Abstract
Genetic factors have been implicated in stroke risk but few replicated associations have been reported. We conducted a genome-wide association study (GWAS) in ischemic stroke and its subtypes in 3,548 cases and 5,972 controls, all of European ancestry. Replication of potential signals was performed in 5,859 cases and 6,281 controls. We replicated reported associations between variants close to PITX2 and ZFHX3 with cardioembolic stroke, and a 9p21 locus with large vessel stroke. We identified a novel association for a SNP within the histone deacetylase 9 (HDAC9) gene on chromosome 7p21.1 which was associated with large vessel stroke including additional replication in a further 735 cases and 28583 controls (rs11984041, combined P = 1.87×10−11, OR=1.42 (95% CI) 1.28-1.57). All four loci exhibit evidence for heterogeneity of effect across the stroke subtypes, with some, and possibly all, affecting risk for only one subtype. This suggests differing genetic architectures for different stroke subtypes.
Cerebrovascular disease (stroke) is one of the three most common causes of death and the major cause of adult chronic disability (1). Stroke represents an increasing health problem throughout the world as the proportion of elderly increases, and is an important cause of dementia and age-related cognitive decline. While conventional risk factors such as hypertension account for a significant proportion of stroke risk, much remains unexplained (2). Twin and family history studies suggest genetic factors are responsible for some of this unexplained risk (3). Stroke is a syndrome rather than a single disease, and subtypes of stroke are caused by a number of different specific disease processes. About 80% of stroke is ischemic; the three most common ischemic stroke subtypes are large vessel, cardioembolic and small vessel (lacunar) stroke. Genetic epidemiological studies show heterogeneity between stroke subtypes, the large vessel subtype being more strongly associated with family history (4). SNPs associated with atrial fibrillation were found only to be significantly associated with cardioembolic stroke (5,6), and a 9p21 variant initially associated with coronary artery disease and atherosclerosis only associated with large vessel stroke (7). This suggests that different genetic variants can predispose to different subtypes of ischemic stroke.
To date there have been few genome wide association studies (GWAS) in ischemic stroke and few replicable associations have been identified (8). To further understand the genetic basis of ischemic stroke, we undertook a GWAS as part of the Wellcome Trust Case Control Consortium 2 (WTCCC2). We hypothesised that associations might be present only with specific stroke subtypes. To investigate this, cases were classified into stroke subtypes according to the pathophysiological TOAST classification (9), using clinical assessment as well as brain and vascular imaging where available (see Online Methods). Association analyses were performed on all ischemic stroke combined (including individuals not further classified by stroke subtype), and also the three major stroke subtypes: large vessel, small vessel and cardioembolic stroke. Discovery samples were of European ancestry and were genotyped on Illumina arrays (see Online Methods). Following quality control, the discovery set consisted of 3,548 cases (2,374 British, 1,174 German) and 5,972 controls (5,175 British WTCCC2 common controls, and 797 German controls) genotyped on an overlapping set of 495,851 autosomal SNPs (Table 1 and Online Methods). Within the British and German data, cases and controls were well matched for ancestry (see Online Methods and Supplementary Figure 1). We therefore performed association analysis separately in the two groups and combined them using a fixed effect meta-analysis approach. A two-stage replication study was performed in 5,859 cases (3,863 European, 1,996 American) and 6,281 controls (4,554 European, 1,727 American) all of self-reported European ancestry. (Table 1 and Online Methods). Full details of the cohorts are available in the Supplementary material. and Supplementary Table 1.
Table 1. Post quality control breakdown of case and control by cohort and ischaemic stroke subtype.
