible to the solvent, which indicates that the bromodomain and the PHD finger have independent functions in protein-protein interactions even in the tandem motif9,14. However, these structural features contrast sharply with the distinct integrated nature of the structure of the KAP1 tandem PHD finger–bromodomain (Fig. 2a, left).
Figure 2.
Interdomain interactions in tandem-domain motifs. (a) Structural comparison of interdomain interactions in the PHD finger–bromodomain units in human KAP1 (left) and BPTF (middle), and in the double bromodomain modules in human TAFII250 (right). The second bromodomain in each tandem module are green, and each structure is oriented with respect to helix αZ of this bromodomain in mauve. (b) Comparison of two-dimensional 1H-15N HSQC spectra of the KAP1 proteins. Left, a spectral comparison between the wild-type PHD finger–bromodomain (black) and the enterokinase-susceptible mutant (red). Middle, comparison of the KAP1 mutant before (black) and after (red) enterokinase cleavage at the linker sequence. Right, comparison of the KAP1 mutant after enterokinase cleavage (red) and the KAP1 PHD finger alone (blue). (c) Differences of the backbone amide NMR resonances of the KAP1 PHD finger between the PHD finger alone and the PHD finger–bromodomain unit, as highlighted in the protein structure in front and top views. The residues are color coded according to the extent of chemical shift difference (Supplementary Fig. 2c): red, >2.0 ppm; magenta, 1.0–2.0 ppm; pink, 0.5–1.0 ppm; and gray, <0.5 ppm.
To characterize the structural integration of the PHD finger–bromodomain, we engineered an enterokinase cleavage site into the interdomain linker by mutating the sequence LKEEDGSLSL (residues 675–684) to LKDDDDKLSL. The two-dimensional 1H-15N HSQC (heteronuclear single quantum correlation) spectrum of the mutated KAP1 superimposes nearly perfectly to that of the wild type, except for the resonances of a few residues at the cleavage site (Fig. 2b, left). Although yielding only fragments corresponding to the PHD finger and the bromodomain (Supplementary Fig. 2a,b online), enterokinase treatment of mutated KAP1 resulted in both loss and perturbations of many protein peaks in the NMR spectra (Fig. 2b, middle). Notably, the remaining NMR signals match those of the PHD finger alone (Fig. 2b, right), with the exception of a few signals of different terminal residues in the corresponding protein constructs. Thus, the lost signals are those of the bromodomain and could possibly be due to nonspecific enterokinase cleavage at several sites in the protein or aggregation of the cleaved protein that forms a species of large molecular weight. The former is not a likely cause, as indicated by our SDS-PAGE analysis of the protein upon enterokinase treatment (Supplementary Fig. 2b); if it were, the cleaved KAP1 bromodomain fragments would yield strong NMR signals in the HSQC spectrum. Given the hydrophobic nature of αZ, the latter is more likely. Indeed, when expressed alone in Escherichia coli, the bromodomain was unstable in solution (data not shown). On the other hand, these results show that the stable PHD-finger fold is established via coordination of two zinc atoms. This is supported by the NMR observation that only the residues located at the domain interface show major chemical shift differences between the PHD finger alone and the tandem-domain motif (Fig. 2c and Supplementary Fig. 2c). Taken together, these results argue that the tandem PHD finger–bromo-domain of KAP1 is one integrated structural unit and probably functions cooperatively.
Structural features of the bromodomain and the PHD finger of KAP1
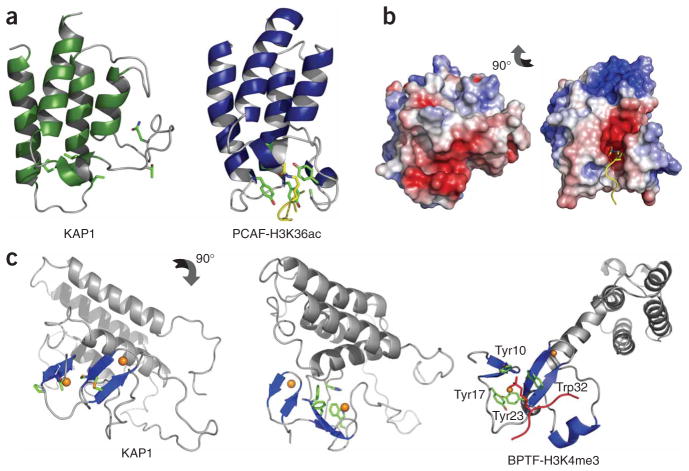
The bromodomain functions as an acetylly-sine binding domain in the epigenetic control of gene transcription10. The acetyllysine of a bromodomain target protein is bound in a pocket between the ZA and BC loops, wherein the acetyl group is typically recognized through hydrogen-bonding interactions of the amide nitrogen and carbonyl oxygen of the ace tyl group with three highly conserved residues in the domain family corresponding to Tyr760 in the ZA loop and Tyr802 and Asn803 in the BC loop of the p300/CBP-associated factor (PCAF) bromodomain (Fig. 3a). Notably, although the four-helical bundle fold is conserved, the ZA and BC loops—except for the three conserved acetyllysine binding residues—vary in length and amino acid composition, which contributes to distinct ligand binding specificity by different bromodomains. However, in the KAP1 bromodomain, these three conserved residues are substituted with glutamine, leucine and threo-nine, respectively, which have completely different functionality (Fig. 3a). Moreover, because of the direct packing of the PHD finger, the ZA loop is positioned in a different orientation with respect to the BC loop compared to that of the PCAF bromodomain. Consequently, the characteristic pocket between the ZA and BC loop in the canonical bromodomain of PCAF, CBP or BPTF is distorted in the atypical bromodomain of KAP1 (Fig. 3b and Supplementary Fig. 3a,b online). These distinct structural features, however, are consistent with our observation that the tandem PHD finger–bromodomain of KAP1 does not bind to lysine-acetylated peptides derived from histones H3 or H4, as shown by NMR binding.
