Abstract
Hepatitis C virus (HCV) leads to progressive liver disease and hepatocellular carcinoma. Current treatments are only partially effective, and new therapies targeting viral and host pathways are required. Virus entry into a host cell provides a conserved target for therapeutic intervention. Tetraspanin CD81, scavenger receptor class B member I, and the tight-junction proteins claudin-1 and occludin have been identified as essential entry receptors. Limited information is available on the role of receptor trafficking in HCV entry. We demonstrate here that anti-CD81 antibodies inhibit HCV infection at late times after virus internalization, suggesting a role for intracellular CD81 in HCV infection. Several tetraspanins have been reported to internalize via motifs in their C-terminal cytoplasmic domains; however, CD81 lacks such motifs, leading several laboratories to suggest a limited role for CD81 endocytosis in HCV entry. We demonstrate CD81 internalization via a clathrin- and dynamin-dependent process, independent of its cytoplasmic domain, suggesting a role for associated partner proteins in regulating CD81 trafficking. Live cell imaging demonstrates CD81 and claudin-1 coendocytosis and fusion with Rab5 expressing endosomes, supporting a role for this receptor complex in HCV internalization. Receptor-specific antibodies and HCV particles increase CD81 and claudin-1 endocytosis, supporting a model wherein HCV stimulates receptor trafficking to promote particle internalization.
INTRODUCTION
Hepatitis C virus (HCV) is a member of the Flaviviridae family and an important human pathogen that leads to progressive liver disease and is a leading indication for liver transplantation. At present, there is no HCV vaccine, and the only approved therapy, interferon and ribavirin, has limited efficacy. Unsurprisingly, there is an international effort to develop new antiviral agents and vaccines that are effective across all major HCV genotypes. A number of drugs targeting HCV replicase enzymes are in development; however, recent trials show a rapid appearance of drug-resistant viruses (for reviews, see references 1 and 52). The essential and conserved nature of the entry step in the HCV life cycle offers an attractive target for therapeutic intervention.
Virus entry into a host cell is defined by specific interaction(s) with cell surface proteins or receptors that confer host and cellular tropism (66). Recent advances in the development of in vitro systems to study the HCV life cycle have demonstrated an essential role for tetraspanin CD81 (54), scavenger receptor BI (SR-BI) (58), and tight-junction protein occludin (3, 42, 55) and several members of the claudin family (21, 47, 76) in virus entry. Low-density lipoprotein receptor and cell surface glycosaminoglycans, including heparan sulfate, have been reported to play a role in the initial attachment of HCV to the cell surface (2, 49). Coexpression of human CD81, SR-BI, occludin, and claudin-1 renders nonliver cells permissive for HCV entry, demonstrating that these four proteins constitute the minimal viral receptor requirement (18, 55). CD81 and SR-BI bind HCV encoded E1E2 glycoproteins with high affinity (54, 58), and antibodies targeting these molecules neutralize virus infection after cell attachment, suggesting a role for CD81 and SR-BI in the lateral diffusion and endocytosis of HCV particles (12). In contrast, there is limited evidence for tight-junction protein association with HCV, which may reflect an indirect role for these proteins in the virus internalization process.
Many viruses enter cells with their cognate receptors by using constitutive endocytic trafficking pathway(s); for example, Moloney murine leukemia virus internalizes with murine cationic amino acid transporter (38), Nipah virus internalizes with ephrin B2 (17), Poliovirus internalizes with CD155 (16), and some coronaviruses endocytose with their major receptor aminopeptidase N (24). Some viruses have been reported to stimulate receptor trafficking and endocytosis: severe acute respiratory syndrome coronavirus triggers angiotensin converting enzyme 2 endocytosis (70) and herpes simplex virus induces nectin-1/virus complex internalization (63). At present, limited information is available on the role of receptor trafficking in HCV entry and whether virus engagement promotes receptor endocytosis. In the present study we demonstrate that anti-CD81 monoclonal antibodies (MAbs) can inhibit HCV infection at late times after virus internalization, suggesting an intracellular site of antibody neutralization and a role for endosomal CD81 in virus infection. Ligation of CD81 with antibodies or HCV particles promotes receptor endocytosis in a clathrin- and dynamin-dependent process. Live cell imaging demonstrates antibody-primed CD81 and claudin-1 coendocytosis and fusion with Rab5 expressing early endosomes, supporting a role for this receptor complex in virus internalization and fusion with endosomal membranes.
MATERIALS AND METHODS
Cell lines, antibodies, and reagents.
Huh-7.5 cells (Charles Rice, The Rockefeller University, New York, NY) (9), Huh-7 Lunet cells (Thomas Pietschmann; Twincore, Hanover, Germany) (7), and 293T cells (American Type Culture Collection) were propagated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS)–1% nonessential amino acids (Invitrogen, California) and grown in a humidified atmosphere at 37°C in 5% CO2. Huh-7.5 cells were transduced to express AcGFP.CD81, as previously reported (26). Huh-7 Lunet cells were transduced to express wild-type CD81 and a CD81 mutant lacking the C-terminal cytoplasmic domain (CD81ΔC) (6) (a gift from Martin Hemler, Harvard, Cambridge, MA). Claudin-1 expression in Huh-7.5 cells was siRNA silenced (Dharmacon) using Lipofectamine RNAiMAX according to the manufacturer's instructions.
Anti-CD81 MAbs 2s66 and 2s131 were generated after immunization of mice with recombinant full-length CD81 (32). Purified mouse immunoglobulin was a gift from M. Goodall (University of Birmingham). Anti-CD81 JS81 (BD Biosciences) and 1.3.3.22 (Santa Cruz) MAbs were purchased from their respective suppliers. Anti-CD81 affinity for recombinant MBP-CD81 large extracellular loop (LEL) (19) was assessed using the Biacore 3000 system (Biacore AB). Recombinant MBP-CD81 was immobilized to a CM5 chip using a standard amine coupling method (Biacore, GE Healthcare) and increasing concentrations of anti-CD81 MAbs flowed over the chip surface (3.9 to 500 μM) at 5 μl/min at 25°C for 50 min and dissociation recorded for 60 min. To control for nonspecific protein interactions, all MAbs were flowed over an “empty” channel. The binding affinity (Kd) for each MAb was quantified by fitting a one-site binding (hyperbola) curve to the data. Anti-claudin-1 clone 421203 was purchased from R&D Systems. Anti-EEA1 clone1G11 was obtained from Abcam. Anti-tubulin clone T9026 was purchased from Sigma. Alexa-594 conjugated transferrin (Tfn-594) and secondary Alexa 488/594-labeled goat anti-mouse, goat anti-rabbit, and goat anti-rat immunoglobulins were obtained from Invitrogen. Species-specific secondary antibodies were used to detect the colocalization of anti-receptor antibodies, and their specificities were confirmed in each staining experiment.
Virus genesis, infection, and anti-CD81 neutralization.
Cell culture-derived HCV (HCVcc) J6/JFH was generated as previously described (40). Briefly, RNA was transcribed in vitro from full-length genomes (RiboMax t7 kit; Promega) and electroporated into Huh-7.5 cells. Viral stocks were generated by two sequential passages through Huh-7.5 cells with a specific infectivity in the range of 1/500 to 1/780 (21). Supernatants were collected, pooled, and stored at −80°C. Infected cells were detected by methanol fixation and staining for HCV-encoded NS5A with MAb 9E10 (a gift from Charles Rice and Tim Tellinghuisen Rockefeller University); bound antibody was detected with anti-mouse IgG-Alexa 488 and quantified by enumerating NS5A-expressing cells.
Huh-7.5 cells were seeded at 1.5 × 104 cells/cm2, and the following day the medium was replaced with ice-cold 3% FBS-DMEM containing J6/JFH, followed by incubation on ice for 30 min. The cells were washed, the medium was replaced with prewarmed 3% FBS-DMEM and, at 0, 20, 60, and 120 min, anti-CD81 MAbs, at a concentration known to neutralize >95% virus infectivity (5 μg/ml) or proteinase K (PK; 50 μg/ml) were added in serum-free DMEM. PK-treated cells were subsequently incubated with 3% FBS-DMEM to inactivate its activity, as previously reported (59). At 48 h postinfection, the cells were fixed, and the infectivity was quantified (59). Virus escape from anti-CD81 or PK was calculated relative to the maximum neutralization at 120 min, and the time taken for 50% of the virus (t1/2) to escape treatments was determined.
