Abstract
Lifelong self-renewal of the adult intestinal epithelium requires the activity of stem cells located in mucosal crypts. Lgr5 and Bmi1 are two molecular markers of crypt-cell populations that replenish all lineages over time and hence function as stem cells. Intestinal stem cells require Wnt signaling, but the understanding of their cellular niche is incomplete. Lgr5-expressing crypt base columnar cells (CBCs) reside deep in the crypt, mingled among mature Paneth cells that are well positioned for short-range signaling. Partial lineage ablation previously had implied that Paneth cells are nonessential constituents of the stem-cell niche, but recently their absence was reported to interfere with Lgr5+ CBCs, resurrecting an appealing idea. However, previous mouse models failed to remove Paneth cells completely or permanently; defining the intestinal stem-cell niche requires clarity with respect to the Paneth cell role. We find that Lgr5+ cells with stem-cell activity cluster in future crypts early in life, before Paneth cells develop. We also crossed conditional Atoh1−/− mice, which lack Paneth cells entirely, with Lgr5GFP mice to visualize Lgr5+ CBCs and to track their stem-cell function. In the sustained absence of Paneth cells, Lgr5+ CBCs occupied the full crypt base, proliferated briskly, and generated differentiated progeny over many months. Gene expression in fluorescence-sorted Lgr5+ CBCs reflected intact Wnt signaling despite the loss of Paneth cells. Thus, Paneth cells are dispensable for survival, proliferation, and stem-cell activity of CBCs, and direct contact with Lgr5-nonexpressing cells is not essential for CBC function.
Stem cells in selected adult tissues, such as the bone marrow, skin, and digestive tract, play a vital role in replenishing multiple cell types throughout life, and their unique and potent capacity for self-renewal is replicated in cancer (1). These stem cells occupy specialized niches and respond to the local environment (2). The functions of such niches range from delivering trophic signals that control cell proliferation and prevent stem-cell depletion to preventing unrestrained cell replication (3). Defining the cellular and molecular constituents of adult stem-cell niches therefore is an important challenge in biology and medicine.
Intestinal stem cells reside in mucosal crypts and generate four distinct cell types. Enterocytes, goblet cells, and enteroendocrine cells line deep crypts in the colon and villi that project into the small bowel lumen; Paneth cells lie at the crypt base in the small intestine, increasing in number from duodenum to ileum, but are absent from the colon (4). Two small intestine crypt-cell populations are able to generate all four cell types over extended periods: Lgr5-expressing crypt base columnar cells (CBCs), which lie deep in the crypt, interspersed among Paneth cells (5), and Bmi1-expressing cells that occupy several crypt tiers, most notably the +4 position (6). Although recent evidence suggests that each of these cell populations can engender the other (7–9), CBCs fulfill all criteria for adult tissue stem cells, similar to Lgr5-expressing cells in the stomach (10) and hair follicles (11). In the intestine, Lgr5 gene expression responds to Wnt signaling (5), which controls essential stem-cell properties (12, 13), but the source of Wnt ligands and the requisite cellular constituents of the stem-cell niche are unclear.
Mature Paneth cells secrete microbicidal peptides, enzymes, and growth factors (14), and their tissue location in small intestine crypts suggests a possibly key role in the stem-cell niche. Using transgenic CR2-tox175 mice, which express diphtheria toxin from the mouse Cryptdin2 promoter to destroy Paneth cells, investigators found that crypt proliferation and differentiation were preserved (15). However, Paneth cell loss in this model was incomplete; significant numbers persisted in older mice, and the unavailability of stem-cell markers hindered precise elucidation of stem functions in this context. Recent reexamination of the role of Paneth cells in the Lgr5+ CBC niche in Gfi1−/− (16), conditional Sox9−/− (17, 18), and CR2-tox175 mice led to the conclusion that Lgr5+ cells require the presence of adjacent Paneth cells (19). Importantly, Paneth cell loss in all these animal models was incomplete or temporary; also, the means used to remove Paneth cells may have affected CBCs directly. To overcome these limitations, we crossed Lgr5GFP-IRES-CreER knockin (5) and Villin-CreER transgenic (20) mice to conditional-null Atoh1flx/flx mice (21), a mutant strain that totally and permanently eliminates all intestinal secretory lineages, including Paneth cells. By visualizing Lgr5+ CBCs directly and using long-term lineage tracing to monitor stem cell progeny in the unambiguous and sustained absence of Paneth cells, we show that this differentiated lineage is dispensable for CBC survival, proliferation, stem cell activity, and response to Wnt signaling. In agreement with these findings, Lgr5+ cells cluster in future crypts and show stem-cell activity early in gut maturation, before Paneth cells develop.
Results
Lgr5+ Cells Localize in Intestinal Intervillus Regions Before Birth and Exhibit Stem-Cell Properties in the Absence of Paneth Cells.
