Background: Proteins with redox-sensitive thiols confer rapid response to changes in redox conditions and/or oxidant levels.
Results: Quantitative redox proteomics unveiled steady-state thiol oxidation states of ∼5% of yeast proteins and revealed physiologically relevant redox- and peroxide-sensitive proteins in yeast.
Conclusion: Redox-sensitive thiols appear structurally distinct from peroxide-sensitive thiols.
Significance: Pathways are fine-tuned by protein oxidation under both non-stress and oxidative stress conditions.
Keywords: Metabolism, Oxidative Stress, Redox Regulation, Thiol, Yeast
Abstract
To understand and eventually predict the effects of changing redox conditions and oxidant levels on the physiology of an organism, it is essential to gain knowledge about its redoxome: the proteins whose activities are controlled by the oxidation status of their cysteine thiols. Here, we applied the quantitative redox proteomic method OxICAT to Saccharomyces cerevisiae and determined the in vivo thiol oxidation status of almost 300 different yeast proteins distributed among various cellular compartments. We found that a substantial number of cytosolic and mitochondrial proteins are partially oxidized during exponential growth. Our results suggest that prevailing redox conditions constantly control central cellular pathways by fine-tuning oxidation status and hence activity of these proteins. Treatment with sublethal H2O2 concentrations caused a subset of 41 proteins to undergo substantial thiol modifications, thereby affecting a variety of different cellular pathways, many of which are directly or indirectly involved in increasing oxidative stress resistance. Classification of the identified protein thiols according to their steady-state oxidation levels and sensitivity to peroxide treatment revealed that redox sensitivity of protein thiols does not predict peroxide sensitivity. Our studies provide experimental evidence that the ability of protein thiols to react to changing peroxide levels is likely governed by both thermodynamic and kinetic parameters, making predicting thiol modifications challenging and de novo identification of peroxide sensitive protein thiols indispensable.
Introduction
Reactive oxygen species (ROS),4 such as hydrogen peroxide (H2O2), superoxide anions (·O2−), and hydroxyl radicals (·OH) are generated constantly during aerobic respiration as products of NADPH oxidases and various other enzymes (1). Organisms have developed a number of enzymatic and non-enzymatic strategies to preserve an overall reducing redox environment within the cell while at the same time allowing for the presence of low levels of ROS necessary for physiological signaling processes (2). Once ROS concentrations exceed the antioxidant capacity of the cell, however, oxidative damage to DNA, lipids, and proteins can occur, generating a cellular stress condition generally termed oxidative stress (3). Oxidative stress conditions appear to be involved in numerous pathological conditions, including cancer, neurodegenerative diseases, and diabetes (4, 5), illustrating the importance of maintaining cellular redox homeostasis.
Over the past few years, an increasing number of proteins have been identified that are not damaged by ROS but use reversible ROS-mediated thiol modifications to regulate their function (6). Like other posttranslational modifications, oxidative thiol modifications are fully reversible, and the extent to which cysteines are modified appears to depend on the amounts and types of ROS present, the reactivity of the cysteine thiols involved, and the kinetic and thermodynamic stability of the respective thiol modifications (1). Although many proteins with redox-sensitive cysteines have been identified in the past, it is still an unresolved question as to the extent to which proteins and pathways are affected by oxidative stress conditions in the cell. One confounding factor was that most proteomic approaches, although successful in detecting proteins with oxidized thiols, were unable to identify which cysteines are affected and to quantify the extent to which the cysteines were modified. Proteins were typically defined as oxidation-sensitive when a significant increase in thiol oxidation status was observed upon oxidative stress treatment or in the absence of specific oxidoreductases (7). However, depending on the oxidation status of a protein under non-stress conditions, such increase in oxidation could entail as much as 100% of the total protein population if 50% of the protein was thiol oxidized under steady-state conditions, or as little as 2%, if only 1% of the representative protein thiol was found to be oxidized before the stress treatment.
Here, we set out to globally quantify the in vivo thiol status of yeast proteins in the cytosol and subcellular compartments of exponentially growing yeast cells before and after oxidative stress conditions, using our mass spectrometry-based thiol trapping technique OxICAT (8). By using this approach, we identified numerous proteins and several pathways in yeast that are prone to oxidation, determined proteins that are involved in cellular antioxidant defense response, and applied bioinformatics tools to gain insights into the structural properties that define the oxidation status of protein thiols in vivo.
EXPERIMENTAL PROCEDURES
Strains and Cell Growth
Saccharomyces cerevisiae strain EG103 (DBY746; MATα, leu2–3 112 his3Δ1 trp1–289a ura3–52) was cultivated in synthetic complete medium (0.67% yeast nitrogen base with complete amino acid dropout solution, 2% glucose) at 30 °C. For purification of nuclei and vacuoles, EG103 was grown in yeast protein dextrose (1% yeast extract, 2% bactopeptone, 2% glucose). The medium was supplemented with 3% glycerol for purification of mitochondria.
Organelle Purification
The purification of mitochondria was based on that described by Meisinger et al. (9). Enrichment of vacuolar proteins and nuclear proteins was performed according to Haas (10) and Dove et al. (11), respectively.
H2O2 Stress Treatment
EG103 cells were cultivated until an A600 of 0.2 was reached. Then, 500 μm hydrogen peroxide (H2O2) was added for 15 min. 12 ml of cell culture either before or after addition of H2O2 was harvested on TCA (final concentration 10%) and incubated on ice for 30 min for subsequent OxICAT analysis. Four independent replicates of the experiment were performed.
Differential Thiol Trapping
The thiol trapping protocol established by Leichert et al. (8) was followed with few modifications to adjust for yeast cell lysis, as described in supplemental “Methods.”
ICAT Labeling of Organelle-specific Proteins
200 μg of the organelle proteins (nuclei, vacuoles, mitochondria) were precipitated with 10% TCA on ice for 30 min. TCA precipitates were centrifuged (13,000 × g, 30 min, 4 °C), washed and dissolved in 160 μl DAB buffer (6 m urea, 0.5% sodium dodecyl sulfate, 10 mm EDTA, 200 mm Tris-HCl, pH 8.5) supplemented with 1.25 mm of the thiol reductant tris(2-carboxyethyl)phosphine. The sample was split into equal volumes and added to the contents of one vial of light or heavy isotope-coded affinity-tagged (ICAT) reagent (Applied Biosystems), respectively. The samples were incubated under constant shaking at 1300 rpm for 2 h at 37 °C to label all cysteines. Excess ICAT reagent was removed by acetone precipitation as described (8). Tryptic digest of the ICAT-labeled peptides was conducted. After their purification using streptavidin columns, light- and heavy-labeled peptides were combined in a 1:1 ratio and subjected to biotin tag cleavage and LC-MS/MS. Labeling with both ICAT reagents was conducted to aid in our automated detection of ICAT pairs.
LC-MS/MS Analysis and Data Analysis
Liquid chromatography and mass spectrometry was performed at the Michigan Proteome Consortium. A 1100 Series nano-HPLC system (Agilent) and the Applied Biosystems 4800 Maldi TOF/TOF Analyzer was used (12). Data analysis was conducted as described, focusing on those peptides that are labeled with both light- and heavy-labeled ICAT reagents. All peroxide-sensitive peptides were confirmed using the visualization tools in msInspect (12).