Pseudoviruses expressing a luciferase reporter were generated as previously described (30). Briefly, 293T cells were transfected with a 1:1 ratio of plasmids encoding HIV provirus expressing luciferase and HCV strain H77 E1E2 envelope glycoproteins (HCVpp), murine leukemia virus envelope glycoprotein (MLVpp), vesicular stomatitis virus G glycoprotein (VSV-Gpp), or empty vector (Env−pp). Supernatants were harvested 48 h posttransfection and filtered, and virion-associated p24 was determined using a commercial assay (Aalto Bioreagents). Infection was quantified by measuring the luciferase activity (relative lights units [RLU]), and the specific infectivity was determined by subtracting the mean Env−pp signal from the HCVpp, MLVpp, or VSV-Gpp values.
Cell surface biotinylation endocytosis assay.
Huh-7.5 cells were seeded at 1.5 × 104 cells/cm2 and the following day serum starved for 60 min and, where indicated, incubated with Dynasore (80 μM) for 30 min, prior to adding Sulfo-NHS-SS-biotin (Pierce) at 0.5 mg/ml for 30 min at 4°C. The cells were washed and maintained at 4°C or transferred to 37°C for 60 min in the presence or absence of antibodies or viral pseudoparticles, as indicated. After the cells were returned to 4°C, they were washed with ice-cold buffer (50 mM Tris [pH 7.5], 100 mM NaCl [pH 8.6]) and biotin cleaved from surface-labeled proteins by incubation with the cell-impermeable reducing agent 2-mercaptoethanesulfonic acid at 15 mg/ml (MesNA; Sigma) for 30 min or incubation with ice-cold buffer in the absence of MesNA to determine the total protein expressed at the cell surface. The cells were lysed in lysis buffer (1% Brij97, 10 mM Tris [pH 7.5], 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 0.01% sodium azide). Biotinylated proteins were immunoprecipitated from defined quantities of cell lysate with streptavidin-coupled Dynabeads (Invitrogen). Precipitates and whole-cell lysates were separated by SDS-PAGE, transferred onto polyvinylidene difluoride membrane, and probed for CD81, claudin-1, or tubulin expression by enhanced chemiluminescence (GeneFlow). Densitometric analysis was performed using ImageJ software and the percentage of internalized CD81 or claudin-1 presented relative to the total CD81 or claudin-1 expressed at the cell surface.
Quantitative imaging of CD81 and claudin-1 endocytosis.
Parental Huh-7.5 cells or those transduced to express AcGFP.CD81 were seeded on glass coverslips at 1.5 × 104 cells/cm2 and the following day incubated with anti-CD81 or anti-claudin-1 MAbs in 3% FBS-DMEM–0.5% bovine serum albumin (BSA) at 37°C. The cells were fixed in 3.6% electron microscope-grade formaldehyde (EM-F; TAAB, United Kingdom), and protein-antibody complexes were detected using Alexa Fluor-conjugated antibody diluted in phosphate-buffered saline–0.05% saponin–0.5% BSA for 60 min at room temperature. The cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen) for 5 min, and coverslips were mounted onto glass slides (ProLong Gold Antifade; Invitrogen). The cells were imaged by laser scanning confocal microscopy (LSCM; Zeiss LSM510) using a ×100 oil immersion objective lens. To avoid any bias in the selection of cells for quantitation, the cells were initially identified by their DAPI nuclear stain and imaged for CD81 or claudin-1 expression. Microscope settings were optimized for each fluorescent protein to obtain the highest signal to noise ratio while controlling for cross talk. z-sections were collected, and the central section found to give optimal resolution. Consequently, a single 1-μm z-section across the cell center were selected for quantitation. CD81 and claudin-1 localization at the plasma membrane and intracellular locations were quantified by using the overlay and intensity frequency tools included with the Zeiss LSCM software. Cellular protein expression (arbitrary fluorescence units [AFU]) was calculated by combining the number of fluorescent pixels and multiplying this value by the average intensity. The fluorescence intensity at the plasma membrane and the intracellular locations were determined by extracting the regions of interest from the whole-cell image. The results are expressed as a percentage of the total fluorescence and represent the mean intracellular fluorescence intensity ± the standard error of the mean (SEM) of between 10 and 20 cells. Statistical analyses were performed using nonparametric one-way analysis of variance (ANOVA; Kruskal-Wallis test) or the Student t test in Prism 4.0, where necessary corrections for multiple comparisons were made. Intracellular colocalization was determined using ImageJ colocalization finder plug-in software, and the overlap coefficient is presented as a percentage of the colocalized pixels averaged from analyzing 10 cells.
Mechanism of CD81 endocytosis.
Huh-7.5 cells were transfected with green fluorescent protein (GFP) control plasmid or GFP-tagged transdominant mutants of Eps15 (GFP-EH29; Alexandre Benmerah, Institute Cochin, Paris, France) and Dynamin2 (K44A-GFP; Mark Niven, The Mayo Clinic, Rochester, MN) using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). GFP-, GFP-EH29-, and K44A-GFP-expressing cells were seeded onto glass coverslips at 1.5 × 104 cells/cm2 and incubated in serum-free medium for 60 min at 37°C; where indicated, naive Huh-7.5 cells were preincubated with Dynasore (80 μM) or dimethyl sulfoxide (DMSO) control for 60 min at 37°C, followed by anti-CD81 or Tfn-594 in the continued presence or absence of Dynasore for the indicated time intervals. To assess the role of Rho GTPases and epidermal growth factor (EGF) in CD81 endocytosis, Huh-7.5 cells were serum starved overnight and then incubated with C3 transferase (Cytoskeleton) at 5 μg/ml for 16 h, Rac-1 inhibitor (Calbiochem) at 100 μM for 60 min, EGF at 1 μg/ml, or Erlotinib at 10 μM in serum-free medium for 60 min, respectively, prior to adding anti-CD81 2s66 at 2 μg/ml for 60 min at 37°C. The cells were immediately fixed with EM-F and CD81, visualized, and quantified as detailed above.
Live cell imaging of CD81 and claudin-1 trafficking.
Parental Huh-7.5 and those transfected to express GFP-Rab5 (Alexandre Benmerah) were seeded in glass-bottom culture dishes at 1.5 × 104 cells/cm2. The following day, the cells were incubated for 30 min at 37°C with fluorescence-labeled anti-CD81 2s66 and anti-claudin-1 labeled with the ATTO fluorophores 647N and 565, respectively (ATTO-TEC, Seigen, Germany). The cells were washed and placed in phenol red-free 3% FBS-DMEM supplemented with 20 mM HEPES. Real-time LSCM was performed (inverted Zeiss 780) of a 1-μm z-section through the center of the cell using a ×100 oil immersion objective lens at 37°C over 5 min, and the images were processed using Zeiss software.
HCV-induced receptor endocytosis.
Huh-7.5 cells expressing AcGFP.CD81 were seeded at 1.5 × 104/cm2 on glass coverslips and incubated the following day with lentiviral pseudoparticles or HCVcc J6/JFH for 1 h at 37°C at an approximate multiplicity of infection of 0.3. J6/JFH was incubated with 100 μg of anti-HCV IgG or control IgG/ml for 1 h at 37°C or at 65°C for 1 h, prior to inoculating the cells. The cells were fixed in EM-F using MAbs, intracellular CD81/claudin-1 imaged, and quantified as described above. In parallel plates, unbound J6/JFH was removed by washing, and the medium was replenished with 3% FBS-DMEM for 48 h, after which the virus infection was quantified by enumerating NS5A-expressing cells.
RESULTS
Anti-CD81 MAbs neutralize HCV infection after virus internalization.