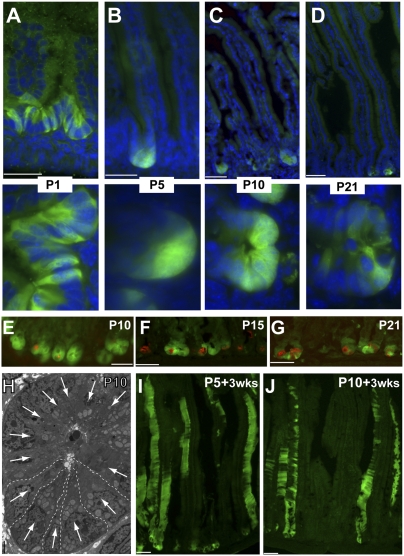
Because Lgr5 marks a stem-cell population in intestinal (5) and distal stomach (10) epithelia and in hair follicles (11), Lgr5+ cell properties have been characterized in detail in normal adult tissues (22), but their emergence has not been examined in detail during intestine development. We examined the distribution of Lgr5+ intestinal cells in fetal and newborn mice, using native GFP expression to monitor Lgr5+ cells in Lgr5GFP-IRES-CreER mice, which express GFP in Lgr5+ CBCs in a mosaic fashion (5). At embryonic day 15 (E15), when the pseudostratified intestinal epithelium has formed the first villi lined with columnar cells, no GFP expression was evident in the duodenum, the proximal segment of the intestine where Lgr5+ cells are most abundant in adults (Fig. S1 A and B). On postnatal day 1 (P1), Lgr5+ cells already were sharply localized to the intervillus zone of proliferating cells (Fig. 1A), suggesting that developing progenitors acquire Lgr5 expression and not that ubiquitous Lgr5 expression becomes restricted. Lgr5+ cells remained confined to the intervillus regions and emerging crypts at postnatal days 5 (P5), 10 (P10), 15 (P15), and 21 (P21) (Fig. 1 B–D and Fig. S1C), as they are in adults. Although GFP+ intervillus cells were interspersed with GFP− cells at P1 (Fig. S1D), this intermingling was rarely the case at P5 or P10, when the deep intervillus space of GFP+ crypts contained only GFP+ cells (Fig. 1 B and C); this distribution is distinct from the intermingled pattern of Lgr5+ and Paneth cells at P21 (Fig. 1D) and thereafter. Consistent with previous reports that recognizable Paneth cells appear gradually after birth (23), immature cryptdin-expressing Paneth cells were infrequent at P5 or P10 (Fig. 1E) and were observed to mingle among GFP+ cells only at P15 and P21 (Fig. 1 F and G); only 20% of P5 intervillus regions expressed the Paneth-cell marker Crs4c1. Lysozyme staining revealed more mature Paneth cells at P15 and fuller maturation by P21 (Fig. S1E). Cells at the intervillus base uniformly exhibited ultrastructural characteristics attributed to CBCs (5): a broad base, narrow apical cytoplasm, wedge-shaped basal nucleus, numerous supranuclear mitochondria, and absence of secretory granules (Fig. 1H and Fig. S2A). Only 10 of 100 intervillus regions showed one or more cells with immature Paneth-like granules (Fig. S2B).
Fig. 1.
Characterization of Lgr5+ CBCs during development. (A–D) Native GFP staining in Lgr5GFP-CreER mouse duodenum at P1 (A), P5 (B), P10 (C), and P21 (D). Lgr5+ cells localize to intervillus regions from the earliest stages. High-magnification images in the lower row emphasize the clustering of GFP+ cells without intervening GFP− cells at P5 and P10. (E–G) Immunostaining with cryptdin Crs4c1 antibody in Lgr5GFP-CreER mouse duodenum at P10 (E), P15 (F), and P21 (G) showing progressive appearance of Paneth cells. (H) Electron micrograph of an intervillus region at P10 showing clustering of CBCs with characteristic ultrastructural features: a broad base, narrow apical cytoplasm, wedge-shaped basal nucleus (white arrows), supranuclear mitochondria, and absence of secretory granules. Dashed lines delineate five adjacent CBCs. (I and J) YFP staining throughout intestinal villi in Lgr5GFP-CreER:Rosa26YFP mice 3 wk after a single tamoxifen injection administered at P5 (H) or P10 (I) indicates stem-cell activity of Lgr5+ cells at these early time points, giving rise to villus epithelial cells over several renewal cycles. (Scale bars: 50 μm in A–G, I, and J; 2 μm in H).
To test whether early Lgr5+ intervillus cells manifest stem-cell properties in the absence of Paneth cells, we crossed Lgr5GFP-IRES-CreER and Rosa26YFP reporter mice (24) and then administered single doses of tamoxifen (0.05 mg/g body weight) to Lgr5GFP-IRES-CreER;Rosa26YFP pups at P5 and 10. This treatment resulted in YFP expression in villus cells within 5 d (Fig. S1F) and uniform YFP expression in villi 3 wk later (Fig. 1 I and J). Taken together, these data demonstrate that Lgr5+ cells cluster in intervillus regions without intervening cells and behave like self-renewing progenitors early in gut maturation; thus, Paneth cells are dispensable in the development of functioning Lgr5+ stem cells.