All: all ischaemic stroke, LVD: large vessel stroke, SVD: small vessel stroke, CE: cardioembolic stroke. Note that not all strokes are classified into a subtype.
| All strokes | LVD | CE | SVD | Controls | ||
|---|---|---|---|---|---|---|
| DISCOVERY | Munich | 1174 | 346 | 330 | 106 | 797 |
| UK1 | 2374 | 498 | 460 | 474 | 5175 | |
|
| ||||||
| Total | 3548 | 844 | 790 | 580 | 5972 | |
|
| ||||||
| STAGE 1 REPLICATION - EUROPEAN | Krakow | 1214 | 152 | 362 | 170 | 551 |
| Leuven | 418 | 63 | 154 | 52 | 650 | |
| Lund | 428 | 21 | 139 | 97 | 465 | |
| Munich2 | 54 | 19 | 16 | 5 | 310 | |
| UK3 | 1749 | 306 | 303 | 490 | 2578 | |
|
| ||||||
| Total | 3863 | 561 | 974 | 814 | 4554 | |
|
| ||||||
| STAGE 2 REPLICATION - US | Boston | 533 | 150 | 206 | 56 | 522 |
| Cincinnati | 438 | 67 | 106 | 90 | 257 | |
| GEOS | 419 | 37 | 90 | 54 | 498 | |
| ISGS | 606 | 121 | 156 | 111 | 450 | |
|
| ||||||
| Total | 1996 | 375 | 558 | 311 | 1727 | |
|
| ||||||
| STAGE 1 + STAGE 2 REPLICATION | Total | 5859 | 936 | 1532 | 1125 | 6281 |
|
| ||||||
| DISCOVERY + REPLICATION | Total | 9407 | 1780 | 2322 | 1705 | 12253 |
The UK discovery cohort was made of three British cohorts from London, Oxford and Edinburgh and used the shared WTCCC2 controls.
The Munich replication samples comprised some samples planned for the discovery GWAS where there was insufficient DNA for GWAS but sufficient for replication. It used controls from a German cohort enrolled in the PROCARDIS trial.
The UK replication cohorts included samples from Aberdeen, Glasgow and Imperial as well as some samples planned for the discovery GWAS where there was insufficient DNA for GWAS but sufficient for replication (see methods). The UK replication cohorts used shared POBI controls genotypes as part of the WTCCC2.
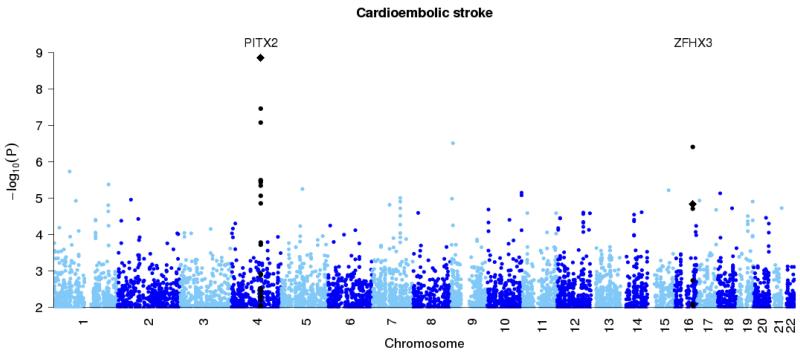
Table 2 shows results at previously reported loci and Figure 1 shows the association analysis results across the autosomes. We replicated an association between cardioembolic stroke and variants close to the PITX2 gene and also a SNP in the ZFHX3 gene, both of which were initially associated with atrial fibrillation, a well recognised risk factor for stroke (5,6,10). We also replicated a previously reported association between large vessel stroke and the 9p21 region (7). As we, and others, already reported (11,12), we did not confirm the previously published association between all stroke and variants in the 12p13 region (13, 14).
Table 2. Association signals at the newly associated locus (upper tier) and at loci previously reported as associated with stroke or one of the stroke subtype (lower tier).
All: all ischaemic stroke, LVD: large vessel stroke, CE: cardioembolic stroke. For PITX2 and ZFHX3, results are given for one SNP reported in the literature and the SNP that showed the strongest association signal in the discovery samples. For the 9p21 region, the SNP reported in the literature is also the one showing the strongest association signal in the discovery cohort. There is some overlap between samples in this study and previous published studies of associations with above loci (5,6,10,13). A SNP in PRKCH, associated with stroke in Japanese populations (23) was very rare (<0.5%) in our Caucasian population so we had no power to perform an analysis of association with this SNP.