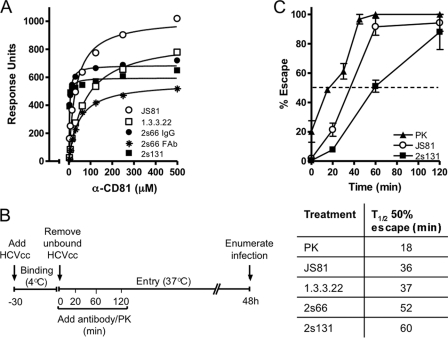
Several reports have studied the time for HCV to escape the neutralizing activity of anti-receptor antibodies to investigate the temporal involvement of cellular molecules in the virus entry process (13, 21, 23, 37, 75). The majority of these reports have used the commercially available anti-CD81 MAbs JS81 and 1.3.3.22 that bind conformation-dependent epitopes in the large extracellular loop (LEL) (29). We recently generated two MAbs 2s66 and 2s131 that recognize a linear epitope in CD81 (amino acids 173 to 192) and demonstrate a significantly higher affinity for CD81 LEL than JS81 or 1.3.3.22 (2s66, Kd = 3.33 μM; 2s131, Kd = 2.09 μM; JS81, Kd = 31.69 μM; and 1.3.3.22, Kd = 76.2 μM) (Fig. 1A). To ascertain whether the affinity or epitope specificity of CD81 MAbs plays a role in their mode of neutralization, we compared the time for HCV to escape the neutralizing activity of these MAbs. HCVcc J6/JFH was bound to Huh-7.5 cells on ice for 30 min, unbound virus removed by washing and entry initiated by transferring the cells to 37°C. Anti-CD81 MAbs were added at a concentration that inhibited >95% of virus infectivity at the indicated times, and the half-time (t1/2) for HCV to escape their neutralizing activity was assessed (Fig. 1B). HCV J6/JFH escaped the neutralizing activity of 2s66 and 2s131 at significantly later times (52 and 60 min, respectively) than MAbs JS81 and 1.3.3.22 at 36 and 37 min, respectively (Fig. 1C). To independently assess HCV internalization, we measured the time for HCV to escape the proteolytic activity of the serine protease, PK. J6/JFH acquired resistance to PK after only 18 min (t1/2) (Fig. 1C), a finding consistent with our earlier report (59). The observation that anti-CD81 MAbs could neutralize virus infection up to 60 min after internalization is initiated and long after PK has ceased to have an effect suggests intracellular sites for anti-CD81 MAb neutralization and a role for endosomal CD81 in virus infection.
Fig 1.
Anti-CD81 MAbs neutralize HCV postinternalization. (A) Anti-CD81 binding affinity for MBP-CD81 was assessed by surface plasmon resonance. The data are representative of two independent experiments. (B) Schematic illustration of experimental design. (C) Time course of J6/JFH escape from proteinase K (PK), JS81, and 2s131. The time for J6/JFH to escape 50% of the neutralizing effects of PK and anti-CD81 MAbs (t1/2) was calculated relative to the maximum inhibition observed at 120 min and is presented in the accompanying table. The data presented are from a single experiment and are representative of two independent experiments.
CD81 endocytosis.
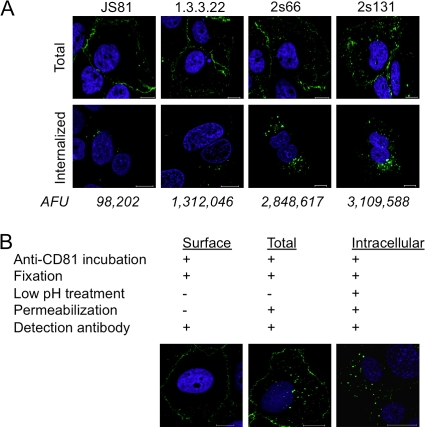
The majority of CD81 in Huh-7.5 hepatoma cells localizes to the plasma membrane with a limited pool of intracellular protein detected (Fig. 2A, upper panel). To study CD81 endocytosis, Huh-7.5 cells were incubated with a saturating concentration of anti-CD81 MAbs JS81, 1.3.3.22, 2s66, or 2s131 at 37°C for 60 min. Cell surface-bound MAb was removed by a low pH treatment, the cells were fixed and permeabilized, and intracellular CD81-MAb complexes were visualized by confocal microscopy. Significantly greater amounts of intracellular 2s66 and 2s131 were detected compared to JS81 and 1.3.3.22 (Fig. 2A, lower panel), which is consistent with our earlier neutralization data showing later escape times for MAbs 2s66 and 2s131 (Fig. 1C). To confirm we were studying intracellular CD81, we demonstrated that saponin permeabilization was required to visualize CD81 puncta and low pH treatment designed to remove cell surface-bound MAb had minimal effect on secondary antibody detection of intracellular CD81 (Fig. 2B).
Fig 2.
CD81 internalization. (A) The total (fixed and permeabilized [upper panel]) or internalized (live cell staining [lower panel]) CD81 in Huh-7.5 cells was evaluated with the anti-CD81 MAbs JS81, 1.3.3.22, 2s66, or 2s131 at 2 μg/ml for 60 min. To observe the internalized CD81 after live cell staining, surface-bound MAb was removed by low-pH treatment, and the cells were fixed and permeabilized with EM-formaldehyde and saponin, respectively. The average intracellular fluorescence intensity (arbitrary fluorescence unit [AFU]) per cell was calculated from 10 cells. (B) Cell surface-expressed CD81 was visualized by staining live Huh-7.5 cells with anti-CD81 MAb 2s131 (2 μg/ml) for 1 h at 37°C and fixed, and bound antibody was detected with anti-mouse immunoglobulin Alexa 488. Total CD81 representing the cell surface and intracellular pools was detected by permeabilizing the cells with 0.05% saponin postfixation. Low-pH treatment of antibody-treated Huh-7.5 cells removed the cell surface-bound MAb and enabled the sole detection of intracellular antibody after the fixation and permeabilization of cells. Nuclei were stained with DAPI, and single 1-μm z-sections from the central region of the cell were obtained by laser scanning confocal microscopy.
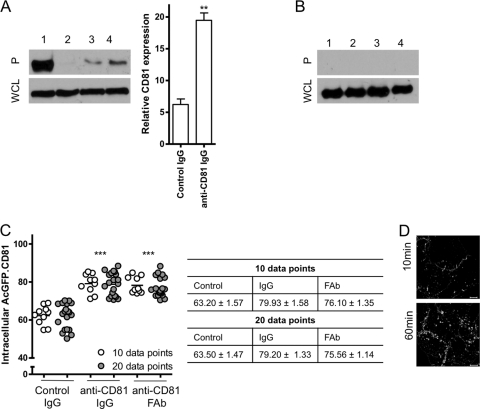
To confirm that antibody ligation promoted CD81 endocytosis, we surface labeled Huh-7.5 cells with Sulfo-NHS-SS-biotin on ice according to published protocols and maintained the cells on ice to prevent endocytosis or incubated at 37°C for 1 h in the presence of irrelevant mouse immunoglobulin or anti-CD81 2s66. Subsequent incubation of the cells with MesNA cleaves biotin from surface-labeled proteins, while internalized proteins are protected. Total cell surface-labeled proteins or biotinylated proteins that have entered the cell are detected by streptavidin capture, and CD81 was visualized by Western blotting. Incubation with anti-CD81 2s66 significantly stimulated CD81 endocytosis (19.5%), whereas control IgG treated cells showed a low level of constitutive endocytosis (6%) (Fig. 3A). Under identical conditions we failed to precipitate the intracellular protein tubulin, confirming specific labeling of cell surface proteins (Fig. 3B).
Fig 3.