Lgr5+ CBCs Occupy the Paneth Cell Zone in the Absence of Paneth Cells.
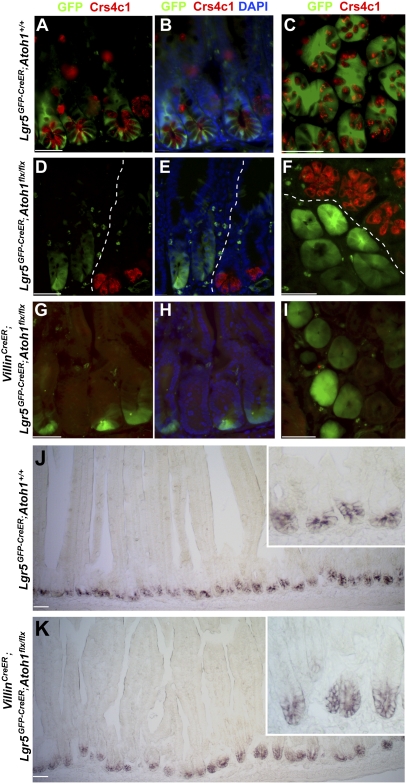
A recent study reported on the requirement for Paneth cell-mediated Wnt signaling in Lgr5+ cell function in epithelial organoid cultures and genetically engineered mice (19). In these mouse models, the decline in stem-cell numbers generally followed that of Paneth cell numbers, but no model eliminated Paneth cells completely (19). We therefore reexamined the requirement for Paneth cells in a mouse model with complete and unambiguous loss of Paneth cells in affected crypts. The intestine- and neural-restricted transcription factor Atoh1 is necessary to produce intestinal secretory cells, including Paneth cells (21, 25). We crossed Lgr5GFP-IRES-CreER mice to Atoh1flx/flx mice, which carry a conditional floxed null allele, and studied the compound mutants 3 wk after administration of tamoxifen. Because Paneth cells have a long life (26), and a few cells persisted after 3 wk (Fig. S3A), we also examined mice 3 mo after tamoxifen injection. Atoh1+/+ mice showed the expected alternating pattern of Lgr5+ (GFP+) CBCs and Crs4c1-expressing Paneth cells (Fig. 2 A–C). No Paneth cells were present after 3 mo in Atoh1-deleted crypts, in contrast to adjacent crypts that lacked Lgr5-GFP expression and had thereby avoided Cre-mediated Atoh1 recombination (Fig. 2 D–F). These data confirm efficient Atoh1 and Paneth cell loss in numerous intestinal crypts. Lgr5+ CBCs not only were intact but occupied the whole base in such Paneth cell-depleted crypts, as revealed by uniform GFP staining (Fig. 2 D–F).
Fig. 2.
Lgr5+ CBCs occupy the Paneth cell zone in the absence of Paneth cells. (A–I) Costaining of Lgr5-GFP (green) and cryptdin Crs41c (red) in control Lgr5GFP-CreER;Atoh1+/+ mice (A–C) shows a typically alternating arrangement of Lgr5+ and Paneth cells, whereas uniformly GFP-expressing Lgr5+ cells are present in Lgr5GFP-CreER;Atoh1flx/flx mice (D–F) 3 mo after tamoxifen injections and in VillinCreER;Lgr5GFP-CreER;Atoh1flx/flx mice (G–I) 3 wk after tamoxifen exposure. Images in A, B, D, E, G, and H show longitudinal sections; images in C, F, and I show cross-sectional views. The images in B, E, and H correspond to those in A, D, and G, respectively, with DAPI stain added to visualize all nuclei. Dotted lines in D–F demarcate Paneth cell-depleted and Paneth cell-replete areas as a result of clonally mosaic Cre expression in Lgr5GFP-CreER intestines. (J and K) In situ hybridization analysis of Olfm4 showing intact Olfm4+ stem cells occupying the full base of Paneth cell-depleted crypts in VillinCreER;Lgr5GFP-CreER;Atoh1flx/flx mice 1 d after five daily tamoxifen injections. (Scale bars: 50 μm; Insets show crypts at 10× higher magnification.)