Figure 3.
Structural features of the bromodomain and the PHD finger of KAP1. (a) Ribbon structures of the bromodomains of human KAP1 and PCAF, highlighting the conserved structural fold. Side chains of the highly conserved residues in the PCAF acetyllysine binding pocket are shown. H3K36ac, acetylated lysine 36 of histone H3. (b) Surface electrostatic potential depiction of the KAP1 and PCAF bromodomains. The proteins are shown in a similar orientation. (c) Structure of the KAP1 PHD finger–bromodomain, with side chains of residues corresponding to those forming the aromatic cage in human BPTF (left) and showing a cluster of four aromatic residues of the PHD finger packing against helix αZ of the bromodomain (middle). Right, the crystal structure of the human BPTF PHD finger–bromodomain bound to a histone H3 peptide containing trimethylated lysine 4 (H3 K4me3, in red; PDB code 2FSA). The side chains of the aromatic cage formed by Tyr10, Tyr17, Tyr23 and Trp32 are shown.
The PHD finger in KAP1 is also functionally different from that of BPTF. The latter recognizes methylated lysine 4 of histone H3 (H3K4) by a cage of four aromatic side chains located on the surface of the β-sheet of the PHD finger14 (Fig. 3c, right). This aromatic cage is absent in the corresponding site in KAP1; instead, a four-aromatic-aminoacid cluster in KAP1 lies on the opposite side of the β-sheet packing against αZ of the bromodomain (Fig. 3c, left and middle). However, the KAP1 PHD finger, either alone or in tandem with the bromodomain, does not bind to lysine-methylated or nonmodified peptides derived from methylation sites in histones, such as H3K4, H3K9 and H3K27, as shown in the NMR binding study (data not shown). Moreover, in a dot blot assay we also ruled out possible KAP1 PHD-finger binding to phosphoinositides, binding of which has been reported for inhibitor of growth family, member 2 (ING2) and other PHD fingers38. Collectively, these results show that the PHD finger and the bromodomain of KAP1 do not have typical activities of methyllysine and acetyllysine binding, respectively.
SUMOylation and binding of the KAP1 tandem domain to UBC9
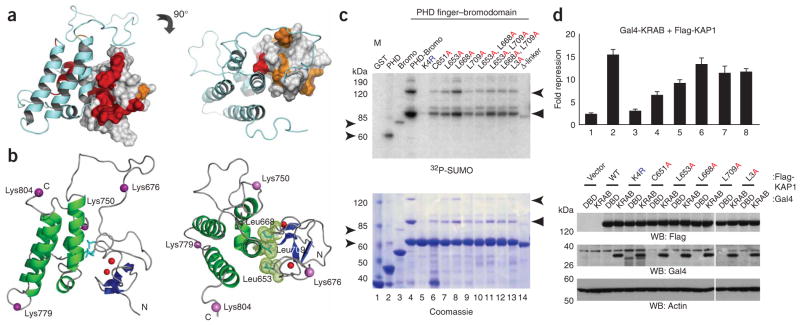
We recently reported that the co-repressive activity of KAP1 is dependent on lysine SUMOylation, which is unique within the TIF1 protein family (Supplementary Fig. 4a,b online), and that the PHD finger of KAP1 functions as a SUMO E3 ligase for KAP1 SUMOylation33. Of six SUMOylation sites mapped in KAP1, four reside within the PHD finger–bromodomain unit (Fig. 4) and two in a region N-terminal to the PHD finger33. Using an in vitro SUMOylation assay and lysine-to-arginine mutations of the PHD finger–bromodomain, we showed that two major SUMOylation sites are located at Lys779 in the BC loop and Lys804 in the loop following αC of the bromodomain (Supplementary Fig. 4c, lanes 6 and 7), and two minor sites reside at Lys676 in the interdomain linker and Lys750 in the AB loop of the bromodomain (Supplementary Fig. 4c, lanes 4 and 5).