| Chr | rsID | Position6 | Candidategene | Strokesubtype | Riskallele | RAF7 | Discovery | Stage 1&2 | Stage3 | Combined |
|---|---|---|---|---|---|---|---|---|---|---|
| P-value | P-value (one-sided) | P-value (one-side) | P-value | |||||||
| OR(95% CI) | OR (95% CI) | OR (95% CI) | OR(95%CI) | |||||||
|
| ||||||||||
| 7p21.1 | rs119840411,2 | 18,998,460 | HDAC9 | LVD | A | 0.09 | 1.07E-05 | 7.90E-05 | 2.25E-04 | 1.87E-11 |
| 1.50 (1.25-1.79) | 1.38 (1.17-1.63) | 1.39 (1.15-1.68) | 1.42 (1.28-1.57) | |||||||
|
| ||||||||||
| 4q25 | rs22007332,3,4 | 111,929,618 | PITX2 | CE | A | 0.10 | 3.64E-06 | 3.99E-04 | - | 5.06E-8 |
| 1.49 (1.26-1.77) | 1.24 (1.09-1.41) | - | 1.32 (1.20-1.46) | |||||||
| rs19065993 | 111,932,135 | A | 0.19 | 3.45E-08 | 3.16E-04 | - | 1.39E-09 | |||
| 1.45 (1.27-1.66) | 1.19 (1.08-1.32) | - | 1.28 (1.18-1.39) | |||||||
|
| ||||||||||
| 9p21.3 | rs23832073 | 22,105,959 | CDKN2A, CDKN2B |
LVD | G | 0.51 | 2.35E-03 | 2.03E-03 | - | 2.93E-05 |
| 1.18 (1.06-1.31) | 1.16 (1.05-1.28) | - | 1.17 (1.09-1.25) | |||||||
|
| ||||||||||
| 12p13.33 | rs118335793,5 | 645,460 | NINJ2 | All | G | 0.75 | 9.65E-01 | 5.25E-01 | - | 9.81E-01 |
| 1.00 (0.92-1.08) | 1.00 (0.94-1.06) | - | 1.00 (0.95-1.05) | |||||||
|
| ||||||||||
| 16q22.3 | rs71933433 | 71,586,661 | ZFHX3 | CE | A | 0.16 | 1.94E-05 | - | - | - |
| 1.36 (1.18-1.57) | - | - | - | |||||||
| rs129324452 | 71,627,389 | G | 0.17 | 3.91E-07 | 4.84E-02 | - | 1.44E-05 | |||
| 1.44 (1.25-1.66) | 1.09 (0.98-1.21) | - | 1.20 (1.11-1.31) | |||||||
Krakow replication samples were not considered because of Hardy-Weinberg test p-value < 5×10−4 in controls.
SNP imputed in GEOS replication samples.
SNP reported in the literature.
ISGS replication samples were not considered because of Hardy-Weinberg test p-value < 5×10−4 in controls.
SNP imputed in the British discovery samples and not genotyped nor imputed in the discovery and replication German controls.
NCBI human genome build 36 coordinates.