To exclude an artifact that might follow from mosaic Cre expression in Lgr5Cre mice, we also crossed Lgr5GFP-IRES-CreER;Atoh1flx/flx mice to the VillinCreER strain (20); in the resulting progeny, VillinCreER drives Atoh1 deletion in all crypts, and Lgr5GFP provides a means to visualize Lgr5+ CBCs in a significant fraction of those crypts. Within 2 wk of tamoxifen injection, VillinCre-mediated Atoh1 deletion removed Paneth cells throughout the intestine, consistent with a requirement for Atoh1 in Paneth cell survival and representing total, sustained Paneth cell loss of unprecedented scope. Fewer than 1 in 500 crypts escaped total Paneth cell depletion; the crypts that avoided total depletion of Paneth cells contained residual long-lived cells from a previous renewal cycle before Cre-mediated Atoh1 deletion (Fig. S3B). In crypts that entirely lacked Paneth cell markers, we again observed uniform GFP staining, demonstrating that Lgr5+ CBCs were intact and lacked intermingled cells (Fig. 2 G–I). In situ hybridization analysis for the independent CBC marker Olfm4 (27) after Villin-Cre–mediated Atoh1 and Paneth cell loss revealed intact Olfm4+ stem cells throughout crypt bases (Fig. 2 J and K), indicating that these cells are bona fide CBCs and are not residual GFP+ descendants. Moreover, compared with control mice, GFP+ cells were modestly increased in number in the absence of Paneth cells (Fig. S3C). To see if CBCs may be affected more adversely by reduced Paneth cell numbers than by their total absence, we modulated each animal model, administering a single 0.1-mg dose of tamoxifen to VillinCreER;Lgr5GFP;Atoh1flx/flx mice or examining Lgr5GFP-IRES-CreER;Atoh1flx/flx mice 2 wk after tamoxifen treatment. These approaches allowed us to examine individual crypts containing small numbers of residual Paneth cells; neither approach to subtotal Paneth cell depletion decreased GFP+ CBCs (Fig. S3C).
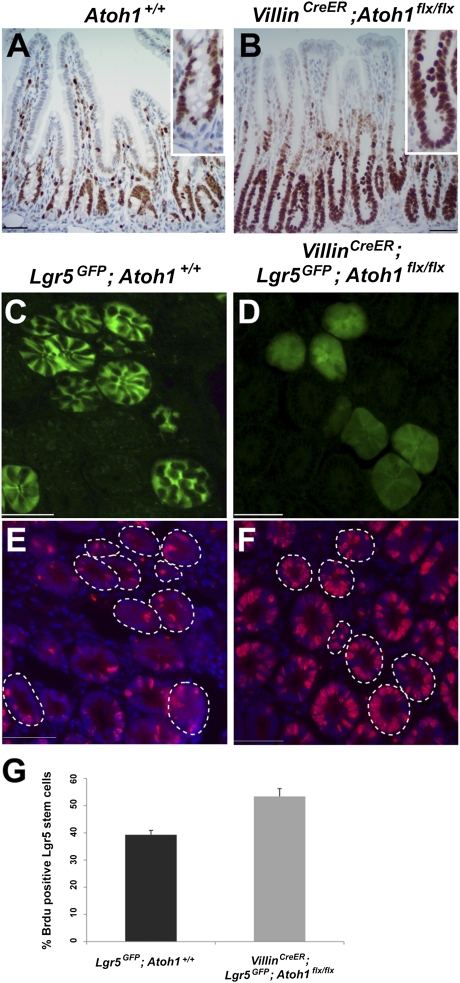
Proliferation of Lgr5+ CBCs Is Increased and Stem-Cell Activity Is Preserved in the Absence of Paneth Cells.
Two weeks after tamoxifen administration, the proliferative marker Ki67 revealed that all crypt cells in VillinCreER;Atoh1flx/flx mice were replicating actively, compared with 82% of cells in wild-type crypts (Fig. 3 A and B). To look specifically at proliferation of Lgr5+ CBCs, we introduced the S-phase marker BrdU into VillinCreER;Lgr5GFP-IRES-CreER;Atoh1flx/flx mice 3 wk after tamoxifen injection, confirming that Paneth cells were eliminated by this time (Fig. S2C). Sequential analysis of native GFP/Lgr5 expression and BrdU immunostaining revealed a 14% absolute increase in the fraction of BrdU+ cells among Lgr5+ CBCs compared with control Lgr5GFP-IRES-CreER intestines (Fig. 3 C–G). To test if this increased proliferation of Lgr5+ CBCs accelerates epithelial turnover, we traced BrdU after a single pulse. Progression of BrdU-stained cells along the crypt–villus axis at 1 h, 1 d, and 2 d was similar in mutant and wild-type mice (Fig. S4A). In agreement with enhanced cell replication, however, we did find 13% and 15% increases in crypt height in the duodenum and ileum, respectively, in Atoh1-depleted intestines (Fig. S4B).
Fig. 3.
Increased proliferation of Lgr5+ CBCs in the absence of Paneth cells. (A) Only occasional Ki67+ CBCs are mingled among Paneth cells in Atoh1+/+ tissue. (B) Ki67+ proliferating cells occupy the Paneth cell zone in VillinCreER;Atoh1flx/flx intestines 2 wk after tamoxifen injection. This difference is highlighted in the Insets. (C–F) Sequential imaging of GFP expression (C and D) and BrdU immunostains (E and F) reveal an increased S-phase fraction among Lgr5+ CBCs in VillinCreER;Lgr5GFP-CreER;Atoh1flx/flx intestines compared with Lgr5GFP-CreER controls. Dotted lines outline the crypts that express GFP in the CBC compartment in mosaic Lgr5GFP-CreER intestines. (Scale bars: 50 μm in A–F.) (G) Quantitation of three independent intestines from each group of mice; at least 250 Lgr5+ CBCs per sample were analyzed. Error bars represent SD.