Figure 4.
Human KAP1 PHD finger–bromodomain acts as one functional unit in SUMOylation and transcriptional repression. (a) The structure of the tandem PHD finger–bromodomain of KAP1 in front and top views, highlighting residues that showed major line-broadening upon UBC9 binding in the NMR spectra (Supplementary Fig. 5a). The affected residues whose peaks disappeared are red and those with a major reduction in peak intensity are gold. (b) Three-dimensional structure of the KAP1 PHD finger–bromodomain, depicting the location of four lysine SUMOylation sites (magenta spheres) and the side chains of three hydrophobic residues, Leu653, Leu668 and Leu709 (light blue) at the interdomain interface. The two zinc atoms in the PHD finger are red. (c) In vitro SUMOylation of the KAP1 PHD finger and bromodomain, separately (lanes 2 and 3) and in the PHD finger–bromodomain module: wild type (lane 4), SUMOylation sites mutant K4R (lane 5), structural PHD mutant (lane 6) or mutants with different substitutions in leucines involved in the interdomain interaction (lanes 7–13). L3A is a triple mutant, L653A L668A L709A, and δ-linker is a deletion of the linker residues 674–694. (d) U2OS K4 cells were co-transfected with Gal4-DBD or Gal4-KRAB, empty vector or Flag-KAP1 wild type or indicated mutant constructs together with 5×Gal4-TK-luc plasmid and a pTK-β-gal normalization vector. The expression of Flag-KAP1 and Gal4-KRAB proteins was confirmed by western blot (below). Data are the mean ± s.d. of at least two experiments performed in triplicate.
To characterize the functional role of the PHD finger–bromodomain in KAP1 SUMOylation, we assessed the tandem-domain interactions with the SUMO E2 ligase UBC9 using NMR. As shown in the 1H-15N HSQC spectra of the PHD finger–bromodomain, a substantial number of KAP1 backbone amide resonances underwent line broadening upon addition of UBC9 fused to glutathione S-transferase (GST-UBC9) but not GST alone, indicative of direct protein-protein interactions (Supplementary Fig. 5a online). Notably, the residues that showed major chemical shift perturbations are localized at the tandem-domain interface (Fig. 4a), consistent with the NMR-mapped location of UBC9 binding sites on the PHD finger alone33 (Supplementary Fig. 5b,c). Although UBC9 recognizes the SUMOylation consensus sequence φKxE/D39,40, we did not observe major perturbations of residues at any of the four SUMOylation sites. Localization of UBC9 binding at the domain-domain interface suggests that tandem-domain association may be coupled to UBC9 binding and facilitated by a flexible interdomain linker. The observation of different KAP1 residues perturbed upon UBC9 binding of the PHD finger alone or the tandem PHD finger–bromodomain (Supplementary Fig. 5b,c) indicates that the two domains would become rearranged to accommodate UBC9–PHD finger–bromodomain association. Viewed from the top of the tandem-domain structure, the four SUMOylation sites (Fig. 4b, magenta spheres) are spatially located on the periphery of the globular structural fold, making the PHD finger–bound E2 conjugating enzyme UBC9 accessible to all four sites. Taken together, these results confirm a coherent nature of the PHD finger–bromodomain association in the tandem unit and their interactions with UBC9.
Functional role of the structural integrity of the tandem domain
To assess the functional importance of its structural integration, we performed in vitro SUMOylation of the tandem PHD finger–bromodomain of KAP1. As shown in Figure 4c, either the PHD finger or the bromodomain alone was a poor substrate for SUMOylation (lanes 2 and 3), whereas the tandem-domain motif showed at least ten-fold stronger SUMOylation (lane 4). The increased level of SUMOylation cannot be ascribed to a possible difference in protein stability, as the PHD finger alone is as stable as the tandem domain. As such, these results strongly suggest that the two domains cooperate. The marked increase in SUMOylation is due to the stimulatory effect of the PHD finger on the bromodomain as the major SUMO conjugation sites, Lys779 and Lys804, are located in the bromodomain (Fig. 4c). Disruption of the PHD finger fold by C651A mutation due to the ablation of zinc coordination resulted in a complete loss of the PHD finger–mediated stimulatory effect on bromodomain SUMOylation (lane 6), which is consistent with the inability of the PHD C651A mutant to bind to UBC9 (ref. 33).