Bioinformatic Analysis
pKa values and disulfide bond propensities for the identified cysteines were predicted using PROPKA (version 2.0) (13). Secondary structure predictions were performed by STRIDE (http://molbio.info.nih.gov/structbio/basic.html), a program that uses hydrogen bond energy and main chain dihedral angles to recognize secondary structural elements in proteins from their atomic coordinates. For both PROPKA and STRIDE, the available crystal structures served as input (supplemental Table S2). To determine the interaction network of thiols with neighboring residues, the AQUAPROT program (14) was revised to define inter-atomic interactions within one protein chain. Relative accessibility of cysteine residues within the available crystal structures was calculated according to Gerstein et al. (15) using a probe size of 1.4 Å and excluding water and ligand atoms. For statistical analysis, we used the non-parametric Kruskal-Wallis one-way analysis of variance test and chi-square test. A p value of < 0.05 was considered statistically significant (GraphPad). Detailed description of the bioinformatic analysis can be found in supplemental “Methods”.
RESULTS AND DISCUSSION
Using OxICAT to Analyze the Yeast Redoxome
A number of proteomic studies have resulted in the identification of bacterial and eukaryotic proteins, which use oxidative modification of highly sensitive cysteine residues to rapidly modulate their function, structure, and/or localization in response to oxidative stress conditions (16–20). As many of these methods are semi-quantitative at best (21), it is difficult to evaluate whether the proportion of the population of a protein affected by oxidative stress conditions is sufficiently large to make a physiological impact likely. It is also rather unclear which proteins are modified oxidatively in the absence of oxidative stress. We therefore decided to use the quantitative thiol trapping method OxICAT to first determine the oxidation status of protein thiols under non-stress conditions. We focused on total cellular proteins, including those in the cytoplasm as well as in major subcellular compartments, including mitochondria, nuclei, and vacuoles.
We cultivated yeast cells in minimal media until mid-logarithmic growth was reached. We then lysed the cells using acid trapping, a commonly used method to minimize non-specific air oxidation, and applied our OxICAT technique (22). OxICAT is based on the modification of in vivo-reduced cysteines with the isotopically light 12C version of the thiol-specific isotope-coded affinity tag ICAT (supplemental Fig. S1A). This modification is followed by the reduction of reversibly oxidized cysteine thiols and their labeling with the 9-Da heavier 13C-labeled version of ICAT. This sequential labeling generates chemically identical proteins that only differ in the specific mass of their ICAT-labeled cysteines (9 Da of additional mass per in vivo-oxidized cysteine). After tryptic digest and affinity purification, LC is used to separate the ICAT-labeled peptides. It is followed by MS and MS/MS to identify the peptides and to quantify the ratio of light- and heavy-labeled peptides as a measure of their in vivo oxidation status. By using this ratio approach, OxICAT is impervious to changes in protein expression and turnover.
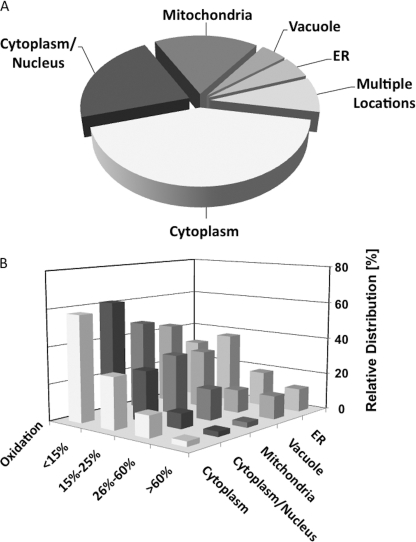
One major problem in analyzing the redox status of organelle-specific proteins is acid quenching, the first step in most thiol-trapping experiments. Although necessary to preserve the in vivo thiol oxidation status, it inevitably destroys all subcellular compartments and results in the dilution of organelle-specific proteins with other cellular proteins, thus limiting their identification. Therefore, time-consuming organelle purification is usually conducted, which commonly alters the redox status of the organelle (23). To increase the number of organelle-specific peptide identifications within our acid-quenched cell lysates, we decided to expand our OxICAT protocol. We first prepared protein extracts of purified organelles. We reduced all protein thiols and labeled them with equal amounts of light and heavy ICAT to aid in our automated detection of ICAT pairs (supplemental Fig. S1B). We then analyzed the ICAT-labeled organelle-specific peptides using LC/MS analysis (Fig. 1, left panel) and greatly enhanced the number of organelle specific peptides that we could identify. To assess their in vivo redox status, we then located the corresponding peptides in our acid-trapped total cell lysates utilizing their specific m/z values and their precise elution profile in the LC run as identifying markers (Fig. 1, right panel). By combining these approaches, we determined the in vivo oxidation status of >390 different thiol-containing peptides representing 290 individual proteins in four independent replicates (supplemental Table S1). Based on the annotated protein localization provided in the yeast database (Saccharomyces Genome Database), 43% of our identified proteins reside in the cytoplasm, 18% are purely mitochondrial, and an additional 8% have isoforms in both compartments (Fig. 2A). These results are in excellent agreement with general protein distribution studies in yeast, which demonstrated that ∼50% of all proteins are localized to the cytosol and ∼20% to the mitochondria (24). In addition, we identified a significant number of proteins that localize to both nucleus and cytoplasm (17%) and found about 10–15 proteins that are present exclusively in either nuclei, ER/Golgi, or vacuoles (Fig. 2A and supplemental Table S1).
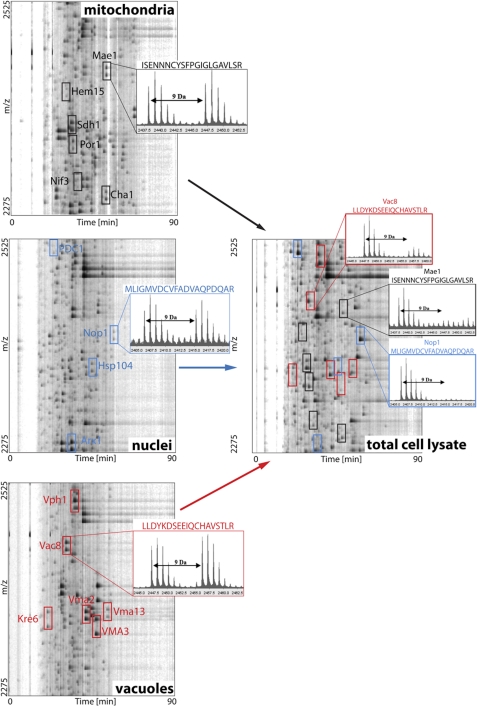
FIGURE 1.
Organelle-specific protein enrichment for enhanced peptide identification in acid-trapped cell lysates. Organelle-specific proteins were enriched by organelle isolation, fully reduced, and labeled with either light or heavy ICAT reagent. Then, light- and heavy-labeled peptides were mixed in a 1:1 ratio, and LC-MS/MS was conducted on each organelle sample. A graphical representation of LC/MS runs of ICAT-labeled mitochondrial, nuclear, and vacuolar peptides in the range of m/z = 2275–2525 Da is shown (left panels). In each panel, up to six different peptides that were identified by MS/MS analysis are indicated by colored boxes. For representative purposes, one peptide per organelle was selected and the respective mass spectrum is shown. The two mass peaks correspond to the light and heavy ICAT-labeled peptides mixed in a 1:1 ratio. The right panel shows the corresponding m/z range of total yeast cell lysate after differential thiol trapping using OxICAT. The organelle-specific peptides were identified within the total cell lysate by their m/z value and position in the HPLC run and are highlighted. The corresponding mass spectra indicate their in vivo redox status in exponentially growing yeast cells.
FIGURE 2.
The redox baseline, localization, and oxidation states of yeast proteins under non-stress conditions. Identified yeast proteins (392 total) were plotted according to their predominant cellular localization based on annotations in the S. cerevisiae Genome Database (A) and their in vivo oxidation status during exponential growth as determined by OxICAT (B; see supplemental Table S1). Most nuclear proteins are assigned both cytoplasmic and nuclear localizations and were thus grouped together. All other proteins that were found in more than one compartment are grouped under multiple locations.