Risk allele frequency computed in the British discovery controls
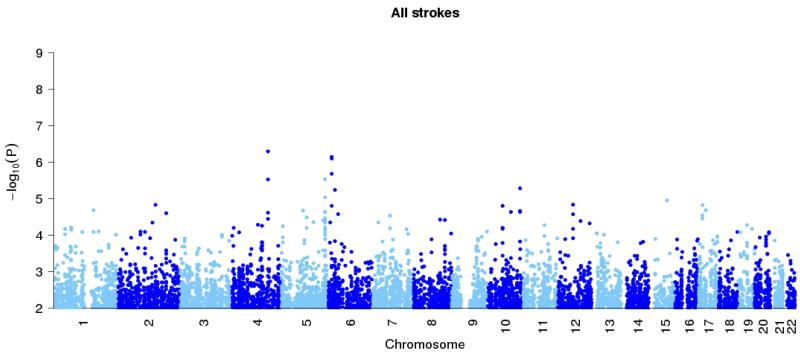
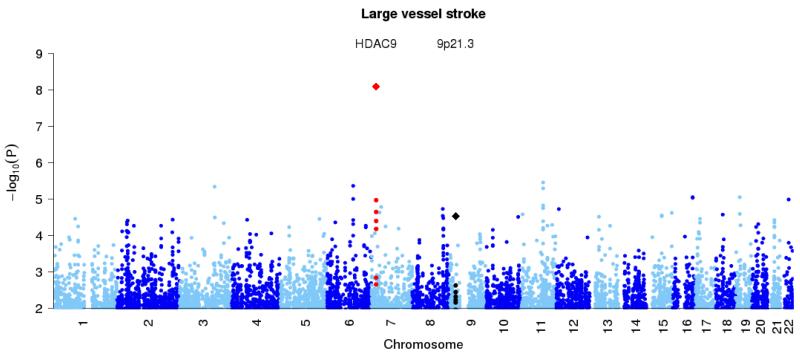
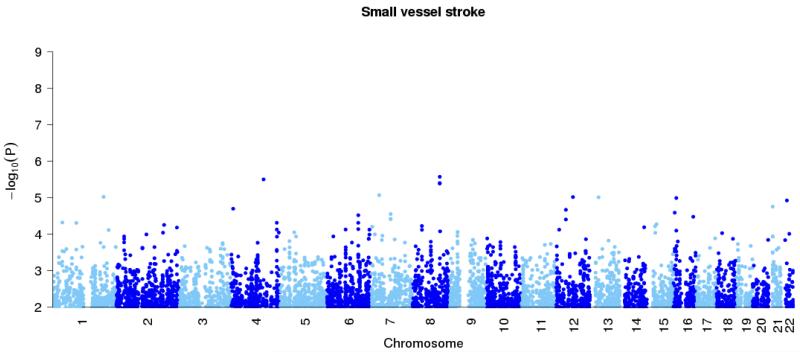
Figure 1. Genome-wide association results at autosomal SNPs in combined British and Germany discovery samples.
All ischemic stroke (top panel), large vessel stroke (second panel), small vessel stroke (third panel) and cardioembolic stroke (final panel). Loci previously reported in the literature for particular stoke subtypes (Table 2) are shown in black, with the new HDAC9 locus shown in red. In addition the combined p-values for discovery study and stages one and two of the replication study, at the top SNPs for these loci, are marked with diamonds.
Thirty-eight previously unreported loci showed potential association for all stroke or one of the stroke subtypes in the discovery samples, and we further investigated these loci in the European replication samples by genotyping 43 SNPs covering these loci as well as 7 SNPs to cover the previously reported loci (Supplementary Table 2). Thirteen of these previously unreported loci and the previously reported loci were taken forward to replication in the American samples with genotyping of 20 SNPs covering these regions (Supplementary Table 3). Most replication samples were genotyped using Sequenom assays; for those previously typed with GWAS chips we used genotype imputation where the SNP was not directly typed (see Supplementary Tables 2 and 3 and Online Methods). A SNP at chromosome 7p21.1 (rs11984041) showed evidence of association with large vessel stroke in the discovery data (P=1.07×10−5) and in the joint European and US replication data in the same direction (one-sided P=7.9×10−5). As a further check, we investigated this SNP in three further collections of large vessel cases and matched controls (735 cases, 28583 controls in total), which we refer to as Stage 3 replication (see Online Methods for details). The Stage 3 data also showed evidence in the same direction (one-sided P=2.25×10−4). Together, the combined discovery and three-stage replication data provide strong evidence for association (P=1.87×10−11) and suggest each copy of the A allele increases risk of large vessel stroke by approximately 1.4 fold (Table 2 and Figure 2). This SNP is within the final intron of the gene HDAC9. The risk allele (A) frequency was 9.29% and 8.78% in the UK and German discovery controls respectively.