To further characterize the functional role of the interdomain association, we made alanine mutations at three leucine residues, Leu653, Leu668 and Leu709, located at the interdomain interface, and tested the effects of these mutations on in vitro KAP1 SUMOylation (Fig. 4c) and KAP1 transcriptional repression in a transient gene reporter assay (Fig. 4d). Although these conservative amino acid substitutions should not cause major structural changes, alanine mutation of Leu653 in the PHD finger or Leu709 in the bromodomain showed detrimental effects on KAP1 SUMOylation to a level similar to that of the C651A mutation (Fig. 4c, lanes 7 and 9 versus lane 6), or about 65% and 40% of the C651A mutation effects on in vivo transcription-repression activity, respectively (Fig. 4d, lanes 5 and 7 versus lane 4). It is important to note that, when compared to the mutation effects on in vitro SUMOylation of the PHD finger–bromodomain, the less profound reduction observed in in vivo repression activity in these measurements is probably due to the use of full-length KAP1, which contains two additional SUMOylation sites located N-terminal to the PHD finger, and the fact that SUMOylation of these sites occurs in the absence of or upon structural perturbation of the PHD-bromodomain module33. Indeed, only when all six SUMOylation sites in KAP1 were mutated (the K6R mutant) did we observe a nearly complete loss of repression activity (Fig. 4d, lane 3), indicating that SUMO modification has a major role in KAP1 repression function.
On the basis of the NMR structure and the UBC9 binding data as described above, we argue that the tandem-domain interactions and their UBC9 binding are coupled and that amino acid mutations at the interdomain interface can affect PHD finger–bromodomain association and/or their binding to UBC9. Thus, the effects of single, double or triple leucine-to-alanine mutations on KAP1 SUMOylation probably result from a net balance of these two related factors. The effects of single leucine-to-alanine mutations can indicate their relative functional importance in KAP1 SUMOylation through contributions to domain-domain interactions and UBC9 binding (such as Leu653 and Leu709 versus Leu668), whereas a double or triple mutant does not necessarily produce an additive effect when compared to the corresponding single or double mutant (Fig. 4c). Along the same line of reasoning, we predicted and confirmed that deletion of the linker would have a major impact on SUMOylation (Fig. 4c, lane 4 versus 14). As the linker contains only one minor SUMOylation site at Lys676 (Supplementary Fig. 4c, lane 4), a nearly complete loss of SUMOylation of the linker deletion mutant does not result from elimination of a SUMOylation site, but rather from a major structural perturbation of the tandem-domain motif (data not shown). Taken together, these results argue that the tandem PHD finger–bromodomain is an integrated structural and functional unit that is important for KAP1 SUMOylation and co-repression activity.
DISCUSSION
Our structure and molecular functional characterization of the PHD finger–bromodomain from human KAP1 reveals how a uniquely integrated structural architecture enables this tandem protein module to function as a SUMO E3 ligase for KAP1 SUMOylation. Unlike other SUMO E3 ligases—such as the RING/U-box and HECT domain E3s39, which commonly function to bring together an activated E2 SUMO and a substrate to promote SUMO conjugation—the KAP1 PHD finger–bromodomain is an intramolecular SUMO E3 ligase, thus ensuring ordered and stoichiometric site-specific SUMOylation at several lysine residues within the bromodomain and in the region N-terminal to the PHD finger, which together are required for KAP1’s co-repression activity33,41,42.
The location of the C-terminal SUMOylation sites in KAP1 is of particular functional interest. For example, Lys676 and Lys750 are close to the interdomain linker, whereas Lys779 and Lys804 reside in the BC loop and in a loop C-terminal to the last helix αC of the bromodomain, respectively. Although the underlying mechanisms of SUMO-mediated KAP1 repression activity together with other chromosomal proteins remain to be elucidated, it is worth noting that, in a canonical bromodomain, the BC loop typically participates in interactions with lysine-acetylated proteins10, and Lys804 is close to the C-terminal Ser824 phosphorylation site, which has recently been reported to be important for KAP1 function in modulating chromatin relaxation in response to DNA double-strand breaks in an ATM-dependent manner43.
Lysine SUMOylation has emerged as an important post-translational modification in the regulation of protein functions39,40. Because of the large size of SUMO, lysine SUMOylation can alter a target protein’s conformation or modulate its interactions with other cellular proteins in a SUMO-dependent manner. Recent studies show that SUMOylation of transcription factors has a direct role in transcriptional control and subnuclear compartmentalization44. Indeed, KAP1 SUMOylation has recently been shown to recruit the SETDB1 histone methyltransferase and the NuRD remodeling complex by binding to SUMO-interacting motifs, leading to chromatin remodeling and modifications at the site of a target gene33. Although, as shown in our in vitro study, the KAP1 PHD finger and the bromodomain do not interact with methylated lysine, acetylated lysine or even un-modified histone peptides (consistent with its lack of key residues required for such interactions), the tandem PHD finger–bromodomain may function uniquely as an integrated structural template. As such, the PHD finger–bromodomain in KAP1 coordinates in a SUMO-dependent manner the assembly of a large protein complex consisting of both chromatin-modification and chromatin-remodeling activities, thereby modulating transcriptional silencing in the chromatin context.