Defining the Redox Baseline, Steady-state Thiol Redox Proteome of Yeast
Analysis of the thiol oxidation status of the identified thiol-containing peptides revealed that the majority of the identified cysteines are reduced in vivo. We found that about 60% of protein thiols are oxidized to <15%, and >80% of protein thiols have oxidation states between 0 and 25% (Fig. 2B and supplemental Table S1). This result confirmed previous studies, which suggested that yeast cells maintain a reducing environment during exponential growth (17). Of the remaining protein thiols, ∼5% were oxidized to >60% under steady-state growth conditions (Table 1). This group of highly oxidized proteins includes proteins with known disulfide bonds, such as the cytosolically localized Cu,Zn-Sod (Sod1), which requires formation of an intramolecular disulfide bond between Cys-58 and Cys-147 for activity (25). We identified both cysteine-containing Sod1 peptides (amino acids 45–69; amino acids 138–154) and showed that the respective cysteines are oxidized to 69 and 72% in vivo (supplemental Fig. S2A), suggesting that the majority of Sod1 molecules are oxidized in vivo. As reducing conditions prevail in the cytoplasm, the Sod1 molecules that have reduced cysteines might constitute newly synthesized Sod1 molecules as well as those in transit to the intermembrane space (26). Another highly oxidized protein that we identified is the mitochondrial Tim10, which chaperones hydrophobic proteins inserted at the mitochondrial inner membrane. Tim10 is known to contain two CX3C motifs, with Cys-44/Cys-61 and Cys-40/Cys-65 each forming disulfide bridges (27). We identified that 93% of the peptide containing both Cys-44 and Cys-61 were labeled with two heavy ICAT labels, showing that the Cys-44/Cys-61 cysteine pair is almost completely oxidized in vivo (supplemental Fig. S2B). Apart from these known disulfide-containing proteins, we also identified several other proteins with highly oxidized cysteines, most of which have not been previously shown to contain oxidative thiol modifications (Table 1). This group contains mitochondrial proteins such as the translational activator for a F1F0 ATP synthase subunit Atp22 and the tRNA methlytransferase Trm1, vacuolar proteins, such as the zinc-containing protein Zps1 and aminopeptidase Y (Ape3), as well as several cell wall-associated proteins. We found that proteins with oxidized cysteines are slightly overrepresented in vacuoles, ER, and the cell wall (Fig. 2B).
TABLE 1.
Significantly thiol-oxidized (>25%) yeast proteins
| Swiss-Prot | Gene (Cys) | Protein name | Oxidationa | Ref. |
|---|---|---|---|---|
| % | ||||
| Cell wall | ||||
| O13547 | CCW14 (42, 51, 53)b | Cell wall protein 14 | 93 ± 4 | |
| Q12512 | ZPS1 (123, 130)b | ZPS1 | 96 ± 4 | Ref. 20 |
| Mitochondria | ||||
| Q05933 | ARC18 (47) | Actin-related 2/3 complex subunit 3 | 56 ± 7 | |
| P50273 | ATP22 (672) | Translational activator ATP22 | 75 ± 6 | |
| P14540 | FBA1 (158) | Fructose bisphosphate aldolasec | 26 ± 2 | Refs. 18, 33, 34, 46, 47 |
| P38631 | FKS1 (1056) | 1,3-β-Glucan synthase component | 31 ± 5 | |
| P16622 | HEM15 (123) | Ferrochelatase | 27 ± 4 | |
| P49367 | LYS4 (340) | Homoaconitase | 35 ± 5 | |
| P36013 | MAE1 (220) | Malic enzyme | 29 ± 5 | |
| P36060 | MCR1 (263) | NADH-cytochrome b5 reductase 2 | 34 ± 5 | |
| Q12283 | MCT1 (198) | Malonyl CoA-acyl carrier protein transacylase | 27 ± 5 | |
| Q03028 | ODC1 (230) | Mitochondrial 2-oxodicarboxylate carrier 1 | 29 ± 6 | |
| P33302 | PDR5 (1050) | Pleiotropic ABC efflux transporter | 30 ± 6 | |
| P07256 | QCR1 (312) | Cytochrome b-c1 complex subunit 1 | 26 ± 3 | |
| P07257 | QCR2 (137) | Cytochrome b-c1 complex subunit 2 | 29 ± 6 | |
| Q99258 | RIB3 (133) | 3,4-Dihydroxy-2-butanone 4-phosphate synthasec | 45 ± 6 | |
| Q00711 | SDH1 (296) | Succinate dehydrogenase flavoprotein | 31 ± 3 | Ref. 48 |
| P32568 | SNQ2 (1098) | Protein SNQ2c | 30 ± 7 | |
| P87108 | TIM10 (44, 61)b | Mitochondrial import protein TIM10 | 93 ± 5 | Ref. 27 |
| P15565 | TRM1 (333) | tRNA methyltransferased | 65 ± 13 | |
| Cytosol | ||||
| P47079 | CCT8 (336) | T-complex protein 1 subunit θ | 47 ± 10 | Ref. 49 |
| P00549 | CDC19 (418) | Pyruvate kinase 1 | 46 ± 7 | Refs. 17, 18, 28 |
| P04802 | DPS1 (255) | Aspartyl-tRNA synthetased | 28 ± 4 | |
| P20081 | FPR1 (55) | FK506-binding protein 1e | 76 ± 6 | |
| P53849 | GIS2 (49, 52) (67, 70) (118, 121)b | Translational activator GIS2 | 66 ± 7, 52 ± 6; 70 ± 5 | Ref. 28 |
| P41277 | GPP1 (211) | dl-glycerol 3-phosphatase 1/2d | 27 ± 7 | Ref. 18 |
| P25373 | GRX1 (27, 30)b | Glutaredoxin 1 | 80 ± 10 | |
| P11986 | INO1 (19) | Inositol 1-phosphate synthase | 34 ± 4 | |
| Q12447 | PAA1 (51, 55)b | Polyamine N-acetyltransferase 1 | 26 ± 8 | |
| P40525 | RPL34B (44, 47)b | 60 S ribosomal protein L34-B | 43 ± 7 | |
| P51402 | RPL37B (34, 37)b | 60 S ribosomal protein L37-B | 38 ± 4 | Ref. 28 |
| P14796 | RPL40A (15), (20) | 60 S ribosomal protein L40 | 26 ± 3, 66 ± 8 | |
| P49631 | RPL43B (40, 43)b | 60 S ribosomal protein L43 | 46 ± 6 | |
| P39939 | RPS26B (74, 77)b | 40 S ribosomal protein S26-B | 46 ± 9 | |
| P41057 | RPS29A (24), (39) | 40 S ribosomal protein S29-A | 47 ± 9; 46 ± 9 | |
| P41058 | RPS29B (24), (39) | 40 S ribosomal protein S29-B | 41 ± 7; 55 ± 13 | |
| P05759 | RPS31 (45, 50)b | 40 S ribosomal protein S31 | 46 ± 4 | |
| P26783 | RPS5 (87) | 40 S ribosomal protein S5 | 39 ± 8 | |
| P07284 | SES1 (370, 373)b | Seryl-tRNA synthetase | 26 ± 4 | |
| P00445 | SOD1 (58), (147) | Cu,Zn-superoxide dismutasee | 69 ± 4; 72 ± 7 | Refs. 28, 50 |
| P38788 | SSZ1 (81) | Ribosome-associated complex subunit SSZ1 | 29 ± 8 | |
| P00359 | TDH2/3 (150, 154)b | Glyceraldehyde 3-phosphate dehydrogenase 2/3 | 26 ± 3 | Refs. 17–19, 34, 46, 47, 49, 51 |
| P04801 | THS1 (268) | Threonyl-tRNA synthetasee | 26 ± 4 | |
| P29509 | TRR1 (142, 145)b | Thioredoxin reductase 1 | 39 ± 6 | Ref. 49 |
| P22515 | UBA1, (600) | Ubiquitin-activating enzyme E1d | 27 ± 3 | |
| P25654 | YCR090C (124) | UPF0587 protein YCR090Cd | 34 ± 8 | |
| P25491 | YDJ1 (143, 146)b, (185, 188)b | Hsp40 co-chaperone | 64 ± 5; 46 ± 4 | |
| P53111 | YGL157W (240) | Putative oxidoreductase YGL157Wd | 31 ± 6 | |
| P52553 | YKE2 (82) | Prefoldin subunit 6 | 88 ± 10 | |
| Nucleus | ||||
| P28000 | RPC19 (89) | DNA-directed RNA polymerase I/III RPAC2 | 29 ± 6 | |
| ER/Golgi/plasma membrane | ||||
| P32476 | ERG1 (174) | Squalene mono-oxygenase | 27 ± 4 | |
| P32472 | FPR2 (36) | FK506-binding protein 2 | 79 ± 3 | |
| P27809 | KRE2 (124) | Glycolipid 2-α-mannosyltransferase | 33 ± 3 | |
| P32486 | KRE6 (479, 481)b | β-Glucan synthesis-associated KRE6 | 92 ± 3 | |
| P39926 | SSO2 (122) | Protein SSO2 | 39 ± 4 | |
| Vacuoles | ||||
| P37302 | APE3 (187) | Aminopeptidase Y | 76 ± 7 | Ref. 20 |
| P27614 | CPS1 (368) | Carboxypeptidase S | 33 ± 3 | |
| P09232 | PRB1 (361) | Cerevisin | 28 ± 5 | |
a Values denote the average oxidation of at least four independent replicates ± S.D.
