Abstract
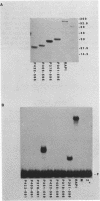
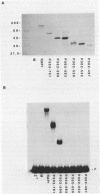
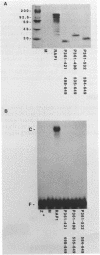
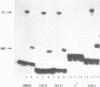
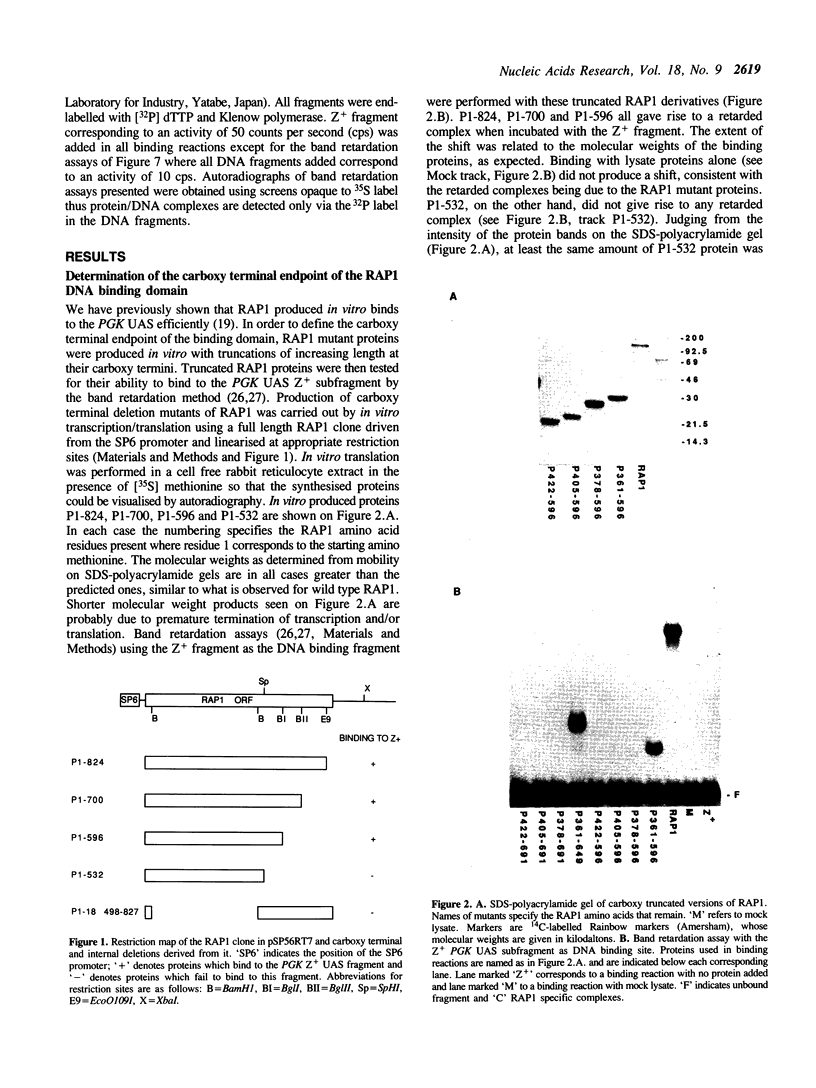
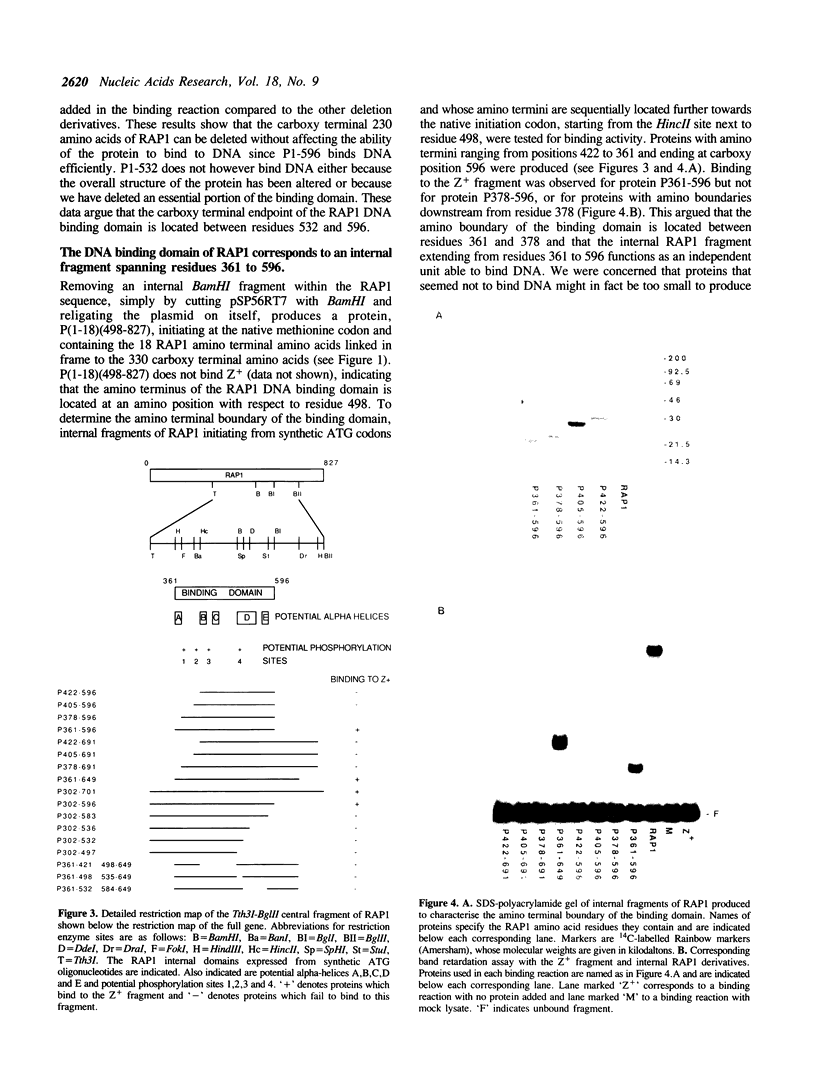
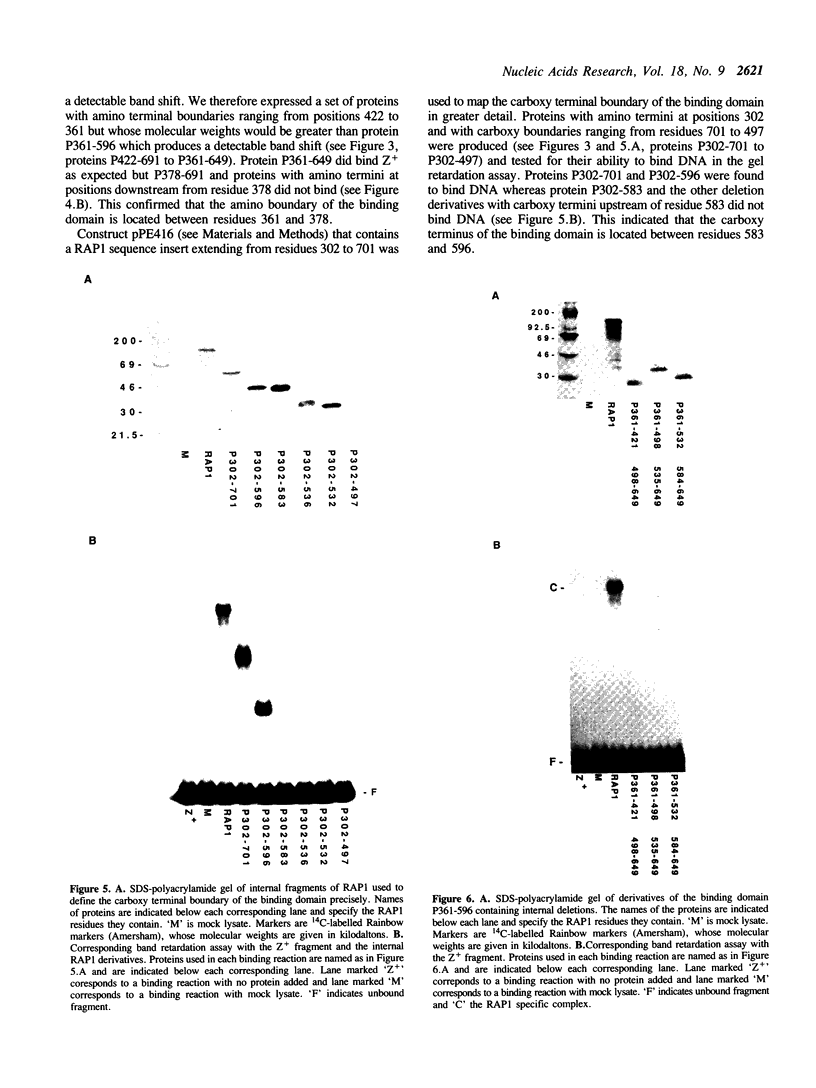
The 827 amino acid yeast RAP1 protein interacts with DNA to regulate gene expression at numerous unrelated loci in the yeast genome. By a combination of amino, carboxy and internal deletions, we have defined an internal 235 amino acid fragment of the yeast RAP1 protein that can bind efficiently to the RAP1 binding site of the PGK Upstream Activation Sequence (UAS). This domain spans residues 361 to 596 of the full length protein and lacks any homology to the DNA binding 'zinc finger' or 'helix-turn-helix' structural motifs. All the RAP1 binding sites we have tested bind domain 361-596, arguing that RAP1 binds all its chromosomal sites via this domain. The domain could not be further reduced in size suggesting that it represents the minimal functional DNA binding domain. The relevance of potential regions of secondary structure within the minimal binding domain is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Kawasaki G. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J Mol Appl Genet. 1982;1(5):419–434. [PubMed] [Google Scholar]
- Bemis L. T., Denis C. L. Identification of functional regions in the yeast transcriptional activator ADR1. Mol Cell Biol. 1988 May;8(5):2125–2131. doi: 10.1128/mcb.8.5.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Butler G., McConnell D. J. Identification of an upstream activation site in the pyruvate decarboxylase structural gene (PDC1) of Saccharomyces cerevisiae. Curr Genet. 1988 Nov;14(5):405–412. doi: 10.1007/BF00521261. [DOI] [PubMed] [Google Scholar]
- Capieaux E., Vignais M. L., Sentenac A., Goffeau A. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J Biol Chem. 1989 May 5;264(13):7437–7446. [PubMed] [Google Scholar]