Quantification of CD81 internalization. CD81 endocytosis was examined by surface biotinylation. Total surface-biotinylated protein (lane 1), biotin-labeled protein internalized at 4°C (lane 2), or biotin-labeled protein internalized at 37°C for 1 h in the presence of irrelevant mouse immunoglobulin (lane 3) or anti-CD81 2s66 (lane 4) (2 μg/ml) was captured by streptavidin pulldown. Precipitates (P, upper panel) and whole-cell lysates (WCL, lower panel) were subjected to SDS-PAGE, and the reactivity with anti-CD81 (A) or anti-tubulin (B) was assessed by Western blotting. The data are representative of two separate experiments from which the intracellular CD81 relative to total surface biotinylated CD81, determined by densitometric analysis, is combined. The data were compared using a Student t test (**, P < 0.01). (C) Huh-7.5 cells expressing AcGFP.CD81 were incubated with control mouse immunoglobulin or equimolar amounts of 2s66 or Fab for 1 h at 37°C. After fixation, 10 or 20 cells were imaged, and the intracellular CD81 fluorescence was quantified as a percentage of the total fluorescence ± the SEM. The data were compared by using a nonparametric ANOVA (Dunn's test, Kruskal-Wallis, ANOVA [***, P < 0.001]) and revealed no significant differences between 10 and 20 data points. (D) Huh-7.5 cells were incubated with anti-CD81 MAb 2s131 (2 μg/ml) for 10 or 60 min at 37°C and then fixed and permeabilized with saponin (0.05%); surface and intracellular bound MAb was visualized with anti-mouse immunoglobulin-Alexa 488. Similar observations were made with anti-CD81 MAb 2s66.
We noted that antibody/biotin removal from the cell surface was on occasion incomplete. Therefore, to further study the mechanism of CD81 internalization, we developed an imaging-based approach to quantify plasma membrane and intracellular CD81, allowing the use of fluorescently tagged transdominant-negative proteins/antibodies. Initial experiments demonstrated that a 1-h treatment with MAb 2s66 increased intracellular localization of AcGFP.CD81 by 16.8% (Fig. 3C). Comparable results were obtained from analyzing 10 or 20 cells, and the data showed a Gaussian distribution, demonstrating an adequate sampling of the cell population (Fig. 3C).
The increased detection of intracellular CD81 observed with MAbs 2s66 and 2s131 compared to JS81 and 1.3.3.22 (Fig. 2A) may reflect differences in binding affinity or epitope-specific effects of MAb ligation on CD81 trafficking. The observation that 2s66 FAb bound CD81 LEL with a reduced Kd of 44 μM compared to 3.33 μM for the complete IgG molecule (Fig. 1A) allowed us to study the role of ligand affinity on CD81 endocytosis. Both IgG and FAb antibody preparations at equimolar concentrations induced comparable levels of AcGFP.CD81 internalization (16.8 and 15.8%, respectively) (Fig. 3C), demonstrating that bivalent antibody induced cross-linking is not required to promote CD81 internalization, and ligand-induced CD81 internalization may not solely be driven MAb binding affinity. Brazzoli et al. reported that antibody ligation of CD81 promoted protein movement to cell-cell contacts in Huh-7 cells, suggesting a mechanism for CD81 tethered virus to interact with tight-junction-associated claudin-1 and occludin proteins (11). In contrast, we found CD81 to preferentially localize at cell-cell contacts in naive untreated cells (Fig. 2A) and failed to observe any significant effect of MAb engagement on CD81 localization at cell-cell contacts (Fig. 3D).
Mechanism of CD81 internalization.
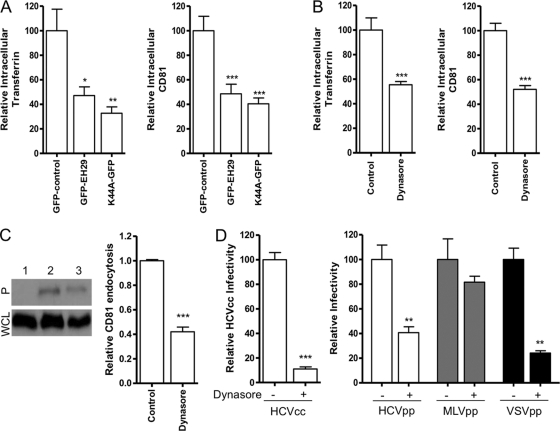
HCV has been reported to enter cells in a clathrin-dependent manner (8, 48). To assess the clathrin dependency of CD81 endocytosis, we transfected Huh-7.5 cells to express GFP or GFP-tagged transdominant mutants of Eps15 (EH29), a regulatory protein essential for the formation of clathrin-coated pits (4), or Dynamin2 (K44A), a protein necessary for the scission of vesicles from the plasma membrane (46) (reviewed in reference 57). Both mutants reduced Alexa 594-labeled transferrin uptake (Fig. 4A), confirming clathrin-dependent endocytosis. CD81 internalization was reduced by 51.5 and 59.5% in GFP-EH29- and K44A-GFP-expressing cells, respectively, compared to control GFP-transfected cells (Fig. 4A), demonstrating a clathrin- and dynamin-dependent internalization process. In agreement with these data, treating Huh-7.5 cells with the dynamin inhibitor Dynasore (34, 44, 50, 65) reduced both transferrin and CD81 uptake (Fig. 4B). Decreased CD81 endocytosis in the presence of Dynasore was confirmed by surface biotinylation (Fig. 4C). Furthermore, Dynasore reduced HCVcc, HCVpp, and VSV-Gpp infection (33) but had minimal effect(s) on MLVpp infection (38) (Fig. 4D). We conclude that CD81 internalizes via a clathrin- and dynamin-dependent process.
Fig 4.
CD81 internalization is clathrin and dynamin dependent. (A) Huh-7.5 cells expressing GFP-control, GFP-EH29, or K44A-GFP were incubated with Alexa 594-labeled transferrin (Tfn-594) or anti-CD81 2s131 (2 μg/ml) for 1 h at 37°C. Intracellular transferrin and CD81-Ab fluorescence in 10 selected GFP-EH29- and K44A-GFP-expressing cells was measured and is presented relative to the intracellular fluorescence in control GFP-expressing cells. (B) Serum-starved Huh-7.5 cells were treated with Dynasore (80 μM) or DMSO prior to incubation with Tfn-594 or 2s131 for 1 h at 37°C. After fixation, bound antibody was visualized using anti-mouse immunoglobulin-Alexa 488; the intracellular transferrin and CD81-Ab fluorescence in a sample population of 10 individual cells was determined and is presented relative to the intracellular fluorescence in control untreated cells. (C) Biotin-labeled proteins internalized after incubation of the cells at 4°C (lane 1) or 37°C for 1 h in the presence of 2 μg of anti-CD81 2s66/ml after pretreatment with DMSO control (lane 2) or 80 μM Dynasore (lane 3) were captured by streptavidin pulldown. Precipitates (P, upper panel) and whole-cell lysates (WCL, lower panel) were subjected to SDS-PAGE, and the reactivity with anti-CD81 was assessed by Western blotting. The data are representative of two separate experiments from which the relative changes, as determined by densitometric analysis, are combined. The data were compared using a Student t test (***, P < 0.001). (D) Huh-7.5 cells were serum starved and incubated with Dynasore prior to infection with HCVcc J6/JFH, HCVpp-H77, MLVpp, or VSV-Gpp. Infectivity is expressed relative to untreated control cells and represents the mean of three replicate infections. The data presented are from a single experiment and are representative of three independent experiments. Transferrin and CD81 internalization relative to control was compared using a nonparametric ANOVA (Dunn's test, Kruskal-Wallis, ANOVA [*, P < 0.05; **, P < 0.01; ***, P < 0.001)].
CD81 endocytosis is independent of the intracellular C-terminal domain.