b Both cysteines were present in the same peptides and oxidized to the same extent.
c Protein found in cytosol.
d Protein found in nucleus.
e Protein found in mitochondria.
In addition to these highly oxidized proteins, ∼15% of all identified peptides harbored cysteine residues with intermediate oxidation levels between 25 and 60% (Table 1). This group of partially oxidized proteins included thioredoxin reductase 1 at 39% ± 6% oxidized. Thioredoxin reductase is a central player of one of the two major cellular redox systems, which use direct thiol-disulfide exchange reactions to maintain protein thiols in a reduced form. We also found the small ribosomal proteins Rps26B, Rps29A/B, and Rps31 to be highly oxidized. Note that these three proteins represent all but one of the small ribosomal proteins that contain more than one cysteine residue. We also identified four large ribosomal proteins with significantly oxidized thiols. All four proteins either contain a CX2C motif (Rpl34B, Rpl37B, Rpl43B) or a CX3C motif (Rpl40A) (Table 1 and supplemental Table S2). Other proteins that we found to be significantly oxidized include proteins involved in glycolysis such as GAPDH (i.e. Tdh2/3) (26%), fructose bisphosphate aldolase (26%), and pyruvate kinase (46%) (Table 1). It is of note that several of these partially oxidized proteins have been previously identified to contain reactive cysteines and are substrates of thioredoxin reductase in vitro (Table 1) (28). These proteins are excellent candidates to harbor allosteric disulfide bonds, which control the function of the proteins depending on the oxidation status of their cysteines (29). These results suggest that many important cellular pathways are directly influenced by the redox environment of the cell.
Identification of Peroxide-sensitive Yeast Proteins
To investigate the effects of defined oxidative stress conditions on the thiol oxidation status of yeast proteins, we exposed yeast cells to H2O2 concentrations that transiently inhibit their growth (supplemental Fig. S3A) and performed OxICAT analysis 15 min after treatment. This analysis was expected to help us determine which proteins undergo stable yet reversible thiol modifications in response to peroxide treatment, ascertain the extent of their in vivo oxidation, and establish their subcellular distribution. We compared the oxidation status of our 390 previously identified cysteine-containing peptides before and after oxidative stress treatment. Proteins were considered to be peroxide-sensitive only if their cysteine residues were at least 1.5-fold more oxidized after H2O2 treatment than before treatment and if the oxidized species constituted 20% or more of the total protein (12). We found that 44 of our previously identified peptides, representing 41 different proteins, fit these criteria (Table 2). This result confirmed earlier observations, which showed that stable H2O2-mediated cysteine oxidation is only detected in a distinct subset of proteins (1, 17). This result does not exclude that other protein thiols in our identified peptides are not also rapidly oxidized by peroxide. However, if oxidized at all, these modifications appear to be rapidly re-reduced by cellular redox systems. Although such nonspecific and presumably very transient thiol oxidation might contribute to the general redox buffering capacity of cells (30), it will not likely affect specific cellular processes. In contrast, proteins with stable oxidative modifications at structurally or functionally important cysteine residues such as those identified in this study, will inevitably alter the pathways that they are involved in, particularly if a sufficiently large protein population is affected.
TABLE 2.
Identified yeast proteins with H2O2-mediated thiol modifications
| Swiss-Prot | Gene | Cys | Protein name | Locationa | Oxidationb |
Oxidation sensitivityc | |
|---|---|---|---|---|---|---|---|
| Non-stress | H2O2 | ||||||
| % | |||||||
| Protein translation | |||||||
| P11745 | RNA1d | 180 | Ran GTPase-activating protein 1 | C, N | 14 ± 3 | 47 ± 10 | |
| P07284 | SES1d | 370/373 | Seryl-tRNA synthetase | C | 26 ± 4 | 42 ± 6 | |
| P09436 | ILS1d | 119 | Isoleucyl-tRNA synthetase | C | 25 ± 5 | 38 ± 7 | |
| P15180 | KRS1d | 486 | Lysyl-tRNA synthetase | C | 13 ± 2 | 23 ± 4 | |
| Q06053 | DUS3 | 224 | tRNA-dihydrouridine synthase 3 | C, N | 13 ± 2 | 23 ± 2 | |
| P05754 | RPS8B | 179 | 40 S ribosomal protein S8 | C | 14 ± 2 | 23 ± 4 | Ref. 28 |
| P26781 | RPS11B | 128 | 40 S ribosomal protein S11 | C | 18 ± 2 | 27 ± 5 | |
| P05759 | RPS31d | 45/50 | 40 S ribosomal protein S31 | C | 46 ± 4 | 67 ± 4 | |
| Q12672 | RPL21B | 101 | 60 S ribosomal protein L21-B | C | 9 ± 3 | 21 ± 2 | |
| P51402 | RPL37B | 34/37 | 60 S ribosomal protein L37-B | C | 38 ± 4 | 54 ± 8 | |
| P14796 | RPL40A | 39 | 60 S ribosomal protein L40 | C | 18 ± 3 | 36 ± 6 | |
| P02405 | RPL42B | 74 | 60 S ribosomal protein L42 | C | 15 ± 3 | 39 ± 3 | |
| P02994 | TEF1 | 324 | Elongation factor 1-α | C | 11 ± 4 | 20 ± 4 | Refs. 17, 19 |
| P02994 | TEF1 | 409 | Elongation factor 1-α | C | 13 ± 3 | 26 ± 5 | |
| P02994 | TEF1 | 361/368 | Elongation factor 1-α | C | 17 ± 3 | 24 ± 2 | |
| P32324 | EFT2 | 136 | Elongation factor 2 | C | 11 ± 2 | 20 ± 3 | Ref. 28 |
| Amino acid metabolism | |||||||
| P28777 | ARO2 | 221 | Chorismate synthase | C | 13 ± 3 | 29 ± 5 | |