Given the flexibility of the tandem-domain linker, it is conceivable that the PHD finger and the bromodomain of an integrated structure can be connected in reverse order compared to KAP1, as seen in the transcriptional coactivator p300/CBP. Indeed, the tandem bromodomain–PHD finger of p300/CBP shows cooperative nucleosome binding activity27. Note in this case that the bromodomain of p300/CBP is capable of recognizing acetylated lysine residues on histones.
In conclusion, this new structure of the tandem PHD finger–bromodomain of KAP1 yields important mechanistic insights into its function in transcriptional repression. Notably, our study highlights the functional versatility of these two conserved protein structural modules in chromatin-mediated transcriptional regulation. Some PHD fingers and bromodomains are known to act as methyllysine or acetyllysine binding domains, respectively, which can function either individually or sequentially in a tandem motif, as reported in BPTF25,26. Our study reveals a new functionality of the tandem PHD finger–bromodomain as one interactive functional unit in modulating gene transcription in a SUMOylation-dependent manner. Therefore, we expect that such tandem protein modules are likely to hold a key to a new understanding of mechanistic models of epigenetic regulation of gene activation or silencing that is directed by structurally conserved protein domains in combinatorial manners, a subject that requires further structural and functional analyses of the relevant chromosomal protein complexes.
METHODS
Protein expression and purification
The tandem PHD finger and bromodomain of human KAP1 (residues 624–812) was expressed in E. coli BL21(DE3) cells in the pET15b vector (Novagen). We prepared isotope-labeled proteins from cells grown on a minimal medium containing 15NH4Cl, with or without 13C6-glucose, in H2O or 75% 2H2O. The protein was purified by nickel-IDA affinity chromatography followed by thrombin cleavage to remove an N-terminal polyhistidine tag. GST-fused bromodomain–PHD finger of KAP1 were expressed in E. coli in the pGEX4T3 vector (Pharmacia), and purified with glutathione-Sepharose beads. NMR spectra of the purified KAP1 proteins were acquired to ensure proper protein folding.
In vitro SUMOylation
In vitro SUMOylation reactions were carried out at 30 °C for 3 h. Briefly, 2 μg of GST-fused proteins of KAP1 were loaded onto glutathione beads and incubated in the presence of 500 ng of UBC9, 700 ng of human E1 (BostonBiochem) and 2 μg of 32P-labeled SUMO1 in 20 μl reaction mix with buffer containing ATP (LAE Biotech International). After incubation, beads were washed five times with 1 ml of BLB500 buffer, proteins were eluted in 2× Laemmli buffer, resolved by SDS-PAGE and visualized by Coomassie blue staining and subsequent autoradiography. 32P-labeled SUMO1 was prepared from GST–PKA-SUMO1 as described45. We performed in vitro SUMOylation reactions for the tandem PHD finger–bromodomain motifs from three other TIF1 family members and the tandem bromodomain–PHD finger from p300/CBP.
Gene repression assay
U2OS cells were cultured in DMEM supplemented with 10% (v/v) FBS. Transfections were performed on six-well plates in triplicate using FuGene6 (Roche), according to the manufacturer’s instructions. Cells were lysed 36 h after transfection in Tween 20 buffer46. Luciferase Assay System (Promega) and β-galactosidase Assay Reagent (Pierce) were used to measure respective enzymatic activities and subsequent normalization. Nuclei were pelleted, lysed in Triton X-100 buffer46 and used for detection of transfected proteins by western blotting. Lentiviral vector–based KAP1 gene knockdown was performed as described previously47.
NMR spectroscopy
NMR samples contained KAP1 PHD finger–bromodomain of ~0.5 mM in 100 mM phosphate buffer of pH 6.5 containing 200 mM sodium chloride in H2O/2H2O (9/1) or 2H2O. All NMR spectra were acquired at 30 °C on a Bruker 500 MHz, 600 MHz, 800 MHz or 900 MHz NMR spectrometer. The 1H, 13C and 15N resonances of the protein were assigned by triple-resonance NMR spectra collected with a uniformly 13C- and 15N-labeled and 75% deuterated protein37. The distance restraints were obtained from three-dimensional 13C- or 15N-NOESY spectra. Slowly exchanging amides, identified in two-dimensional 15N-HSQC spectra recorded after a H2O buffer was changed to a 2H2O buffer, were used with structures calculated with only NOE distance restraints to generate hydrogen-bond restraints for final structure calculations.
Structure calculations
Structures of the KAP1 PHD finger–bromodomain were calculated using a distance geometry-simulated annealing protocol using the X-PLOR program48. Initial protein structure calculations were performed with manually assigned NOE-derived distance restraints. Hydrogen-bond distance restraints, generated from the H/D exchange data, were added at a later stage of structure calculations for residues with characteristic NOE patterns. ARIA-assigned49 distance restraints agree with structures calculated using only the manually determined NOE distance restraints. For the final 20 lowest-energy NMR structures, no distance or torsional angle restraint was violated by more than 0.5 Å or 5°, respectively (Table 1). Ramachandran plot analysis of the final structures, excluding the linker and the ZA loop, with Procheck-NMR50 showed that 77.4 ± 2.9%, 18.3 ± 2.6%, 3.0 ± 1.5% and 1.4 ± 1.2% of the non-glycine and non-proline residues were in the most favorable, additionally allowed, generously allowed and disallowed regions, respectively. The corresponding values for the residues in the secondary-structure regions of the PHD finger–bromodomain were 88.8 ± 2.7%, 10.7 ± 2.5%, 0.4 ± 0.7% and 0.2 ± 0.5%, respectively.