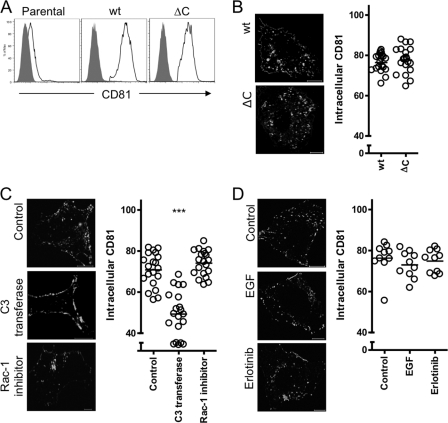
Cell surface receptor endocytosis is often regulated by motifs within the cytoplasmic tail that bind adaptor proteins involved in the formation of clathrin coated pits or caveolae (39). The tetraspanins CD63, CD151, and CD82 have been reported to internalize by diverse cellular routes (41, 56, 71) via an endocytic motif YXXØ in their C-terminal domains (10, 27). However, CD81 does not contain this endocytic motif. To determine whether the C-terminal cytoplasmic domain is required for CD81 endocytosis, a mutant protein lacking this region (CD81ΔC) was expressed in Huh-7 Lunet cells that express low levels of endogenous CD81 detectable by flow cytometry (Fig. 5A). We confirmed cell surface expression of exogenous wild-type and CD81ΔC proteins and observed comparable levels of internalized CD81 after 2s66 MAb treatment (Fig. 5A and B), suggesting a role for partner proteins to regulate CD81 endocytosis. The GTPases Rho and Rac-1 were reported to play a role in HCV entry (11); however, their mode of action was not defined. To study their involvement in CD81 endocytosis, Huh-7.5 cells were incubated with inhibitors of Rho (C3 transferase) or Rac-1. Inhibition of Rho significantly reduced antibody stimulated CD81 endocytosis compared to control cells (Fig. 5C), whereas Rac1 inhibition had no observable effect, supporting a role for Rho GTPase in CD81 endocytosis. Both inhibitors reduced HCVpp infection as previously published (11).
Fig 5.
C-terminal-independent and Rho GTPase-dependent CD81 endocytosis. (A) Parental and Huh-7 Lunet cells transduced to express wild-type (wt) CD81 or a mutant lacking the C-terminal tail (CD81ΔC) were stained for cell surface CD81 expression in the presence of 0.01% sodium azide with anti-CD81 2s131 (2 μg/ml) (black line) or isotype control (solid gray) and then analyzed by flow cytometry. (B) Huh-7 Lunet cells expressing wt or CD81ΔC were incubated with anti-CD81 MAb 2s131 (2 μg/ml) for 1 h at 37°C, fixed, and permeabilized, and the bound antibody was visualized. Representative images are shown, and intracellular antibody fluorescence is presented as a percentage of the total fluorescence ± the SEM in a sample population of 20 cells, where each circle represents an individual cell. (C) Huh-7.5 cells were incubated with C3 transferase (5 μg/ml, 16 h) or Rac-1 inhibitor (100 μM, 1 h), followed by anti-CD81 2s131 (2 μg/ml) for 1 h at 37°C. The cells were fixed, and antibody was visualized as described in the text. Intracellular antibody fluorescence of 20 cells is presented as a percentage of the total fluorescence ± the SEM, where each circle represents an individual cell. CD81 internalization in the presence of C3 transferase or Rac-1 inhibitor relative to control was compared using a nonparametric ANOVA (Dunn's test, Kruskal-Wallis, ANOVA [***, P < 0.001]). (D) Huh-7.5 cells were serum starved overnight and incubated with EGF or Erlotinib for 1 h at 37°C prior to incubation with anti-CD81 2s66 (2 μg/ml) for 1 h at 37°C. The cells were fixed and permeabilized, and antibodies were visualized as described in the text. The intracellular antibody fluorescence of 10 cells is presented as a percentage of the total fluorescence ± the SEM, where each circle represents an individual cell. The images shown are from a single experiment and are representative of two independent experiments.
EGF receptor (EGF-R) was recently reported to play a role in HCV entry by promoting particle internalization (43). Reports that EGF-Rs associate with the tetraspanins CD9, CD81, and CD82 (51) and that EGF promotes CD82 endocytosis (71) prompted us to investigate the effects of EGF-R activation on CD81 endocytosis. We confirmed the stimulatory and inhibitory effect of EGF and Erlotinib, respectively, on HCVpp entry in Huh-7.5 cells (data not shown). However, treating cells with EGF or receptor kinase inhibitor Erlotinib had no effect on CD81 endocytosis (Fig. 5D), demonstrating that EGF-R does not promote HCV entry by regulating CD81 endocytosis.
CD81 and claudin-1 coendocytosis.
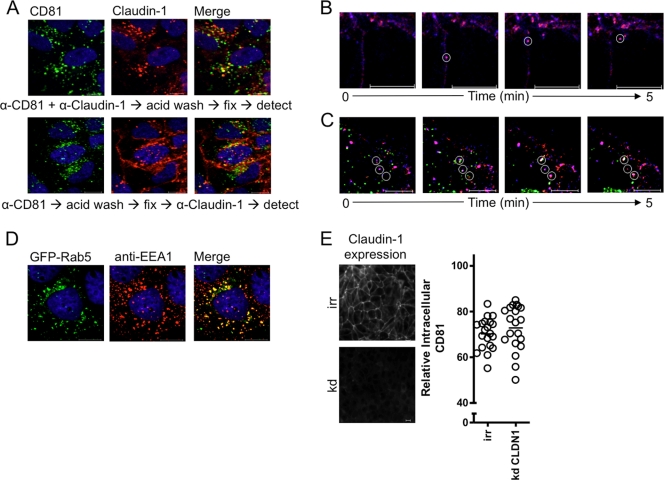
We (26) and others (73) previously reported that CD81 associates with claudin-1 and the receptor complex plays a role in HCV entry (25). Tight-junction proteins such as claudin-1 are known to readily endocytose via stimulus-specific endocytic pathways (67, 74). To ascertain whether CD81 and claudin-1 coendocytose, Huh-7.5 cells were incubated with anti-CD81 and anti-claudin-1 MAbs, surface-bound antibody was removed by a low pH treatment, and the cells were fixed and permeabilized prior to the addition of a secondary antibody. We observed that 46% of the intracellular CD81 colocalized with claudin-1 internalized from the plasma membrane (Fig. 6A). Similarly, incubating Huh-7.5 cells with anti-CD81 MAb alone, followed by low pH and permeabilization steps and anti-claudin-1 staining, demonstrated intracellular vesicular costaining for CD81 and claudin-1 (Fig. 6A), suggesting the coendocytosis of both receptor molecules. To confirm CD81/claudin-1 coendocytosis, Huh-7.5 cells were incubated with directly labeled fluorescent anti-CD81 2s66*647N and anti-claudin-1*565 MAbs at 37°C for 30 min, unbound MAbs were removed by washing, and the cells were imaged over a 5-min time period. We observed CD81 and claudin-1 trafficking at the plasma membrane prior to their coendocytosis (Fig. 6B) (see Movie S1 in the supplemental material). HCV-cell fusion is reported to occur within early endosomes (48, 53). To study the localization of internalized CD81 and claudin-1, Huh-7.5 cells were transfected with a GFP-tagged early endosomal marker Rab5 (GFP-Rab5), and recently internalized CD81 and claudin-1 proteins were visualized with fluorescence-labeled antibodies. Real-time imaging showed CD81- and claudin-1-containing vesicles trafficking to and colocalizing with Rab5 expressing early endosomes (Fig. 6C) (see Movie S2 in the supplemental material), demonstrating the recruitment of both receptors to early endosomes. Costaining GFP-Rab5 expressing vesicles with anti-EEA1 confirmed correct targeting of GFP-Rab5 to early endosomes (Fig. 6D). To ascertain whether claudin-1 expression is essential for CD81 endocytosis, we siRNA silenced claudin-1 expression and monitored the effects of MAb 2s66 on CD81 endocytosis. There was no difference in anti-CD81 stimulated endocytosis in claudin-1 silenced cells (Fig. 6E), suggesting CD81 endocytosis is not regulated by claudin-1 expression per se.
Fig 6.