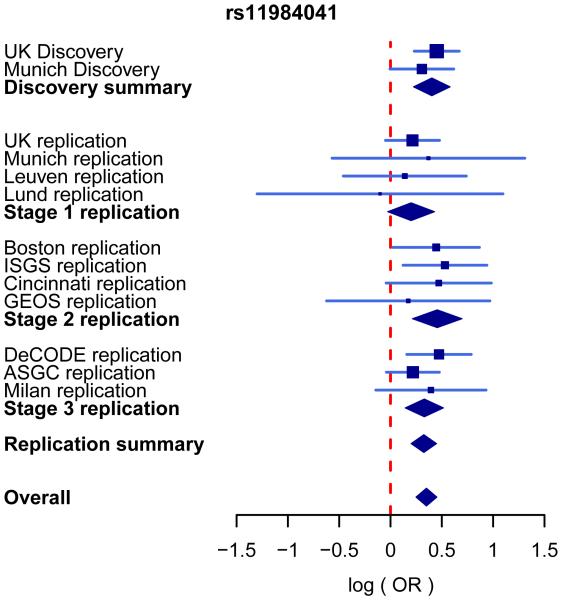
Figure 2. Forest plot for the associations between rs11984041 and large vessel stroke in discovery and replication collections.
The blue lines show the 95% confidence intervals of the log odds-ratio (OR) for each cohort, with the area of each square proportional to the inverse of the standard error. The diamonds indicate the 95% confidence interval for, from top to bottom, the discovery summary (combined British and German discovery collections), combined across collections within each of the three replication stages, the replication summary (combined across all three replication stages) and the overall summary (all discovery and replication collections combined). Evidence was combined across collections via an inverse-variance weighted fixed-effect meta-analysis. There was no evidence of heterogeneity of effect across collections (P = 0.92).
Standard statistical tests of association between rs11984041 and each of cardioembolic and small vessel stroke are not significant (discovery plus 2-stage replication p = 0.12, OR = 1.10, 95% CI = 0.98 – 1.23, and p = 0.06, OR = 1.13, 95% CI = 1.00 – 1.28 respectively). A non-significant result could simply be due to a lack of power: lack of significance in itself cannot rule out an effect in these subtypes. We investigated this potential genetic heterogeneity further by formally comparing different statistical models for the effect of the SNP on the different stroke subtypes. The models we compared were: (i) a model in which the variant has no effect on risk for any of the subtypes (“null” model); (ii) a model in which the SNP has the same effect on each subtype (“same effects” model); (iii) three models, in each of which the SNP has an effect on one subtype, and no effect for the other two subtypes (“LVD”, “SVD” and “CE” models respectively for the effect only in large vessel, small vessel, and cardioembolic stroke); and (iv) a “correlated effects” model allowing different, but correlated, effects for each subtype. We undertook the model comparison in a Bayesian statistical framework (see Online Methods for details), for our new association around HDAC9, as well as for the previously reported associations we confirmed as listed in Table 2. The results, based on the discovery and the first two stages of the replication, are shown in Figure 3.
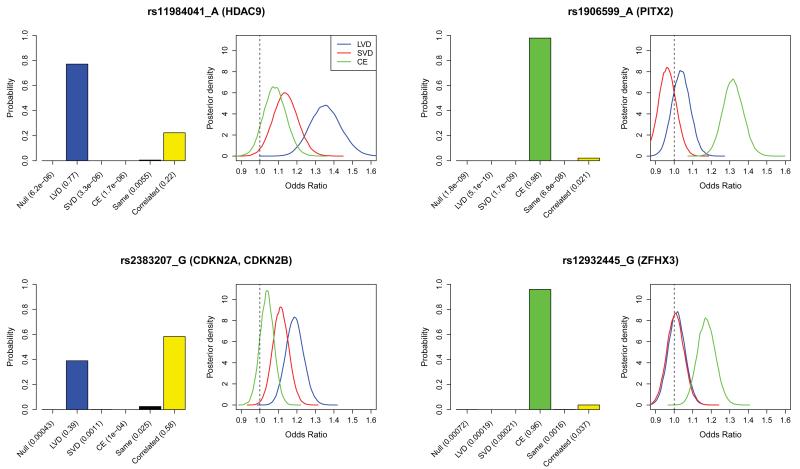
Figure 3. Genetic heterogeneity of different stroke sub-types for the 4 loci with significant associations: HDAC9, PITX2, 9p21(CDKN2A/CDKN2B) and ZFHX3.