Table 1.
Structural statistics of 20 final NMR structures of the KAP1 PHD finger–bromodomain
| PHD finger–bromodomain | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOEs | 3,497 |
| Intra-residue | 1,387 |
| Inter-residue | 2,110 |
| Sequential (|i – j | = 1) | 742 |
| Medium-range (|i – j | < 4) | 684 |
| Long-range (|i – j | > 5) | 684 |
| Protein-zinc restraints | 22 |
| Hydrogen bonds | 57 |
| Total dihedral angle restraints | |
| φ | 75 |
| ψ | 75 |
| Structure statisticsa,b | |
| Violationsc (mean ± s.d.) | |
| Distance constraints (Å) | 0.47 ± 0.02 |
| Dihedral angle constraints (°) | 0.90 ± 0.25 |
| Max. dihedral angle violation (°) | 1.37 |
| Max. distance constraint violation (Å) | 0.51 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.016 ± 0.0003 |
| Bond angles (°) | 1.33 ± 0.02 |
| Impropers (°) | 1.13 ± 0.04 |
| Average pairwise r.m.s. deviation (Å) | |
| Heavy | 1.03 ± 0.15 |
| Backbone | 0.51 ± 0.12 |
Protein residues 624–812, excluding the linker and the ZA loop.
20 final structures were used in r.m.s. deviation calculations, as well as in pairwise r.m.s. deviation calculations.
None of the final structures have distance restraint violations >0.5 Å or dihedral angle violations >5°.
Supplementary Material
Supplementary Fig. 1. Structure-based sequence alignment of the tandem PHD finger-bromodomain motifs. Sequences of tandem PHD finger-bromodomain or bromodomain-PHD finger motifs are aligned based on available structures and protein sequence conversation. Sequence numbers are shown before and after the corresponding protein sequences. Absolutely and highly conserved residues in PHD fingers or bromodomains are colored in red and blue, respectively. Residues are conserved in subgroups of PHD fingers or bromodomains are highlighted in black. Residues in the bromodomains of the human PCAF and CBP or the PHD finger of human BPTF that interact with acetyl-lysine or methyl-lysine, as defined in experimental structural analyses, are underlined.
Supplementary Fig. 2. Characterization of inter-domain interactions within the tandem PHD finger-bromodomain of KAP1. (a) Location of the enterokinase cleavage site (red box) indicated in the protein structure that was constructed by changing the underlined residues in the sequence LKEEDGSLSL (residues 675–684) to LKDDDD-KLSL using site-directed mutagenesis. (b) SDS-PAGE illustrating a time course of the enterokinase cleavage of the KAP1 PHD finger-bromodomain. Although the major products are the GST-PHD finger and the bromodomain, a minor cleavage also occurs within the bromodomain. Because of the low molecular weight of the PHD finger (~6 kDa), the GST fusion tandem protein module was used to demonstrate the specificity of enterokinase cleavage on SDS-PAGE. (c) Differences of NMR backbone amide resonances of the KAP1 PHD finger between its free form and the tandem motif with the bromodomain, as shown with the protein sequence.
Supplementary Fig. 3. Comparison of the structural features of bromodomains. (a) Ribbon structures of the bromodomains from human transcriptional proteins KAP1, BPTF, PCAF and CBP, highlighting the conserved bromodomain fold. Side-chains of the highly conserved amino acid residues located in the acetyl-lysine binding pocket are depicted in the latter three proteins, and color-coded by atom-type. The corresponding residues in KAP1 (by the sequence alignment, see Supplementary Fig. 1) are also shown. (b) Surface electrostatic potential representations of human KAP1, BPTF, PCAF and CBP bromodomains. The proteins are shown in a similar orientation, which is a 90° rotation from that of the corresponding structures in a.