- Chambers A., Stanway C., Kingsman A. J., Kingsman S. M. The UAS of the yeast PGK gene is composed of multiple functional elements. Nucleic Acids Res. 1988 Sep 12;16(17):8245–8260. doi: 10.1093/nar/16.17.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A., Tsang J. S., Stanway C., Kingsman A. J., Kingsman S. M. Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Mol Cell Biol. 1989 Dec;9(12):5516–5524. doi: 10.1128/mcb.9.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989 Nov 24;246(4933):1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Halfter H., Kavety B., Vandekerckhove J., Kiefer F., Gallwitz D. Sequence, expression and mutational analysis of BAF1, a transcriptional activator and ARS1-binding protein of the yeast Saccharomyces cerevisiae. EMBO J. 1989 Dec 20;8(13):4265–4272. doi: 10.1002/j.1460-2075.1989.tb08612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Huet J., Cottrelle P., Cool M., Vignais M. L., Thiele D., Marck C., Buhler J. M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985 Dec 16;4(13A):3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd H. K., Roberts J. W. Upstream regulatory sequences of the yeast RNR2 gene include a repression sequence and an activation site that binds the RAP1 protein. Mol Cell Biol. 1989 Dec;9(12):5359–5372. doi: 10.1128/mcb.9.12.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2401–2405. doi: 10.1073/pnas.84.8.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Leader D. P., Katan M. Viral aspects of protein phosphorylation. J Gen Virol. 1988 Jul;69(Pt 7):1441–1464. doi: 10.1099/0022-1317-69-7-1441. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Wilson N. M., Petracek M. E., Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989 Oct;16(4):225–239. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- Machida M., Uemura H., Jigami Y., Tanaka H. The protein factor which binds to the upstream activating sequence of Saccharomyces cerevisiae ENO1 gene. Nucleic Acids Res. 1988 Feb 25;16(4):1407–1422. doi: 10.1093/nar/16.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Araki R., Teranishi Y. Identification of an upstream activating sequence and an upstream repressible sequence of the pyruvate kinase gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):442–451. doi: 10.1128/mcb.9.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden J. E., Stanway C., Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol Cell Biol. 1986 Dec;6(12):4335–4343. doi: 10.1128/mcb.6.