Bar plots show the posterior probabilities on the models of association: no effect in any subtype (Null), same effect in all subtypes, correlated effects across subtypes or subtype specific effects (see main text). Models are a priori assumed to be equally likely. Bayes factors, which compare the evidence (marginal likelihood) between any pair of models, can be calculated as the ratio of the posterior probability assigned to each model as reported under each bar of the plot. Accompanying density plots show the marginal posterior distribution on the odds ratio of the risk allele for each stroke subtype assuming a model of correlated effects (see methods for specification of priors). These analyses were performed using both discovery and replication samples (stage 1 & 2).
For rs11984041 at HDAC9 there is very strong evidence against the null model and both the SVD and CE models (unsurprisingly given we ascertained this SNP on the basis of evidence for an effect in LVD) and also strong evidence against the model in which the SNP has the same effect in each subtype, thus demonstrating genetic heterogeneity across stroke subtypes at this SNP. The greatest posterior weight rests on the model in which there is only an effect for large vessel disease, with some weight on the correlated effects model, and in this model the posterior distributions on effect size for SVD and CE stroke are concentrated on much smaller effect sizes than for LVD.
In our data, heterogeneity is also seen at rs2383207 in the 9p21 region, a locus associated with heart disease and related phenotypes, and previously associated with large vessel stroke. Most support is for the model in which the effect sizes for the three stroke subtypes are correlated but there is also substantial weight on the model in which there is only an effect for large vessel stroke. The same analyses in our data for the top SNPs in the regions previously associated with cardioembolic stroke (PITX2 region, rs1906599, and ZFHX3 regions, rs12932445) show strong support for the model in which these SNPs only affect risk for cardioembolic stroke. Together these analyses provide compelling evidence for heterogeneity of genetic effects between stroke subtypes.
The association with rs11984041, in the gene HDAC9, implicates a novel region of the genome in an individual’s susceptibility to stroke. Any association with stroke could be mediated via associations with intermediate cardiovascular risk factors that themselves increase large vessel stroke risk. Our study design does not allow a direct assessment of this, as such risk factors were not available for control individuals. However, to date no associations have been reported between rs11984041 or correlated SNPs and hypertension (15), hyperlipidaemia (16), or diabetes (17) from large-scale GWAS of these risk factors.
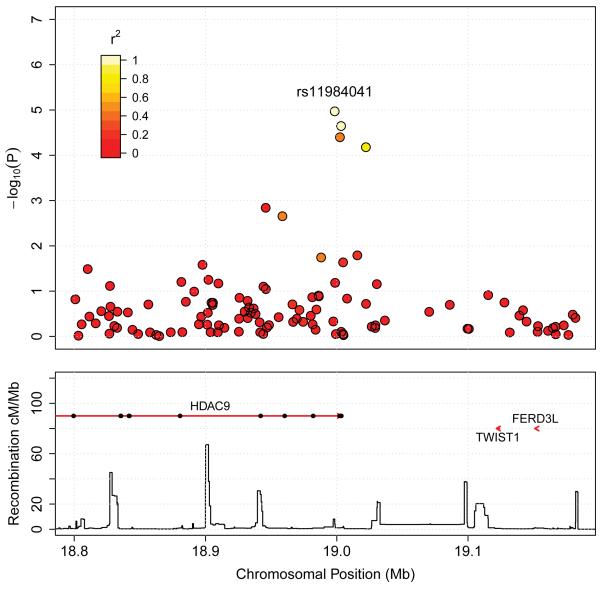
Association of genetic variants surrounding HDAC9 are represented in Figure 4. All variants showing an association signal reside within a peak between two recombination hotspots and encompass the tail end of HDAC9. The downstream genes TWIST1 and FERD3L are physically relatively close to the identified peak and cannot be excluded as possible mechanisms via which genetic variants may exert cis-effects on the large vessel stroke phenotype. HDAC9 is a member of a large family of genes that encode proteins responsible for deacetylation of histones, and therefore regulation of chromatin structure and gene transcription (18). HDAC9 is ubiquitously expressed, with high levels of expression in cardiac tissue, muscle and brain (19). Although known as histone deacetylases, these proteins also act on other substrates (20) and lead to both upregulation and downregulation of genes (21).