Supplementary Fig. 4. Sumoylation reaction of the tandem PHD finger-bromodomain of KAP1. (a) Linear range of in vitro sumoylation reaction. Comparison of the time course of in vitro sumoylation of the KAP1 PHD-bromodomain wild type versus the triple leucine-to-alanine mutant (L653A/L668A/L709A). The reaction condition was similar to that described in Fig. 4b. (b) SUMO modification of the PHD-bromodomain module is unique for KAP1, as illustrated in in vitro sumoylation of GST-fusion proteins of PHD-bromodomain modules from all four members of the TIF1 family and the bromodomain-PHD motif of CBP. The sumoylation reaction was carried out as follows: GST proteins were immobilized on glutathione beads and incubated with recombinant E1, Ubc9, and 32P-labeled SUMO1 for 2 hours. Reaction components were washed out from the beads, bound GST proteins were analyzed by Coomassie blue staining (bottom) and autoradiography (top). The positions of the major mono- and di-sumoylated forms are shown with arrowheads. (c) Identification of major sumoylation sites K779 and K804 in the KAP1 bromodomain. In vitro sumoylation of GST-KAP1 PHD finger-bromodomain, triple and quadruple sumoylation site mutants as indicated on the top right. The sumoylation reaction was carried out as described above in b. The positions of the sumoylated forms are shown with arrowheads.
Supplementary Fig. 5. Comparison of Ubc9 binding to the KAP1 tandem PHD finger-bromodomain and to the PHD finger alone. (a) Superposition of 2D 1H-15N HSQC spectra of the KAP1 PHD finger-bromodomain in the free form (black signals) and in the presence of GST-Ubc9 (left panel, red signals), or GST (right panel, red signals). The molar ratio of KAP1:GST is about 1:1.5. (b) The structure of the tandem PHD finger-bromodomain of KAP1 (front and top views, upper and lower panels) highlighting amino acid residues that exhibited major line-broadening upon Ubc9 binding. The affected residues are colored in red for those whose peaks disappeared and orange for those with a major reduction in peak intensity. (c) Amino acid residues of the PHD finger alone that exhibited major line-broadening upon Ubc9 binding are depicted in the structure of the tandem PHD finger-bromodomain of KAP1 (front and top views, upper and lower panels), for the purpose of comparison to a. The affected residues are color-coded as described in b.
Acknowledgments
We wish to acknowledge the use of the NMR facilities at the New York Structural Biology Center for this study. K.L.Y. was supported by a Terry Fox Foundation postdoctoral fellowship from the National Cancer Institute of Canada. F.J.R. was supported by grants from the US National Institutes of Health (CA095561 and CA092088), The Samuel Waxman Cancer Research Foundation, The Pardee Foundation and The Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health. M.-M.Z. was in part supported by funds from the Dr. Golden and Harold Lamport Chair, and grants from the US National Institutes of Health (CA087658) and the US National Science Foundation (#0517352).
Footnotes
Accession codes Protein Data Bank: Coordinates for the solution structure of the KAP-1 PHD finger–bromodomain have been deposited with accession code 2RO1.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
L.Z. determined the protein structure by NMR, K.L.Y. characterized the protein structure-function by NMR; A.V.I. performed in vitro SUMOylation and transcription repression studies; X.W., S.M., and O.P. contributed to molecular cloning, protein purification and characterization of the study. The project was directed by M.-M.Z. and F.J.R. All authors contributed to the preparation of the manuscript.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 6.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 7.Berger J, Bird A. Role of MBD2 in gene regulation and tumorigenesis. Biochem Soc Trans. 2005;33:1537–1540. doi: 10.1042/BST0331537. [DOI] [PubMed] [Google Scholar]
- 8.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 10.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 11.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 12.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 13.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 14.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:1058–1061. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peña PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1111. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramó n-Maiques S, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci USA. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanno T, et al. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 22.Vandemark AP, et al. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 25.Yap KL, Zhou MM. Structure and function of protein modules in chromatin biology. Results Probl Cell Differ. 2006;41:1–23. [PubMed] [Google Scholar]
- 26.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ragvin A, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 28.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Kitagawa H, et al. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113:905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 30.Venturini L, et al. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- 31.Bloch DB, de la Monte SM, Guigaouri P, Filippov A, Bloch KD. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem. 1996;271:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- 32.Capili AD, Schultz DC, Rauscher FJ, III, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov AV, et al. PHD Domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrink M, et al. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 β. Proc Natl Acad Sci USA. 2001;98:1422–1426. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman JR, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 36.Le Douarin B, et al. A possible involvement of TIF1 alpha and TIF1 beta in the epi-genetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 37.Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 38.Gozani O, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 39.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 41.Mascle XH, Germain-Desprez D, Huynh P, Estephan P, Aubry M. Sumoylation of the transcriptional intermediary factor 1β (TIF1β), the co-repressor of the KRAB multifinger proteins, is required for its transcriptional activity and is modulated by the KRAB domain. J Biol Chem. 2007;282:10190–10202. doi: 10.1074/jbc.M611429200. [DOI] [PubMed] [Google Scholar]
- 42.Li X, et al. Role for KAP1 serine 824 phosphorylation and SUMOylation/deSUMOylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 43.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 44.Lin DY, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Yurchenko V, Xue Z, Sadofsky MJ. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol Cell Biol. 2006;26:1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klenova E, Chernukhin I, Inoue T, Shamsuddin S, Norton J. Immunoprecipitation techniques for the analysis of transcription factor complexes. Methods. 2002;26:254–259. doi: 10.1016/S1046-2023(02)00029-4. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 49.Nilges M, O’Donoghue S. Ambiguous NOEs and automated NOE assignment. Prog Nucl Magn Reson Spectrosc. 1998;32:107–139. [Google Scholar]
- 50.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Structure-based sequence alignment of the tandem PHD finger-bromodomain motifs. Sequences of tandem PHD finger-bromodomain or bromodomain-PHD finger motifs are aligned based on available structures and protein sequence conversation. Sequence numbers are shown before and after the corresponding protein sequences. Absolutely and highly conserved residues in PHD fingers or bromodomains are colored in red and blue, respectively. Residues are conserved in subgroups of PHD fingers or bromodomains are highlighted in black. Residues in the bromodomains of the human PCAF and CBP or the PHD finger of human BPTF that interact with acetyl-lysine or methyl-lysine, as defined in experimental structural analyses, are underlined.