CD81 and claudin-1 endocytosis. (A) Huh-7.5 cells were incubated with anti-CD81 2s131 (2 μg/ml) and anti-claudin-1 (2 μg/ml) (upper panel) or with anti-CD81 alone (lower panel) for 1 h, followed by a 5-min low-pH step to remove cell surface-bound MAbs prior to fixation. Intracellular anti-CD81 and anti-claudin-1 MAbs were detected with isotype-specific anti-mouse immunoglobulin-Alexa Fluor conjugates (upper panel). For anti-CD81-treated cells, after fixation and permeabilization, the intracellular claudin-1 was detected, and both primary antibodies were visualized with isotype-specific anti-mouse immunoglobulin-Alexa Fluor conjugates (lower panel). Images are derived from a single experiment representative of three independent experiments. Huh-7.5 cells (B) or Huh-7.5 cells expressing GFP-Rab5 (C) were incubated with anti-CD81 2s66 and anti-claudin-1 tagged with ATTO 647N and ATTO 565, respectively, for 30 min at 37°C. The cells were washed, and real-time confocal imaging of a 1-μm z-section through the center of the cell was performed over a period of 5 min. Static images taken from Movies S3 and S4 in the supplemental material are shown and were derived from a single experiment that is representative of three independent experiments. (D) Huh-7.5 cells transfected to express GFP-Rab5 were fixed and stained with anti-EEA1. (E) Claudin-1 was siRNA silenced in Huh-7.5 cells (see Materials and Methods), and protein expression was confirmed by staining control (irr) or silenced (kd) cells. CD81 endocytosis was determined in cells incubated with anti-CD81 2s66 (2 μg/ml) for 1 h at 37°C. Representative images are shown, and the intracellular antibody fluorescence is presented as a percentage of the total fluorescence ± the SEM in a sample population of 20 cells, where each circle represents an individual cell.
HCV promotes CD81 and claudin-1 endocytosis.
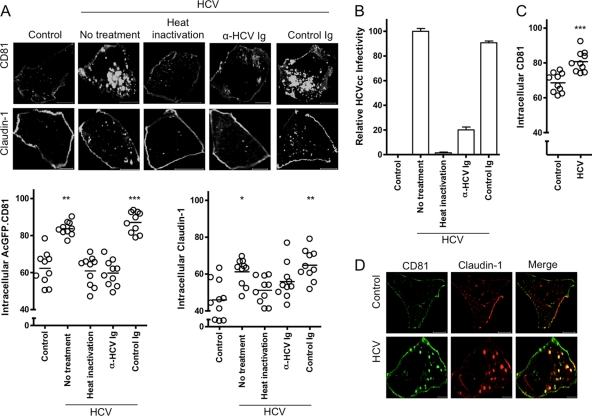
To investigate whether HCV can promote CD81 and claudin-1 endocytosis, Huh-7.5 cells were incubated with cell culture-grown HCV particles (HCVcc), and AcGFP.CD81 and claudin-1 internalization was compared to that in mock-infected (control) cells (Fig. 7A). HCVcc promoted a significant increase in intracellular CD81 (21.4%) and claudin-1 (15.2%), resulting in pixel saturation that may underestimate CD81 endocytosis. Heat inactivation or anti-HCV immunoglobulin inhibited HCVcc infectivity (Fig. 7B) and ablated the effect(s) of virus on CD81 and claudin-1 internalization, suggesting that infectious virus is required to initiate receptor endocytosis. We confirmed HCVcc stimulation of endogenous CD81 endocytosis by antibody detection (Fig. 7C). After HCVcc treatment we observed 89% of intracellular CD81 colocalizing with claudin-1 internalized from the plasma membrane (Fig. 7D).
Fig 7.
HCV promotes CD81 and claudin-1 endocytosis. (A) Huh-7.5 cells expressing AcGFP.CD81 were mock infected (control) or incubated with HCVcc J6/JFH, either alone or pretreated with anti-HCV Ig, control immunoglobulin, or heat inactivated at 37°C for 1 h. The cells were fixed and, where appropriate, claudin-1 detection was performed in saponin-permeabilized cells, and the cells were then imaged as described in the text. Intracellular protein/antibody fluorescence was quantified as a percentage of the total fluorescence ± the SEM in a sample population of 10 individual cells, where each circle represents an individual cell. The data were compared using a nonparametric ANOVA (Dunn's test, Kruskal-Wallis, ANOVA [*, P < 0.5; **, P < 0.01; ***, P < 0.001]). These data are representative of two further experiments which had comparable statistical significance. (B) Infectivity of HCVcc, alone or pretreated with anti-HCV Ig, control immunoglobulin, or heat inactivated, at 37°C for 1 h. The infectivity was measured after 48 h by enumerating NS5A-expressing cells relative to the control and represents the mean of three replicate infections. (C) Huh-7.5 cells were incubated with HCVcc or mock infected (control) for 1 h prior to fixation and antibody detection of CD81 in saponin-permeabilized cells. The cells were imaged as described in the text, and the intracellular antibody fluorescence was quantified as a percentage of the total fluorescence ± the SEM in a sample population of 10 cells, where each circle represents an individual cell. The data were compared using a Student t test (***, P < 0.001). The data presented are from a single experiment and are representative of two independent experiments. (D) Intracellular colocalization of AcGFP.CD81 and antibody detected claudin-1 in mock (control)- or HCVcc-treated Huh-7.5 cells at 37°C for 1 h.
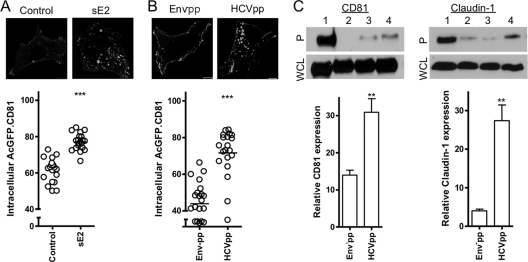
To confirm that this effect was mediated via the virus-encoded glycoproteins, we studied the effect of recombinant soluble HCV E2 glycoprotein (sE2) or lentiviral pseudoparticles expressing HCV E1E2 glycoproteins (HCVpp) or envelope-deficient particles (Env−pp) on AcGFP.CD81 internalization in Huh-7.5 cells. Both sE2 and HCVpp induced a significant increase, 15.6 and 27.8%, respectively, in intracellular AcGFP.CD81 compared to Env−pp-treated cells (Fig. 8A and B). Furthermore, we confirmed HCVpp stimulation of CD81 (30.9%) and claudin-1 (27.4%) endocytosis by surface biotinylation (Fig. 8C). In conclusion, these data illustrate HCV envelope glycoprotein stimulated CD81 and claudin-1 endocytosis.
Fig 8.
HCV envelope glycoproteins promote CD81 and claudin-1 endocytosis. Huh-7.5 cells expressing AcGFP.CD81 were untreated (control) or incubated with recombinant HCV soluble E2 glycoprotein (100 μg/ml) (A) or Env−pp (6 ng) or HCVpp particles (5.4 ng) (B). The cells were fixed and imaged as described in the text. The intracellular protein fluorescence was quantified as a percentage of the total fluorescence ± the SEM in a sample population of 10 cells, where each circle represents an individual cell. The data were compared using a Student t test (***, P < 0.001). These data are representative of two further experiments, which revealed comparable statistical significance. (C) Total surface biotinylated protein (lane 1) and biotin-labeled proteins internalized after incubation of cells at 4°C (lane 2) or at 37°C for 1 h in the presence of Env−pp (lane 3) or HCVpp (lane 4) were captured by streptavidin pulldown. Precipitates (P, upper panel) and whole-cell lysates (WCL, lower panel) were subjected to SDS-PAGE, and the reactivity with anti-CD81 or anti-claudin-1 was assessed by Western blotting. The data are representative of two separate experiments from which the levels of intracellular CD81/claudin-1 relative to the total surface biotinylated CD81/claudin-1, as determined by densitometric analysis, were combined. The data were compared using a Student t test (**, P < 0.01).
DISCUSSION
HCV entry is a multistep process, and there is a limited understanding of the role individual receptors play in virus internalization. We demonstrate that CD81 endocytosis occurs via a clathrin- and dynamin-dependent process in association with claudin-1. Importantly, HCV promotes CD81 and claudin-1 endocytosis, in a manner specifically inhibited by neutralizing anti-HCV immunoglobulin, supporting a direct role for these receptors in virus internalization. Our observation that anti-CD81 MAbs neutralize HCV infection after virus internalization is consistent with an intracellular site of antibody neutralization and a role for CD81 in trafficking virus to the endosomes for subsequent fusion events.