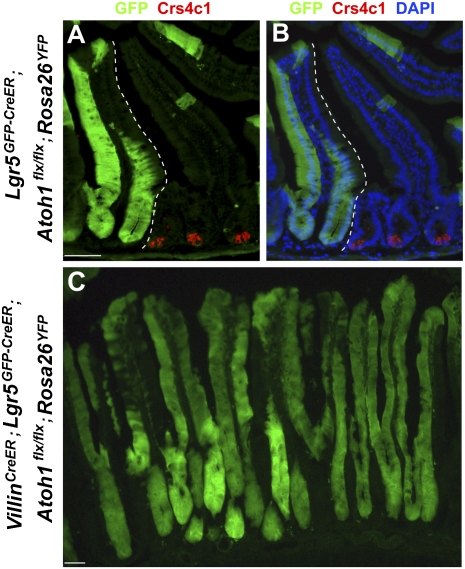
To test long-term stem-cell activity of these proliferating Lgr5+ CBCs, we examined Lgr5GFP-IRES-CreER;Atoh1flx/flx;Rosa26YFP mice 3 mo after tamoxifen administration. These mice revealed Lgr5+ CBC-derived mature cells in entire villi in the complete and persistent absence of Paneth cells (Fig. 4 A and B). Although villus cells derived from Lgr5+ CBCs lacked secretory markers, as expected because of the Atoh1−/− genetic background (25), these data demonstrated the cardinal stem-cell property of self-renewal over a period of at least 3 mo. To exclude again the possibility of artifacts resulting from mosaic Cre expression in Lgr5CreER mice, we asked the same question when Paneth cells were eliminated throughout the intestine in VillinCreER mice. Three months after pulse tamoxifen exposure, VillinCreER;Lgr5GFP-IRES-CreER;Atoh1flx/flx;Rosa26YFP mice also showed YFP staining throughout intestinal villi (Fig. 4C). The duration of these experiments rules out short-term ROSA26 recombination in short-lived mature cells as the basis for mature YFP+ villus cells. Because Villin-Cre marks cells throughout crypts, including Lgr5+ CBCs, this experiment alone does not distinguish between stem-cell activity of Lgr5+ and other putative stem-cell populations. It nevertheless reveals intact long-term self-renewal in the gut epithelium in the absence of Paneth cells and, together with data in Lgr5GFP-IRES-CreER;Atoh1flx/flx;Rosa26YFP mice (Fig. 4 A and B), attributes this Paneth cell-independent stem-cell activity to Lgr5+ CBCs.
Fig. 4.
Stem-cell activity of Lgr5+ CBCs in the absence of Paneth cells. (A and B) YFP staining of Lgr5GFP-CreER;Atoh1flx/flx;Rosa26YFP mice 3 mo after tamoxifen exposure shows stem-cell activity of Lgr5+ cells, which gave rise to all villus epithelial cells over the long term. Staining with Crs4c1 Ab (red) shows persistent Paneth cells in adjacent crypts that lacked Lgr5-Cre expression and absence of Paneth cell in crypts where YFP tracing occurred; the dotted lines demarcate these Paneth cell-depleted and undepleted zones. (C) YFP staining throughout villi in VillinCreER;Lgr5GFP-CreER;Atoh1flx/flx mice 3 mo after tamoxifen administration indicates intact stem-cell activity over 3 mo in the absence of Paneth cells. (Scale bars: 50 μm.)
Cell Replication-Associated Transcriptional Targets of Wnt Signaling Are Intact in the Absence of Paneth Cells.
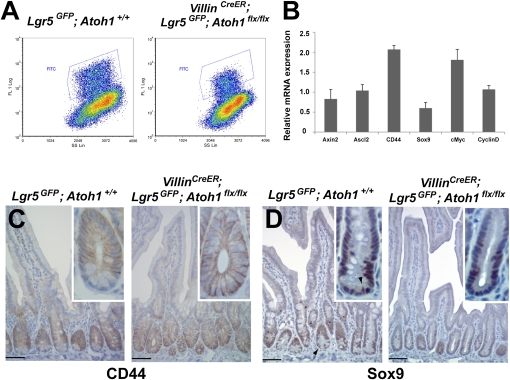
To decipher possible mechanisms for Paneth cell regulation of Lgr5+ CBCs, Sato et al. (19) compared gene expression in isolated CBCs and Paneth cells. Coupled with organoid cultures in vitro, their data implied that Paneth cells may elaborate Wnt ligands to activate Wnt signaling in receptor-expressing Lgr5+ CBCs. To examine Wnt signaling, we sorted GFP+ cells from Paneth cell-depleted VillinCreER;Lgr5GFP-IRES-CreER;Atoh1flx/flx and control VillinCreER;Lgr5GFP-IRES-CreER;Atoh1+/flx crypts 14 d after tamoxifen exposure. We used the purified Lgr5+ CBCs from crypts with and without Paneth cells (Fig. 5A) for quantitative RT-PCR (qRT-PCR) analysis of Wnt target genes. Consistent with enhanced proliferation of Lg5+ CBCs in Paneth cell-depleted crypts, the levels of Wnt target transcripts, including genes associated with cell proliferation such as CD44, Myc, and Ccnd1, were unaffected or even increased compared with wild-type cells (Fig. 5B). Immunohistochemistry further showed expansion of the CD44 expression domain into the crypt base (Fig. 5C), which is fully occupied by Lgr5+ CBCs, and verified the modestly reduced Sox9 expression evident by qRT-PCR (Fig. 5D). Thus, robust crypt cell proliferation in the absence of Paneth cells (Fig. 3) reflects a substantially preserved segment of the Wnt transcriptional response, implying that another source of Wnt signaling must remain active in vivo. To ask if Lgr5+ CBCs themselves might elaborate Paneth-expressed Wnt ligands, thus representing such a source, we tested Wnt3 and Wnt11 mRNA levels in sorted Lgr5+ cells by qRT-PCR. We found no Wnt11 expression, irrespective of Paneth cell exposure, and low Wnt3 levels were reduced further in Lgr5+ cells from Paneth cell-depleted crypts (Fig. S4C). Unless CBCs express another Wnt ligand, these data suggest in vivo Wnt sources other than Lgr5+ or Paneth cells.