| P32449 | ARO4 | 76 | Phospho-2-dehydro-3-deoxyheptonate aldolase | C, N | 9 ± 2 | 26 ± 5 | Ref. 20 |
| P13663 | HOM2 | 259 | Aspartate semialdehyde dehydrogenase | C, N | 21 ± 3 | 31 ± 3 | Ref. 17, 18 |
| P39954 | SAH1d | 231 | Adenosylhomocysteinase | C | 12 ± 2 | 22 ± 5 | |
| P37292 | SHM1 | 103 | Serine hydroxymethyltransferase | M | 14 ± 2 | 20 ± 3 | |
| P00931 | TRP5 | 420 | Tryptophan synthase | C, N | 7 ± 3 | 27 ± 4 | Refs. 17, 18, 20 |
| Carbohydrate metabolism | |||||||
| P00330 | ADH1 | 277/278 | Alcohol dehydrogenase 1 | C | 17 ± 5 | 40 ± 5 | Refs. 19, 52 |
| Q04894 | ADH6 | 163 | NADP-dependent alcohol dehydrogenase | C | 24 ± 3 | 40 ± 6 | |
| P14540 | FBA1d | 112 | Fructose bisphosphate aldolase | C, M | 13 ± 2 | 23 ± 4 | Refs. 19, 52 |
| P09624 | LPD1 | 54 | Dihydrolipoyl dehydrogenase | M | 12 ± 2 | 27 ± 2 | |
| P06169 | PDC1 | 221/222 | Pyruvate decarboxylase isozyme 1 | C, N | 22 ± 4 | 33 ± 6 | Refs. 17, 19 |
| P00358 | TDH2/3 | 150/154 | Glyceraldehyde 3-phosphate dehydrogenase 2/3e | C | 26 ± 3 | 75 ± 5 | Refs. 17–19 |
| P32614 | FRD1 | 454 | Probable fumarate reductase | C, M | 6 ± 2 | 20 ± 3 | |
| P11986 | INO1 | 19 | Inositol 1-phosphate synthase | C | 34 ± 4 | 54 ± 9 | |
| Proteostasis and stress response | |||||||
| P38013 | AHP1 | 120 | Peroxiredoxin type-2 | C | 6 ± 2 | 19 ± 3 | Refs. 17, 28, 52 |
| P38013 | AHP1 | 62 | Peroxiredoxin type-2 | C | 14 ± 2 | 51 ± 2 | |
| P39077 | CCT3d | 518 | TCP-1 subunit γ | C | 14 ± 3 | 28 ± 5 | |
| P42943 | CCT7d | 454 | TCP-1 subunit η | C | 11 ± 3 | 28 ± 5 | |
| P25491 | YDJ1 | 185/188 | Mitochondrial import protein | C | 46 ± 4 | 69 ± 6 | |
| P34223 | SHP1 | 88 | Ubiquitin Regulatory X domain-containing protein 1 | C, N | 13 ± 3 | 20 ± 2 | |
| P14832 | CPR1f | 38 | Peptidyl-prolyl cis-trans isomerase | C | 12 ± 3 | 28 ± 3 | Refs. 16, 28 |
| Other functions | |||||||
| P41338 | ERG10d,f | 358 | Acetyl-CoA acetyltransferase | C | 12 ± 3 | 28 ± 3 | |
| P29704 | ERG9d | 341 | Squalene synthetase | ER | 9 ± 2 | 19 ± 3 | |
| P16550 | APA1 | 306 | AP4A phosphorylase | C, N | 13 ± 2 | 21 ± 4 | |
| P39926 | SSO2 | 122 | Protein SSO2 | ER, G | 39 ± 4 | 65 ± 13 | |
| P18562 | FUR1 | 215 | Uracil phosphoribosyltransferase | C | 11 ± 6 | 19 ± 4 | |
| P53091 | MCM6d | 815 | DNA replication licensing factor | C, N | 20 ± 5 | 45 ± 7 | |
| Q12305 | YOR285W | 98 | Putative thiosulfate sulfurtransferase | M | 21 ± 4 | 49 ± 5 | |
a N, nucleus; M, mitochondria; C, cytoplasm; G, Golgi.
b The average oxidation of at least four independent replicates ± S.D.
c Localization according to the Saccharomyces Genome Database or Huh et al. (53). Shown are references that report proteins that contain oxidation-sensitive cysteines.
d Shown is an essential gene.
e Shown is the peptide identical in both isozymes.
f Two different peptides were identified with the same mass.
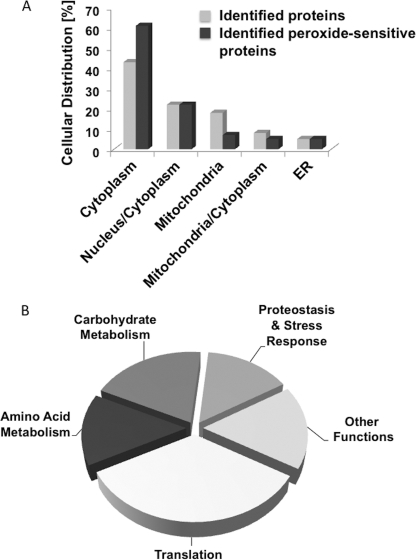
Analysis of the subcellular distribution of peroxide-sensitive proteins revealed that although they are found in most cellular compartments, they are over-represented in the cytoplasm and under-represented in mitochondria (Fig. 3A). Although almost 20% of our identified proteins are located in the mitochondria, only 7% of our identified peroxide-sensitive proteins are located in this organelle. These results imply that cytosolic proteins are either more vulnerable to peroxide mediated oxidation or that mitochondria have evolved more effective antioxidant systems to protect against and quickly reverse oxidative thiol modifications in vivo. Alternatively, exogenously applied peroxide might be unable to reach mitochondria at high enough concentrations to significantly affect the mitochondrial redoxome.
FIGURE 3.
Peroxide-sensitive yeast proteins: subcellular localization and affected pathways. A, relative cellular distribution of the 41 identified H2O2-sensitive yeast proteins as compared with the relative cellular distribution of all 392 identified yeast proteins. B, peroxide-sensitive yeast proteins grouped according to their cellular functions (see Table 2).
Among the 41 oxidation-sensitive proteins that we identified, only eight had been shown to be targets of oxidative thiol modifications in earlier studies (Table 2). In contrast to these previous studies, which identified the proteins as oxidation-sensitive without determining what proportion of the protein population is affected and which cysteine(s) are involved, we have now obtained a precise measurement of their degree of oxidation and the cysteine(s) modified. The remaining 33 proteins had not been identified as being H2O2-sensitive previously. In contrast, several previously identified oxidation-sensitive proteins (e.g. thioredoxin) escaped our detection presumably because our analysis is restricted to those peptides that we reproducibly found in all of our individual replicates (supplemental Table S1). Our work makes it clear that the H2O2 treatment affects key proteins involved in protein translation, amino acid biosynthesis, and carbohydrate metabolism and potentially regulates protein folding and stress responses (Fig. 3B, see below). We noted that peroxide targets the thiol groups of at least 14 proteins whose genes are conditionally essential for growth of yeast cells (Table 2). Altering the functional state of any one of these proteins by oxidative thiol modifications could be responsible for the observed growth inhibition of yeast cells upon treatment with H2O2.