Brazzoli et al. reported that anti-CD81 MAb JS81 induced a lateral movement to cell-cell contact areas in Huh-7 hepatoma cells in a Rho-dependent manner (11). These authors interpreted cell-cell contacts to represent tight junctions and suggested that HCV engagement of CD81 would prime the movement of virus to tight-junction-associated entry factors claudin-1 and occludin in a manner similar to that of coxsackie B virus engagement of decay accelerating factor (5). We did not observe any evidence for antibody or virus promoting CD81 trafficking to cell-cell contacts; however, we confirmed that CD81 endocytosis was Rho dependent. Our data showed an enrichment of CD81 at hepatoma cell-cell contacts in untreated cells, as reported for other tetraspanin proteins (72). In agreement with our data, Coller et al. reported HCV endocytosis at diverse sites along the plasma membrane of Huh-7 hepatoma cells (15). Recent reports using high-resolution single-particle tracking demonstrated the dynamic movement of tetraspanins in live cells (36); using this technique we have demonstrated the mobile nature of CD81 in Huh-7 cells with no evidence of antibody-dependent directed movement to cell-cell contacts.
HCV entry is clathrin dependent (48), and we demonstrate here that CD81 can endocytose via a clathrin- and dynamin-dependent process. Dynamin is intrinsically linked to clathrin- and caveolin-dependent and -independent routes of endocytosis (28). The reported absence of Cav-1 in Huh-7 cells and the inhibitory effect of the Eps15 transdominant mutant (14, 68) suggest a role for dynamin in clathrin-dependent CD81 and HCV endocytosis. Unlike tetraspanins CD63, CD151, and CD82, which have been reported to internalize via an endocytic motif YXXØ in their C-terminal domains (10, 27), CD81 does not contain this motif, and a mutant protein lacking a C-terminal cytoplasmic tail can endocytose and support HCV entry (6). These data suggest a role for CD81-associating proteins in regulating its endocytosis. EGF-R was recently reported to promote HCV entry, and we examined a role for EGF-R in CD81 endocytosis (43, 71). We failed to observe any effect of EGF-R activation or inhibition on CD81 endocytosis, suggesting an alternative mechanism for EGF enhancement of HCV infectivity. Many viruses have been shown to utilize integrins during cell entry (for a review, see reference 61), integrins associate with tetraspanins, and CD81 has been reported to directly associate with α4β1 (60). CD81 may utilize its association with integrins or other known endocytic tetraspanin partner proteins CD63, CD151, or CD82 to internalize. Further work is required to understand the mechanism of CD81 endocytosis in the absence of an endocytic motif.
Silencing claudin-1 expression had no detectable effect on antibody-primed CD81 endocytosis, demonstrating that claudin-1 expression alone does not regulate CD81 endocytosis. Huh-7.5 cells express low levels of receptor active claudin-6 or 9 molecules that associate with CD81 (25) and may provide alternative partner proteins to regulate CD81 endocytosis. Attempts to visualize HCV in association with CD81 or claudin-1, either at the plasma membrane following binding or within intracellular vesicles using antibodies to the structural proteins, were unsuccessful, due in part to the relatively low titer of HCV particles available for these studies. However, it is interesting that both HCVpp and HCVcc preparations stimulated greater levels of CD81 and claudin-1 endocytosis than saturating levels of receptor specific MAbs, suggesting that the virus may be more effective at priming receptor endocytosis.
Antiviral antibodies can neutralize infection by a variety of mechanisms, including the blocking of virus binding to cellular receptors or the inhibition of postentry/prefusion events within the endosome (for reviews, see references 35 and 45). In contrast, less is known about the mechanism and site of action of neutralizing anti-receptor antibodies. Anti-CD81 MAbs have been reported to neutralize cell surface-bound HCV, suggesting a postattachment role for CD81 in the virus internalization process. Our current data suggest that anti-CD81 MAbs can neutralize virus at late times, after escape from PK, proposing intracellular sites for antibody neutralization. HCV has been reported to fuse in early endosomes and Meertens et al. noted a time delay between HCV internalization and endosome fusion that may reflect the recruitment of other cell components or further CD81 molecules to intracellular sites (48). Antibody ligation promotes CD81 internalization, and we propose that treating recently infected cells with anti-CD81 enables the subsequent fusion of internalized antibody and HCV-containing endocytic vesicles, allowing intracellular sites for antibody neutralization. Thus, the internalization of anti-receptor antibodies and their fusion with vesicular virus-receptor complexes is qualitatively different from previous reports on the intracellular targeting of infection by virus-specific antibodies to West Nile virus, tick-borne encephalitis virus, and influenza virus A (20, 62, 64).
Evans et al. reported a role for claudin-1 at a late step in the HCV entry process, using a similar experimental design to that shown in Fig. 1 with a FLAG epitope-tagged claudin-1 molecule and high-affinity anti-FLAG antibodies, demonstrating t1/2 times of 72 and 18 min for anti-FLAG and anti-CD81 antibodies, respectively (21). In contrast, a recent report by Krieger and coworkers demonstrated comparable times for HCV to escape anti-claudin-1; these studies most likely reflect the epitope specificity and affinity of the anti-receptor antibodies under evaluation, the different viral strains, and a hepatoma cell density that alters the HCV internalization rate(s) (37, 59). Our study shows variation in the time for HCV to escape different MAbs targeting CD81, which may be explained by differences in antibody affinity or epitope recognition, although in our study the t1/2 for JS81 was comparable to that reported by Krieger et al. (37). Importantly, we highlight here the complexities inherent in this experimental design and its inadequacies for studying the chronology of receptor usage in HCV entry.
Early endosomes are points of convergence for internalized molecules (22), where endocytosed material can acquire other cellular or signaling components by fusing with endocytic vesicles (31, 69). The localization of CD81/claudin-1 in Rab5-positive early endosomes and the reported 20-min delay between HCV internalization and fusion (48) suggests that HCV/CD81/claudin-1-containing vesicles may converge with other intracellular or endocytosed molecules prior to the fusion of virus and endosomal membranes. Our data support a model wherein HCV infects hepatocytes by priming CD81/claudin-1 endocytosis and exploits the dynamic intracellular trafficking of receptor proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Meredith and Z. Stamataki for critical reading of the manuscript, M. Goodall for the control mouse and anti-CD81 MAbs, R. Bill and J. Mohammed for the CD81 immunogen, C. M. Rice and T. Wakita for J6/JFH, the Huh-7.5 cells, and anti-NS5A 9E10, H. Drummer for MBP-CD81, T. Pietschmann for the Huh-7 Lunet cells, P. Bieniasz for gag-pol plasmid, and S. Young for BIACORE assistance.
This study was supported by the MRC, The Wellcome Trust, and the European Union HEPACIVAC (FP6) and HEPACUTE (FP7) programs.