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Prywes R., Dutta A., Cromlish J. A., Roeder R. G. Phosphorylation of serum response factor, a factor that binds to the serum response element of the c-FOS enhancer. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7206–7210. doi: 10.1073/pnas.85.19.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri P., Bagchi S., Nevins J. R. DNA-binding activity of the adenovirus-induced E4F transcription factor is regulated by phosphorylation. Genes Dev. 1989 May;3(5):620–627. doi: 10.1101/gad.3.5.620. [DOI] [PubMed] [Google Scholar]
- Rhode P. R., Sweder K. S., Oegema K. F., Campbell J. L. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes Dev. 1989 Dec;3(12A):1926–1939. doi: 10.1101/gad.3.12a.1926. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway C. A., Chambers A., Kingsman A. J., Kingsman S. M. Characterization of the transcriptional potency of sub-elements of the UAS of the yeast PGK gene in a PGK mini-promoter. Nucleic Acids Res. 1989 Nov 25;17(22):9205–9218. doi: 10.1093/nar/17.22.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway C., Mellor J., Ogden J. E., Kingsman A. J., Kingsman S. M. The UAS of the yeast PGK gene contains functionally distinct domains. Nucleic Acids Res. 1987 Sep 11;15(17):6855–6873. doi: 10.1093/nar/15.17.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Tavianini M. A., Blumberg H., Young E. T. Localization of a minimal binding domain and activation regions in yeast regulatory protein ADR1. Mol Cell Biol. 1989 Jun;9(6):2360–2369. doi: 10.1128/mcb.9.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura H., Shiba T., Paterson M., Jigami Y., Tanaka H. Identification of a sequence containing the positive regulatory region of Saccharomyces cerevisiae gene ENO1. Gene. 1986;45(1):67–75. doi: 10.1016/0378-1119(86)90133-2. [DOI] [PubMed] [Google Scholar]
- Vignais M. L., Woudt L. P., Wassenaar G. M., Mager W. H., Sentenac A., Planta R. J. Specific binding of TUF factor to upstream activation sites of yeast ribosomal protein genes. EMBO J. 1987 May;6(5):1451–1457. doi: 10.1002/j.1460-2075.1987.tb02386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. K., Gonzalez G. A., Biggs W. H., 3rd, Montminy M. R. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988 Aug 11;334(6182):494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]