Peroxide-sensitive Processes in Vivo
Numerous ribosomal proteins, elongation factors, and tRNA synthetases exhibited a significant increase in oxidative thiol modifications after short term exposure to H2O2 treatment (Table 2). The most heavily targeted proteins involved in protein translation that we identified are the small ribosomal subunit protein Rps31 (67 ± 4% oxidized) and the large ribosomal subunit protein Rpl37A/B (54 ± 8% oxidized). Both proteins have highly conserved CX2C motifs. We also detected oxidation-sensitive cysteines in the elongation factors Eft1 and Eft2, which previously have been characterized to be redox-sensitive (17, 20). This finding is in excellent agreement with recent studies in yeast cells, in which the authors reported the inhibition of protein translation during peroxide stress, caused in part by defects in protein elongation and/or termination (31). By stalling elongating ribosomes, cells not only prevent formation of erroneous proteins but increase their ability to rapidly resume protein synthesis upon return to non-stress conditions (31). As such, down-regulation of protein translation during oxidative stress might provide an adaptive mechanism to increase oxidative stress resistance of cells (32).
Another large group of oxidation-sensitive proteins that we identified with our OxICAT method turned out to be involved in carbohydrate metabolism (Table 2). Of the eight identified proteins, four had previously been shown to harbor cysteines that were qualitatively oxidation-sensitive, including GAPDH (i.e. Tdh2/3) and phosphoglycerate kinase 1. These prior studies suggested that redox regulation of GAPDH involves predominantly S-glutathionylation of its active site Cys-150 (33). However, based on our studies, the majority of oxidized GAPDH molecules appear to be modified at the active site Cys-150 and the conserved nearby Cys-154, whereas only a minor subset of molecules are modified at one cysteine only (supplemental Fig. 3B). This result implies that formation of a Cys-150–Cys-154 disulfide bond is the predominant outcome of peroxide-mediated GAPDH modification in vivo. We found that 75% of the total GAPDH pool is modified at its active site cysteine and hence inactivated after 15 min of peroxide treatment (Table 2), making GAPDH likely primarily responsible for the previously observed down-regulation of glycolysis during oxidative stress (34). This result agrees well with previous observations, which suggested that peroxide treatment re-directs significant amounts of glucose from glycolysis and respiration toward the pentose phosphate pathway. This re-routing of glucose will ensure the continuous production of the reducing equivalent NADPH, which is used by enzymes of the antioxidant defense (e.g. thioredoxin reductase, Trr1) as an electron donor (34). In contrast to previous studies (17), we did not confirm the redox sensitivity of either one of the two cysteines in Tpi1, and we found only one (i.e. Cys-112) of the five cysteines in Fba1 to be moderately oxidation-sensitive. These results suggest that both Tpi1 and Fba1 remain largely active during H2O2 stress. This conclusion makes physiological sense because increasing concentrations of the respective products of Tpi1 and Fba1 will further favor the production of NADPH by the pentose phosphate pathway (35). Other heavily targeted enzymes involved in carbohydrate metabolism include inositol 1-phosphate synthase Ino1 (54 ± 9% oxidized), Pdc1 (33 ± 6% oxidized), Lpd1 (27 ± 2% oxidized), and Adh1 (40 ± 5% oxidation). Partial inactivation of these enzymes likely affects acetyl-CoA and ethanol production, reduces respiration, and potentially minimizes additional ROS production.
We identified several H2O2-sensitive enzymes that affect aromatic amino acid biosynthesis (i.e. Aro4, Aro2, and Trp5) and methionine biosynthesis (Sah1), potentially (and appropriately) ceasing the production of those amino acids that are particularly vulnerable to irreversible oxidative modifications (Table 2). Finally, we found a number of peroxide-sensitive proteins known to be involved in stress defense and proteostasis. We obtained, for instance, a snapshot of the H2O2 scavenging action of alkyl hydroperoxide reductase 1 (supplemental Fig. S3C). Alkyl hydroperoxide reductase 1uses its catalytic Cys-62 for ROS detoxification and the resolving Cys-120 for cysteine regeneration (36). We detected both cysteine-containing peptides in our OxICAT analysis and determined that H2O2 exposure of yeast cells increases their oxidation status by ∼3-fold. In addition, we detected peroxide-sensitive cysteines in several chaperones, including two subunits of the TriC chaperonin complex as well as the DnaJ homologue Ydj1. Significantly 69 ± 5% of the total Ydj1 pool appears to form a disulfide bond between two of its zinc coordinating cysteines (i.e. Cys-185 and Cys-188) in response to peroxide stress, likely preventing Ydj1-mediated ER and mitochondrial protein import until reducing conditions are restored (37).
Structural Features Associated with Redox- or Peroxide-sensitive Thiols
Our quantitative OxICAT analysis revealed that proteins with peroxide-sensitive thiols show the same distribution regarding their steady state thiol oxidation status under non-stress conditions as do peroxide-insensitive protein thiols (supplemental Fig. S4). For instance, ∼60% of our peroxide-sensitive protein thiols have oxidation states of <15%, and only 10% of peroxide-sensitive cysteines have oxidation levels of >30% prior to the stress treatment. Hence, our results indicate that the oxidation status of yeast proteins under non-stress conditions does not serve as a predictor of peroxide sensitivity and that the partial oxidation of proteins under exponential growth conditions is unlikely to be due to elevated peroxide levels alone. To begin to understand whether distinct structural or sequence features of protein thiols might contribute to the observation that some cysteine thiols are partially oxidized under non-stress conditions and yet peroxide-insensitive, whereas others are reduced but form stable peroxide-mediated thiol modifications, we grouped our thiol-containing peptides according to their oxidation status under non-stress conditions and peroxide sensitivity; group I (“reduced”) thiols included all cysteine thiols that are reduced (<15% oxidation) under non-stress conditions and do not form stable peroxide-mediated thiol modifications; group II (“peroxide-sensitive”) thiols included all protein thiols that form stable peroxide-mediated thiol modifications; group III (“oxidized”) thiols included all protein thiols that are partially to fully oxidized (>30%) under non-stress conditions but non-responsive to peroxide treatment (for a complete list of proteins, see supplemental Table S2). Each of these three groups contained 50–70 proteins (i.e. 60–80 thiol-containing peptides), and ∼40% of these have structures available in the Protein Data Bank (supplemental Table S2). We reasoned that by using the primary sequences and tertiary structures as input, we should be able to compare (i) predicted pKa values, (ii) accessibility, (iii) secondary structural context, and (iv) interaction networks that the respective cysteine thiols have with adjacent residues and thereby perhaps gain insight into the structural features that might contribute to the oxidation sensitivity or resistance of individual protein thiols.