Supplementary Fig. 2. Characterization of inter-domain interactions within the tandem PHD finger-bromodomain of KAP1. (a) Location of the enterokinase cleavage site (red box) indicated in the protein structure that was constructed by changing the underlined residues in the sequence LKEEDGSLSL (residues 675–684) to LKDDDD-KLSL using site-directed mutagenesis. (b) SDS-PAGE illustrating a time course of the enterokinase cleavage of the KAP1 PHD finger-bromodomain. Although the major products are the GST-PHD finger and the bromodomain, a minor cleavage also occurs within the bromodomain. Because of the low molecular weight of the PHD finger (~6 kDa), the GST fusion tandem protein module was used to demonstrate the specificity of enterokinase cleavage on SDS-PAGE. (c) Differences of NMR backbone amide resonances of the KAP1 PHD finger between its free form and the tandem motif with the bromodomain, as shown with the protein sequence.
Supplementary Fig. 3. Comparison of the structural features of bromodomains. (a) Ribbon structures of the bromodomains from human transcriptional proteins KAP1, BPTF, PCAF and CBP, highlighting the conserved bromodomain fold. Side-chains of the highly conserved amino acid residues located in the acetyl-lysine binding pocket are depicted in the latter three proteins, and color-coded by atom-type. The corresponding residues in KAP1 (by the sequence alignment, see Supplementary Fig. 1) are also shown. (b) Surface electrostatic potential representations of human KAP1, BPTF, PCAF and CBP bromodomains. The proteins are shown in a similar orientation, which is a 90° rotation from that of the corresponding structures in a.
Supplementary Fig. 4. Sumoylation reaction of the tandem PHD finger-bromodomain of KAP1. (a) Linear range of in vitro sumoylation reaction. Comparison of the time course of in vitro sumoylation of the KAP1 PHD-bromodomain wild type versus the triple leucine-to-alanine mutant (L653A/L668A/L709A). The reaction condition was similar to that described in Fig. 4b. (b) SUMO modification of the PHD-bromodomain module is unique for KAP1, as illustrated in in vitro sumoylation of GST-fusion proteins of PHD-bromodomain modules from all four members of the TIF1 family and the bromodomain-PHD motif of CBP. The sumoylation reaction was carried out as follows: GST proteins were immobilized on glutathione beads and incubated with recombinant E1, Ubc9, and 32P-labeled SUMO1 for 2 hours. Reaction components were washed out from the beads, bound GST proteins were analyzed by Coomassie blue staining (bottom) and autoradiography (top). The positions of the major mono- and di-sumoylated forms are shown with arrowheads. (c) Identification of major sumoylation sites K779 and K804 in the KAP1 bromodomain. In vitro sumoylation of GST-KAP1 PHD finger-bromodomain, triple and quadruple sumoylation site mutants as indicated on the top right. The sumoylation reaction was carried out as described above in b. The positions of the sumoylated forms are shown with arrowheads.
Supplementary Fig. 5. Comparison of Ubc9 binding to the KAP1 tandem PHD finger-bromodomain and to the PHD finger alone. (a) Superposition of 2D 1H-15N HSQC spectra of the KAP1 PHD finger-bromodomain in the free form (black signals) and in the presence of GST-Ubc9 (left panel, red signals), or GST (right panel, red signals). The molar ratio of KAP1:GST is about 1:1.5. (b) The structure of the tandem PHD finger-bromodomain of KAP1 (front and top views, upper and lower panels) highlighting amino acid residues that exhibited major line-broadening upon Ubc9 binding. The affected residues are colored in red for those whose peaks disappeared and orange for those with a major reduction in peak intensity. (c) Amino acid residues of the PHD finger alone that exhibited major line-broadening upon Ubc9 binding are depicted in the structure of the tandem PHD finger-bromodomain of KAP1 (front and top views, upper and lower panels), for the purpose of comparison to a. The affected residues are color-coded as described in b.