Fig. 5.
Intact Wnt target genes in the absence of Paneth cells. (A) Flow cytometry profiles of Lgr5+ CBCs (blue boxes) among total crypt cells in Atoh1+/+ (Left) and VillinCreER; Atoh1flx/flx (Right) mice, each carrying the Lgr5GFP allele. (B) qRT-PCR analysis on Lgr5+ CBCs isolated from control Lgr5GFP-CreER and tamoxifen-treated VillinCreER;Lgr5GFP-CreER;Atoh1flx/flx intestines. Wnt target genes, tested because they are associated with cell proliferation, including CD44, Myc, and Ccnd1, were largely unaffected or even were enhanced. The experiment was done on two independent samples from each group; bars represent mRNA levels relative to the control, which is normalized to 1. Error bars represent SD. (C and D) Immunohistochemical corroboration that Wnt target genes, including proliferation-associated CD44 (C), are preserved in the absence of Paneth cells, although Sox9 (D) levels are subtly reduced. (Arrowhead in D points to a representative cell in the CBC position). Insets show single crypts at high magnification, highlighting the conclusions. (Scale bars: 50 μm.)
Discussion
We critically examined the proposed role of Paneth cells in the Lgr5+ CBC niche, using Atoh1 conditional-null mice to eliminate Paneth cells totally and reliably. We used Lgr5GFP-IRES-CreER mice to visualize CBCs through native GFP staining and, separately, Lgr5CreER and Villin-CreER mouse strains to delete Atoh1 in a mosaic and nonmosaic fashion, respectively. Lgr5Cre-mediated Atoh1 loss removed Paneth cells within 2 mo of tamoxifen exposure, and Villin-Cre-mediated Atoh1 deletion eliminated Paneth cells within 2 wk; both strains confirmed a role for Atoh1 in Paneth cell maintenance. In the ensuing absence of Paneth cells, Lgr5+ CBCs occupied the Paneth cell zone, proliferated robustly, and reconstituted the full villus epithelium. Thus, adult mouse Lgr5+ CBCs survive, replicate, and self-renew without Paneth cells, which therefore are not obligatory constituents of the intestinal stem-cell niche. Moreover, Atoh1-null intestines routinely showed GFP+ CBCs clustered without intervening GFP− cells, indicating that direct cell contact with non-CBCs is dispensable for their function. Last, early in life Lgr5+ cells cluster together in intervillus spaces and give rise to whole villi over several renewal cycles before the appearance of Paneth cells. These findings imply that Paneth cells are not needed to establish the intestinal stem-cell compartment.
These conclusions contrast with the idea that Paneth cells are integral to the intestinal stem-cell niche (19). Our approach differed most notably from previous studies in that Atoh1-null mice show complete and sustained Paneth cell loss, whereas Gfi1−/− and CR2-tox175 mice showed incomplete loss, and removal of Paneth cells in Sox9-mutant mice was transient (19). The Paneth cell zone is occupied by replicating cells in both CR2-tox176 (15) and Gfi1−/−(16) mice. Moreover, because Sox9 is expressed in both Paneth cells and CBCs (17, 18) (Fig. 5D), the requirement attributed to Paneth cells might reflect instead a parallel, cell-autonomous role for Sox9 in Lgr5+ CBCs. In two previous characterizations of efficient, Villin-Cre–driven Sox9 depletion from the whole intestinal epithelium, crypt size and crypt-cell proliferation were enhanced in the face of Paneth cell loss (17, 18); because Villin-Cre expression begins in embryos (20, 28), whereas Ah-Cre was induced in adult mice (29), a difference in timing may explain the different outcomes. Last, mice lacking the transcription factor XBP1 also lack Paneth cells but show intact crypt cell replication (30). Although Lgr5+ CBCs were not examined specifically in the latter strain, these findings collectively support our conclusion that Paneth cells are not essential for proliferation of CBCs or other crypt progenitors.