Footnotes
Published ahead of print 8 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Asselah T, Marcellin P. 2011. New direct-acting antivirals' combination for the treatment of chronic hepatitis C. Liver Int. 31:68–77 [DOI] [PubMed] [Google Scholar]
- 2. Barth H, et al. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 80:10579–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benedicto I, et al. 2009. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J. Virol. 83:8012–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A. 1999. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J. cell science 112(Pt 9):1303–1311 [DOI] [PubMed] [Google Scholar]
- 5. Bergelson JM. 2009. Intercellular junctional proteins as receptors and barriers to virus infection and spread. Cell Host Microbe 5:517–521 [DOI] [PubMed] [Google Scholar]
- 6. Bertaux C, Dragic T. 2006. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J. Virol. 80:4940–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bitzegeio J, et al. 2010. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 6:e1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanchard E, et al. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonifacino JS, Dell'Angelica EC. 1999. Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 145:923–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brazzoli M, et al. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82:8316–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burckhardt CJ, Greber UF. 2009. Virus movements on the plasma membrane support infection and transmission between cells. PLoS Pathog. 5:e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Catanese MT, et al. 2009. Role of SR-BI in HCV entry: kinetics and molecular determinants. J. Virol. 84:34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cokakli M, et al. 2009. Differential expression of Caveolin-1 in hepatocellular carcinoma: correlation with differentiation state, motility and invasion. BMC Cancer 9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coller KE, et al. 2009. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 5:e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coyne CB, Kim KS, Bergelson JM. 2007. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 26:4016–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diederich S, Thiel L, Maisner A. 2008. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology 375:391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dorner M, et al. 2011. A genetically humanized mouse model for hepatitis C virus infection. Nature 474:208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drummer HE, Wilson KA, Poumbourios P. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143–11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edwards MJ, Dimmock NJ. 2001. Hemagglutinin 1-specific immunoglobulin G and Fab molecules mediate postattachment neutralization of influenza A virus by inhibition of an early fusion event. J. Virol. 75:10208–10218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans MJ, et al. 2007. Claudin-1 is a hepatitis C virus coreceptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 22. Gruenberg J. 2001. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell. Biol. 2:721–730 [DOI] [PubMed] [Google Scholar]
- 23. Haberstroh A, et al. 2008. Neutralizing host responses in hepatitis C virus infection target viral entry at postbinding steps and membrane fusion. Gastroenterology 135:1719–1728 [DOI] [PubMed] [Google Scholar]
- 24. Hansen GH, et al. 1998. The coronavirus transmissible gastroenteritis virus causes infection after receptor-mediated endocytosis and acid-dependent fusion with an intracellular compartment. J. Virol. 72:527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harris HJ, et al. 2010. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 285:21092–21102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris HJ, et al. 2008. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J. Virol. 82:5007–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haucke V, Krauss M. 2002. Tyrosine-based endocytic motifs stimulate oligomerization of AP-2 adaptor complexes. European J. Cell Biol. 81:647–653 [DOI] [PubMed] [Google Scholar]
- 28. Henley JR, Krueger EW, Oswald BJ, McNiven MA. 1998. Dynamin-mediated internalization of caveolae. J. Cell Biol. 141:85–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Higginbottom A, et al. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu M, et al. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Husebye H, et al. 2006. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 25:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jamshad M, et al. 2008. Structural characterization of recombinant human CD81 produced in Pichia pastoris. Protein Expr. Purif 57:206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. 2009. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83:440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kirchhausen T, Macia E, Pelish HE. 2008. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 438:77–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klasse PJ, Sattentau QJ. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 83:2091–2108 [DOI] [PubMed] [Google Scholar]
- 36. Krementsov DN, et al. 2010. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 11:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krieger SE, et al. 2010. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology 51:1144–1157 [DOI] [PubMed] [Google Scholar]
- 38. Lee S, Zhao Y, Anderson WF. 1999. Receptor-mediated Moloney murine leukemia virus entry can occur independently of the clathrin-coated-pit-mediated endocytic pathway. J. Virol. 73:5994–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le Roy C, Wrana JL. 2005. Signaling and endocytosis: a team effort for cell migration. Dev. Cell 9:167–168 [DOI] [PubMed] [Google Scholar]
- 40. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 41. Liu L, et al. 2007. Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J. Biol. Chem. 282:31631–31642 [DOI] [PubMed] [Google Scholar]
- 42. Liu S, et al. 2009. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 83:2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lupberger J, et al. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 17:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Macia E, et al. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10:839–850 [DOI] [PubMed] [Google Scholar]
- 45. Marasco WA, Sui J. 2007. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 25:1421–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marks B, et al. 2001. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410:231–235 [DOI] [PubMed] [Google Scholar]
- 47. Meertens L, et al. 2008. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for the hepatitis C virus. J. Virol. 82:3555–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meertens L, Bertaux C, Dragic T. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80:11571–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morikawa K, et al. 2007. The roles of CD81 and glycosaminoglycans in the adsorption and uptake of infectious HCV particles. J. Med. Virol. 79:714–723 [DOI] [PubMed] [Google Scholar]
- 50. Nankoe SR, Sever S. 2006. Dynasore puts a new spin on dynamin: a surprising dual role during vesicle formation. Trends Cell Biol. 16:607–609 [DOI] [PubMed] [Google Scholar]
- 51. Park S-Y, Yoon S-J, Freire-de-Lima L, Kim J-H, Hakomori S-I. 2009. Control of cell motility by interaction of gangliosides, tetraspanins, and epidermal growth factor receptor in A431 versus KB epidermoid tumor cells. Carbohydr. Res. 344:1479–1486 [DOI] [PubMed] [Google Scholar]
- 52. Pawlotsky JM. 2011. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology 53:1742–1751 [DOI] [PubMed] [Google Scholar]
- 53. Perrault M, et al. 2009. Unraveling the details of the entry of the hepatitis C virus into hepatocytic cells by electron microcopy imaging, p 74 16th International Symposium on Hepatitis C and Related Viruses, Nice, France [Google Scholar]
- 54. Pileri P, et al. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941 [DOI] [PubMed] [Google Scholar]
- 55. Ploss A, et al. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pols MS, Klumperman J. 2009. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315:1584–1592 [DOI] [PubMed] [Google Scholar]
- 57. Praefcke GJ, McMahon HT. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell. Biol. 5:133–147 [DOI] [PubMed] [Google Scholar]
- 58. Scarselli E, et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwarz AK, et al. 2009. Hepatoma cell density promotes claudin-1 and scavenger receptor BI expression and hepatitis C virus internalization. J. Virol. 83:12407–12414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Serru V, et al. 1999. Selective tetraspan-integrin complexes (CD81/α4β1, CD151/α3β1, CD151/α6β1) under conditions disrupting tetraspan interactions. Biochem. J. 340(Pt 1):103–111 [PMC free article] [PubMed] [Google Scholar]
- 61. Stewart PL, Nemerow GR. 2007. Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 15:500–507 [DOI] [PubMed] [Google Scholar]
- 62. Stiasny K, Brandler S, Kossl C, Heinz FX. 2007. Probing the flavivirus membrane fusion mechanism by using monoclonal antibodies. J. Virol. 81:11526–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stiles KM, Krummenacher C. 2010. Glycoprotein D actively induces rapid internalization of two nectin-1 isoforms during herpes simplex virus entry. Virology 399:109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thompson BS, et al. 2009. A therapeutic antibody against West Nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 5:e1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thompson HM, McNiven MA. 2006. Discovery of a new “dynasore.” Nat. Chem. Biol. 2:355–356 [DOI] [PubMed] [Google Scholar]
- 66. Thorley JA, McKeating JA, Rappoport JZ. 2010. Mechanisms of viral entry: sneaking in the front door. Protoplasma 244:15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Utech M, Mennigen R, Bruewer M. 2010. Endocytosis and recycling of tight junction proteins in inflammation. J. Biomed. Biotechnol. doi:484910.481155/482010/484987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vainio S, et al. 2002. Dynamic association of human insulin receptor with lipid rafts in cells lacking caveolae. EMBO Rep. 3:95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Visser Smit GD, et al. 2009. Cbl controls EGFR fate by regulating early endosome fusion. Sci. Signal. 2:ra86 doi:10.1126/scisignal.2000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang S, et al. 2008. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 136:8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu C, et al. 2009. CD82 endocytosis and cholesterol-dependent reorganization of tetraspanin webs and lipid rafts. FASEB J. 23:3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yanez-Mo M, Mittelbrunn M, Sanchez-Madrid F. 2001. Tetraspanins and intercellular interactions. Microcirculation 8:153–168 [DOI] [PubMed] [Google Scholar]
- 73. Yang W, et al. 2008. Correlation of the tight junction-like distribution of claudin-1 to the cellular tropism of hepatitis C virus. J. Biol. Chem. 283:8643–8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu D, Turner JR. 2008. Stimulus-induced reorganization of tight junction structure: the role of membrane traffic. Biochim. Biophys. Acta 1778:709–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zeisel MB, et al. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722–1731 [DOI] [PubMed] [Google Scholar]
- 76. Zheng A, et al. 2007. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J. Virol. 81:12465–12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.