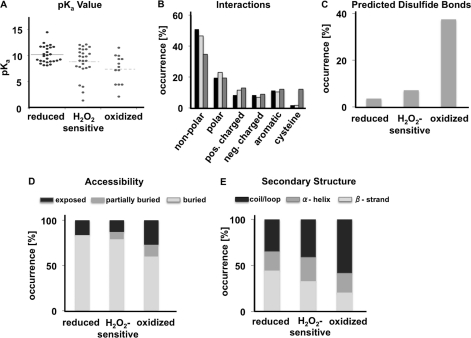
It is well known that the reactivity of cysteines increases with decreasing pKa values (1). We thus compared the pKa values of reduced (group I), peroxide-sensitive (group II), and oxidized (group III) cysteines using the solved protein structures as input and PROPKA (version 2.0) (13) for pKa prediction. As shown in Fig. 4A, we found that reactive group II and III cysteines have, on average, pKa values 1.5 to 2.5 pH units lower than reduced, non-reactive group I cysteines. The pKa value indicates the pH at which half of the population is ionized. As the ionized and thus reactive population decreases 10-fold for each pH unit drop below the pKa, cysteines with pKa values lower than the normal pKa of 8.2 ± 0.2 (38) are more likely to be deprotonated at physiological pH and thus are more reactive (1). Stabilizing interactions between the thiolate anion with neighboring positively charged or aromatic amino acids are thought to contribute to the low pKa values of reactive cysteines (39, 40). Indeed, when we analyzed the network of interactions that our identified protein thiols undergo within a 4 Å radius of their three-dimensional structure (14), we observed a slightly higher occurrence of positively charged amino acids surrounding reactive group II and III cysteines (Fig. 4B). Even more significant was the accumulation of cysteine residues located in the immediate neighborhood of reactive group III cysteines and their nearly complete absence in regions surrounding peroxide-sensitive or non-reactive cysteines. Very similar results were obtained when we compared the distribution of amino acids located within five amino acids upstream and downstream of the respective cysteines (supplemental Fig. S5); we found a highly significant (p < 0.005) accumulation of cysteine residues in the immediate vicinity of group III cysteines. Prediction analysis based on the distance between two vicinal thiols revealed that many group III thiols form disulfide bonds (Fig. 4C). Although this result might be expected for proteins in secretory pathways with known structural disulfide bonds, it is important to note that >85% of our group III protein thiols are localized to mitochondria or the cytosol and are only partially oxidized in vivo. Most surprisingly, however, was the finding that most of our peroxide-sensitive protein thiols lack vicinal cysteines (Fig. 4C). Yet, the presence of two or more cysteines in close proximity has often served as a defining criterion when predicting oxidation-sensitive cysteines in proteins (41). These results suggest that sulfenic acids, which typically form upon peroxide-mediated oxidation, will either undergo long range disulfide bonds that cannot be predicted based on the structure of the reduced protein and/or are kinetically stabilized by yet to be defined structural parameters (42).
FIGURE 4.
Comparison of structural properties between reduced (group I), peroxide-sensitive (group II), and oxidized (group III) cysteine thiols. A, pKa values of cysteine thiols classified as reduced, peroxide-sensitive, or oxidized as predicted by PROPKA (version 2.0). Distribution of pKa values is significantly different among the three sets according to Kruskal-Wallis one-way analysis of variance (p value = 0.01). The median of each group is shown by a solid line. B, distribution of interatomic interactions between the identified cysteine thiols and residues found within a 3 Å distance in reduced (black bars), peroxide sensitive (light gray bars), and oxidized (dark gray bars) groups. C, prediction of disulfide bond-forming propensity according to PROPKA 2.0, which uses a S–S distance criterion of 2.5 Å. D, relative accessibility of cysteine thiols. E, STRIDE analysis of secondary structure elements harboring the identified cysteines. Crystal structures of the respective proteins were used as input (see supplemental Table S2). pos., positively; neg. negatively.
To assess what other structural criteria might be involved in defining the redox and/or peroxide sensitivity of protein thiols, we analyzed the accessibility of the individual cysteine thiols and evaluated the secondary structure environment that they are located in. As shown in Fig. 4, D and E, many of our group III cysteines are located in structurally flexible loop regions of the proteins, Thus, disulfide bonds will likely form without significant structural strain and hence increase the stability of the proteins (43). Depending on the degree of stabilization, which is directly related to the redox potential of the disulfide bond (44), this finding explains why group III protein thiols can accumulate in a partially oxidized form even within the reducing environment of the cytosol. In contrast, reduced cysteine thiols appear to be predominantly buried within the protein structure with >60% located in α-helical or β-sheet structures. Peroxide-sensitive protein thiols did not cluster with either group but seemed to combine features of both reduced and oxidized cysteine thiols. These results suggest that higher accessibility of the cysteine residue does not necessarily translate into more peroxide-sensitive cysteines, particularly as we are focusing on oxidative thiol modifications that are stable over a 15-min time period. Surface accessible cysteines can be quickly oxidized but equally rapidly re-reduced, depending on their redox potential and reactivity with cellular oxidoreductases.
Conclusions
Quantitative redox proteomics not only opens the door to identifying cellular pathways that rapidly and persistently respond to the presence of distinct reactive oxygen species, but it provides the unique ability to address one of the most fundamental questions in redox biology: what makes certain cysteine thiols form stable oxidative thiol modifications, whereas others do not? As outlined above, many previously conducted redox proteomic studies disregarded the steady-state oxidation status of proteins. Cysteine thiols were thus often simply classified as (i) reduced cysteines that remain in the reduced state either on their own or through metal coordination, (ii) oxidized cysteines that form structural disulfide bonds and are hence overrepresented in secreted proteins, and (iii) cysteine thiols that are reduced under non-stress conditions and undergo reversible oxidative modifications in response to the presence of oxidants (41). Our quantitative analysis of the yeast redoxome revealed, however, that yeast cells contain a substantial number of proteins that show thiol oxidation states between 25 and 60% under steady-state conditions and that do not respond to the presence of peroxide with an increase in thiol oxidation. Most of these proteins do not appear to be secreted proteins but are exclusively localized to the reducing environment of the cytosol. This single localization excludes the possibility that isoforms of the respective protein in other, more oxidative stress-prone compartments of the cell might confound the overall oxidation status of the protein. These results strongly suggest that these thiols have redox potentials close to the physiological redox potential, which allows fine-tuning of physiological pathways by small alterations in the cellular redox environment. Yet, as many of these thiols are unresponsive to peroxide treatment, these results also imply that redox sensitivity does not imply peroxide sensitivity. These conclusions are in excellent agreement with previous studies, which demonstrated that oxidants, like peroxide, are very slow to react with thiol groups (20 m−1 s−1) and generally do not alter the cellular redox potential (1), implying that peroxide-mediated thiol modifications might not be induced by direct thiol-disulfide exchange reactions. Our results are consistent with an alternative redox-regulatory mechanism, in which specific kinetic features, rather than thermodynamic properties, characterize peroxide-sensitive protein thiols (1). One such example is OxyR, a peroxide-specific transcriptional regulator in bacteria, which is oxidized rapidly by very low concentrations of peroxide even within a highly reducing redox environment (45). It has been shown that very fast reaction rates between the active site thiol and peroxide of OxyR cause formation of a kinetically stable disulfide bond, which guarantees that OxyR remains oxidized and transcriptionally active for extended periods after peroxide treatment.
In summary, our results help explain why predicting peroxide sensitivity of cysteine thiols has remained a challenge in the past and illustrate the importance of quantitative redox proteomic techniques in the de novo identification of oxidation-sensitive proteins in cells and organisms.
Acknowledgments
LC-MS analyses were performed by the Michigan Proteome Consortium. We are grateful to Kyle Robertson and Greg Bode for database searches, and we thank Dr. James Bardwell for critically reading the manuscript.
This work was supported by National Institutes of Health, NIA Grant AG027349 (to U. J.) and by Postdoctoral Fellowship NIA Training Grant AG000114 (to H. T.). This work was also supported by the European Molecular Biology Organization and Human Frontiers postdoctoral fellowship (to D. R.). The Michigan Proteome was supported in part by funds from the Michigan Life Sciences Corridor.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” Tables S1 and S2, Figs. S1–S5, and additional references.