Mechanisms for microenvironmental regulation of Lgr5+ CBCs are just emerging. Because Wnt signaling is essential for crypt self-renewal (13), and Paneth and Lgr5+ cells respectively express Wnt ligands and receptors (19), we assessed Wnt signaling in Lgr5+ CBCs isolated from Paneth cell-depleted crypts. Axin2 mRNA levels, a reliable indicator (31, 32), were comparable to those in control Lgr5+ CBCs; other Wnt targets associated with cell proliferation also were preserved or enhanced. Together with a high level of cell replication, these proliferation-associated Wnt target transcripts indicate persistent Wnt signaling in CBCs isolated from the mutant crypts. This persistence might occur if Lgr5+ cells can bypass Wnt ligands and activate the pathway further downstream or if there is a source of Wnt other than Paneth cells, either normally or when Paneth cells are missing. Possible sources include CBCs themselves, although they do not express Paneth-specific Wnt 3 or Wnt 11, and underlying mesenchyme. Data showing that GFP+ cells occupy the entire crypt base when Paneth cells are absent argue against a substitute epithelial cell that contacts Lgr5+ cells directly. Recent reports reveal the potential for interconversion between the two intestinal stem-cell populations, Lgr5+ CBCs and Bmi1+ cells located in the +4 crypt tier (7–9). Extending these findings to interpret our data, Rosa-YFP–marked Lgr5+ cells could, in principle, give rise to +4 cells that subsequently drive Paneth cell-independent crypt and villus renewal. In this case, the +4 cells must continually replenish Lgr5+ CBCs that thrive in the absence of Paneth cell support. Future delineation of the relationship between CBCs and +4 cells will resolve these questions.
CBCs express Notch receptors, and Paneth cells express the corresponding ligands (19). Accordingly, Paneth cells are postulated also to signal to CBCs through Notch, a key regulator of crypt cell activities (33, 34), and Paneth cell loss might eliminate Notch signaling in Lgr5+ cells. All intestinal consequences of Notch-pathway inactivity, such as crypt-cell replication arrest and secretory-cell metaplasia, likely occur through induced Atoh1 expression (35–37), and therefore stem-cell deficits resulting from lack of Notch signaling might be masked in the absence of Atoh1. A requirement for Atoh1 solely to mitigate the effects of Notch inactivity in Lgr5+ CBCs, irrespective of Paneth cells, thus offers one explanation for intact stem-cell function in Atoh1−/− intestines. However, findings in mouse models of Paneth cell deficiency and preserved Atoh1 function make this explanation unlikely. Secretory-cell metaplasia, as expected in the absence of Notch signaling (33, 34), is not observed in CR2-tox176 mice, which carry 95% fewer Paneth cells than wild-type mice 4 wk after birth (15), VillinCre;Sox9flx/flx mice (17, 18), or Xbp1−/− (30) mice. Therefore, Notch signaling is not invariably compromised in the face of Paneth cell dearth. We cannot exclude the possibility that crucial CBC functions unrelated to secretory cell metaplasia may require a Paneth cell source of Notch signaling and that the absence of Atoh1 potently masks that need, hence preserving Lgr5+ cells even when the signal is missing.
Paneth cells, which are located immediately near Lgr5+ CBCs, often in a strictly alternating arrangement, provide an ideal niche in principle. Paneth cells might well influence Lgr5+ CBCs, for example by protecting them from harmful microbes or helping accelerate turnover in response to injury. Our results on Paneth cell-depleted crypts in Atoh1−/− adult and young wild-type mice, however, indicate that Paneth cells are dispensable for Lgr5+ cells to survive, replicate, or contribute toward long-term villus cell replacement, a reliable measure of intestinal stem-cell self-renewal.
Materials and Methods
Experimental Animals.
VillinCreER transgenic mice were generously provided by S. Robine (Institut Curie, Paris, France) (20). Lgr5GFP-CreER, Mathflx, and Rosa26YFP mice were purchased from Jackson Laboratories. Animals were housed under specific pathogen-free conditions and handled in accordance with protocols approved by the Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Drug Administration.
Tamoxifen (Invitrogen) was dissolved in 10% (vol/vol) ethanol and sunflower oil. Adult mice were injected i.p. with 1 mg for 5 consecutive d. Pups under the age of 21 d received a single i.p. dose of 0.05 mg/g body weight. Adult mice were killed 1 h, 1 d, or 2 d after i.p. administration of BrdU (50 mg/kg; BD Biosciences).
Immunohistochemistry.
Tissues were fixed overnight in 4% paraformaldehyde at 4 ºC and were washed three times in PBS. For cryo-embedding, fixed tissues were incubated overnight in 30% sucrose in PBS at 4 ºC and then were embedded in Optimal Cutting Temperature compound (OCT; Sakura); sections were cut at 10-μm thickness. For paraffin-embedding, fixed tissues were dehydrated, embedded in paraffin, and sectioned at 5-μm thickness. Antigens were retrieved in 10 mM Na citrate buffer (pH 6) followed by blocking of endogenous peroxidase activity in methanol and 3% H2O2. After blocking with 10% FBS, samples were incubated overnight at 4 ºC with one of the following antibodies: Crs4c1 [1:1,000; gift of Andre Ouellette (University of Southern California, Los Angeles, CA)], lysozyme (1:200; Invitrogen), Ki67 (clone MM1; 1:1,000; Vector Laboratories), BrdU (1:300; AbD Serotec), CD44 (1:500; eBioscience), or Sox9 (1:300; Millipore). Immunohistochemistry samples were washed and treated with biotin-conjugated anti-mouse or anti-rabbit IgG (1:300; Vector Laboratories). Color reactions were developed with Vectastain avidin-biotin complex (ABC kit; VectorLaboratories) and diaminobenzidine substrate (Sigma-Aldrich).
In Situ Hybridization.
After overnight fixation in 4% paraformaldehyde at 4 ºC, intestines were embedded in OCT compound (Sakura). Cut 10-μm sections were treated with10 μg/mL proteinase K (Roche) and hybridized overnight at 64 ºC with digoxigenin-labeled antisense. The riboprobe for Olfm4 was generated from IMAGE clone IRCKp5014N1115Q (Source BioScience imaGenes). Slides were washed in 2× SSC, incubated with alkaline phosphatase-conjugated digoxigenin Ab (1:2,000; Roche) and, to develop the stain, treated with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche).
Crypt Isolation, Lgr5+ CBC Sorting, and qRT-PCR Analysis.
Intestines of control Lgr5GFP-IRES-CreER and VillinCreER;Lgr5GFP-IRES-CreER;Atoh1flx/flx mice were dissected and washed in cold PBS. Villi were removed by scraping with glass microslides (Surgipath). To isolate crypt epithelium, samples were transferred to 5 mM EDTA in PBS (pH 8), followed by three 1-min shakings by hand, a 15-min incubation at 4 ºC, and passage through 70-μm filters (BD Falcon) to collect the flowthrough. After treatment of crypts for 45 min in 3.3% TrypLE (Invitrogen) to disaggregate cells, Lgr5+ CBCs were sorted by flow cytometry. RNA was isolated using TRIzol reagent (Invitrogen) and reverse transcribed using SuperScript enzyme (Invitrogen). cDNA was assessed using SYBR green master mix (Applied Biosystems). Threshold cycle (Ct) values for the test transcripts first were normalized with respect to Gapdh and are expressed as ratios of transcript levels in mutant cells to transcript levels in wild-type cells. Means and SDs for each group (n = 2) were calculated using Microsoft Excel.
Supplementary Material
Acknowledgments
We thank Andre Ouellette for generously providing Crs4c1 Ab; Sylvie Robine for kindly sharing VillinCreER mice; Susumu Ito for assisting with electron microscopy; and Li-Lun Ho, Kodanda Nalapareddy, and Michael Verzi for technical assistance and helpful discussions. This work was supported by National Institutes of Health Grants R01DK081113, R01DK082889, and RC2CA148222 and by an award from the Harvard Stem Cell Institute and was enabled by core facilities supported by the Dana Farber-Harvard Cancer Center Specialized Program in Research Excellence in Gastrointestinal Cancers Grant P50CA127003 and Harvard Digestive Diseases Center Grant P50DK34854.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113890109/-/DCSupplemental.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 4.Bjerknes M, Cheng H. The stem-cell zone of the small intestinal epithelium. I. Evidence from Paneth cells in the adult mouse. Am J Anat. 1981;160:51–63. doi: 10.1002/aja.1001600105. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 6.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda N, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan KS, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 12.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: From endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 14.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 15.Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J Biol Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 16.Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastide P, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori-Akiyama Y, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 19.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 21.Shroyer NF, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 22.Barker N, Clevers H. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology. 2010;138:1681–1696. doi: 10.1053/j.gastro.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Bry L, et al. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc Natl Acad Sci USA. 1994;91:10335–10339. doi: 10.1073/pnas.91.22.10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 26.Ireland H, Houghton C, Howard L, Winton DJ. Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev Dyn. 2005;233:1332–1336. doi: 10.1002/dvdy.20446. [DOI] [PubMed] [Google Scholar]
- 27.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 28.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 29.Ireland H, et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: Effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–1246. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 30.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jho EH, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lustig B, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fre S, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 34.van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 35.Kazanjian A, Noah T, Brown D, Burkart J, Shroyer NF. Atonal homolog 1 is required for growth and differentiation effects of notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology. 2010;139:918–928, 928, e1–e6. doi: 10.1053/j.gastro.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1:18. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim TH, Shivdasani RA. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J Biol Chem. 2011;286:11427–11433. doi: 10.1074/jbc.M110.188797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.