- ROS
- reactive oxygen species
- ICAT
- isotope-coded affinity tag
- SOD
- superoxide dismutase
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Winterbourn C. C., Hampton M. B. (2008) Free Radic. Biol. Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- 2. D'Autréaux B., Toledano M. B. (2007) Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 3. Stadtman E. R., Berlett B. S. (1998) Drug Metab. Rev. 30, 225–243 [DOI] [PubMed] [Google Scholar]
- 4. Cook J. A., Gius D., Wink D. A., Krishna M. C., Russo A., Mitchell J. B. (2004) Semin. Radiat. Oncol. 14, 259–266 [DOI] [PubMed] [Google Scholar]
- 5. Lowell B. B., Shulman G. I. (2005) Science 307, 384–387 [DOI] [PubMed] [Google Scholar]
- 6. Brandes N., Schmitt S., Jakob U. (2009) Antioxid. Redox Signal. 11, 997–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thamsen M., Jakob U. (2011) Curr. Opin. Chem. Biol. 15, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leichert L. I., Gehrke F., Gudiseva H. V., Blackwell T., Ilbert M., Walker A. K., Strahler J. R., Andrews P. C., Jakob U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8197–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meisinger C., Sommer T., Pfanner N. (2000) Anal. Biochem. 287, 339–342 [DOI] [PubMed] [Google Scholar]
- 10. Haas A. (1995) Methods Cell Sci. 17, 283–294 [Google Scholar]
- 11. Dove J. E., Brockenbrough J. S., Aris J. P. (1998) Methods Cell Biol. 53, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumsta C., Thamsen M., Jakob U. (2011) Antioxid Redox Signal 14, 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H., Robertson A. D., Jensen J. H. (2005) Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 14. Reichmann D., Phillip Y., Carmi A., Schreiber G. (2008) Biochemistry 47, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 15. Gerstein M., Richards F. M. (2001) International Tables for Crystallography F, 531–539 [Google Scholar]
- 16. Fratelli M., Demol H., Puype M., Casagrande S., Eberini I., Salmona M., Bonetto V., Mengozzi M., Duffieux F., Miclet E., Bachi A., Vandekerckhove J., Gianazza E., Ghezzi P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3505–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Le Moan N., Clement G., Le Maout S., Tacnet F., Toledano M. B. (2006) J. Biol. Chem. 281, 10420–10430 [DOI] [PubMed] [Google Scholar]
- 18. Leichert L. I., Jakob U. (2004) PLoS Biol 2, e333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Magherini F., Carpentieri A., Amoresano A., Gamberi T., De Filippo C., Rizzetto L., Biagini M., Pucci P., Modesti A. (2009) Cell Mol. Life Sci. 66, 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McDonagh B., Ogueta S., Lasarte G., Padilla C. A., Bárcena J. A. (2009) J. Proteomics 72, 677–689 [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Santamarina S., Boronat S., Espadas G., Ayte J., Molina H., Hidalgo E. (2011) J. Proteomics [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22. Leichert L. I., Jakob U. (2006) Antioxid. Redox Signal. 8, 763–772 [DOI] [PubMed] [Google Scholar]
- 23. Dixon B. M., Heath S. H., Kim R., Suh J. H., Hagen T. M. (2008) Antioxid. Redox Signal. 10, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar A., Agarwal S., Heyman J. A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Cheung K. H., Miller P., Gerstein M., Roeder G. S., Snyder M. (2002) Genes Dev. 16, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hörnberg A., Logan D. T., Marklund S. L., Oliveberg M. (2007) J. Mol. Biol. 365, 333–342 [DOI] [PubMed] [Google Scholar]
- 26. Field L. S., Furukawa Y., O'Halloran T. V., Culotta V. C. (2003) J. Biol. Chem. 278, 28052–28059 [DOI] [PubMed] [Google Scholar]
- 27. Allen S., Lu H., Thornton D., Tokatlidis K. (2003) J. Biol. Chem. 278, 38505–38513 [DOI] [PubMed] [Google Scholar]
- 28. Marino S. M., Li Y., Fomenko D. E., Agisheva N., Cerny R. L., Gladyshev V. N. (2010) Biochemistry 49, 7709–7721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Azimi I., Wong J. W., Hogg P. J. (2011) Antioxid. Redox Signal. 14, 113–126 [DOI] [PubMed] [Google Scholar]
- 30. Hansen R. E., Roth D., Winther J. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shenton D., Smirnova J. B., Selley J. N., Carroll K., Hubbard S. J., Pavitt G. D., Ashe M. P., Grant C. M. (2006) J. Biol. Chem. 281, 29011–29021 [DOI] [PubMed] [Google Scholar]
- 32. Mehta R., Chandler-Brown D., Ramos F. J., Shamieh L. S., Kaeberlein M. (2010) Adv. Exp. Med. Biol. 694, 14–29 [DOI] [PubMed] [Google Scholar]
- 33. Cotgreave I. A., Gerdes R., Schuppe-Koistinen I., Lind C. (2002) Methods Enzymol. 348, 175–182 [DOI] [PubMed] [Google Scholar]
- 34. Shenton D., Grant C. M. (2003) Biochem. J. 374, 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mattiasson B., Hahn-Hägerdal B. (1982) Appl. Microbiol. Biotechnol. 16, 52–55 [Google Scholar]
- 36. Poole L. B. (2007) Subcell. Biochem. 44, 61–81 [DOI] [PubMed] [Google Scholar]
- 37. Caplan A. J., Cyr D. M., Douglas M. G. (1992) Cell 71, 1143–1155 [DOI] [PubMed] [Google Scholar]
- 38. Tajc S. G., Tolbert B. S., Basavappa R., Miller B. L. (2004) J. Am. Chem. Soc. 126, 10508–10509 [DOI] [PubMed] [Google Scholar]
- 39. Rhee S. G., Bae Y. S., Lee S. R., Kwon J. (2000) Sci. STKE 2000, PE1. [DOI] [PubMed] [Google Scholar]
- 40. Zhang Z. Y., Dixon J. E. (1993) Biochemistry 32, 9340–9345 [DOI] [PubMed] [Google Scholar]
- 41. Sanchez R., Riddle M., Woo J., Momand J. (2008) Protein Sci. 17, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roos G., Messens J. (2011) Free Radic. Biol. Med. 51, 314–326 [DOI] [PubMed] [Google Scholar]
- 43. Zhang R. M., Snyder G. H. (1989) J. Biol. Chem. 264, 18472-18479 [PubMed] [Google Scholar]
- 44. Grauschopf U., Winther J. R., Korber P., Zander T., Dallinger P., Bardwell J. C. (1995) Cell 83, 947–955 [DOI] [PubMed] [Google Scholar]
- 45. Aslund F., Zheng M., Beckwith J., Storz G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6161–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reverter-Branchat G., Cabiscol E., Tamarit J., Ros J. (2004) J. Biol. Chem. 279, 31983–31989 [DOI] [PubMed] [Google Scholar]
- 47. Cabiscol E., Piulats E., Echave P., Herrero E., Ros J. (2000) J. Biol. Chem. 275, 27393–27398 [DOI] [PubMed] [Google Scholar]
- 48. Brennan J. P., Wait R., Begum S., Bell J. R., Dunn M. J., Eaton P. (2004) J. Biol. Chem. 279, 41352–41360 [DOI] [PubMed] [Google Scholar]
- 49. Cumming R. C., Andon N. L., Haynes P. A., Park M., Fischer W. H., Schubert D. (2004) J. Biol. Chem. 279, 21749–21758 [DOI] [PubMed] [Google Scholar]
- 50. Furukawa Y., Torres A. S., O'Halloran T. V. (2004) EMBO J. 23, 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baty J. W., Hampton M. B., Winterbourn C. C. (2005) Biochem. J. 389, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Minard K. I., Carroll C. A., Weintraub S. T., Mc-Alister-Henn L. (2007) Free Radic. Biol. Med. 42, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]