Figure 4.
Plot of association signals around rs11984041 for large vessel stroke in the combined British and German discovery samples. SNPs are coloured based on their correlation (r2) with the labelled hit SNP which has the smallest P-value in the region. r2 is calculated from the WTCCC2 control data. The bottom section of each plot shows the fine scale recombination rates estimated from HapMap data, and genes are marked by horizontal red lines. Arrows on the horizontal red lines denote direction of transcription, and black rectangles are exons.
The mechanism by which variants in the HDAC9 region increase large vessel stroke risk is not immediately clear. The specific association with this stroke subtype would be consistent with the association acting via accelerating atherosclerosis. The HDAC9 protein inhibits myogenesis and is involved in heart development (19) although deleterious effects on systemic arteries have not yet been reported. Alternatively it could increase risk by altering brain ischaemic responses and therefore have effects on neuronal survival. The protein has been shown to protect neurons from apoptosis, both by inhibiting JUN phosphorylation by MAPK10 and by repressing JUN transcription. HDAC inhibitors have been postulated as a treatment for stroke (22).
It is not uninformative that a large GWAS (~3,500 cases, ~6,000 controls) failed to find any novel associations for the combined phenotype of ischemic stroke. It may be that the genetic architecture of the disease involves fewer variants of more moderate effect than many other diseases, and/or that these happen not to be well tagged by the Illumina 660-W chip used in the study. On the other hand, as our data demonstrate, all the known loci exhibit genetic heterogeneity across the stroke subtypes, with at least some, and possibly all, affecting only a single subtype. This supports the possibility that distinct subtypes of the disease have differing genetic architectures. However this is based on only four loci and does not exclude the possibility that future loci associated with stroke may predispose to all ischaemic stroke. Clinical classification of disease into subtypes is not perfect. Since errors in classification would reduce power to detect heterogeneity, our findings of homogeneity within classes indirectly reinforces the value of current classification methods. Because GWAS studies to date, including the one reported here, have had relatively small sample sizes for each disease subtype (and hence are underpowered for common variants of small effect), it remains possible, and indeed a priori likely, that the range of effect sizes for each subtype will be similar to those for other common diseases. This suggests that future genetic studies should study adequate sample sizes for particular subtypes of ischaemic stroke, rather than for the disease as a whole.
In summary, in this largest GWAS study of ischemic stroke conducted to date, we identified a novel association with the HDAC9 gene region in large vessel stroke with an estimated effect size which is at the larger end for GWAS loci (OR 1.38, 95% CI 1.22-1.57 from replication data). We also replicated three other known loci, and showed genetic heterogeneity across subtypes of the disease for all four stroke loci. This genetic heterogeneity seems likely to reflect heterogeneity in the underlying pathogenic mechanisms, and reinforces the need for separate consideration of stroke subtypes in the research and clinical context.
Supplementary Material
Acknowledgements
The principal funding for this study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z and WT084724MA). For details of other funding support see Supplementary Material.
We thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, M.L. Perez, C.R. Stribling, S. Taylor and A. Wilk of the Wellcome Trust Sanger Institute’s Sample and Genotyping Facilities for technical assistance. We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02, and of the UK National Blood Service controls funded by the Wellcome Trust. We thank W. Bodmer and B. Winney for use of the People of the British Isles DNA collection, which was funded by the Wellcome Trust.
We thank the following who contributed to collection, phenotyping, sample processing and data management for the different cohorts. Oxford Vascular Study: Annette Burgess, Anila Syed, Nicola Paul. Edinburgh Stroke Study: Martin Dennis, Peter Sandercock, Charles Warlow, Simon Hart, Sarah Keir, Joanna Wardlaw, Andrew Farrall, Gillian Potter, Aidan Hutchison, Mike McDowall. Aberdeen: Alireza Pasdar, Helen Clinkscale. Glasgow: Peter Higgins. ISGS: T. G. Brott, R. D. Brown, S. Silliman, M. Frankel, D. Case, S. Rich, J. Hardy, A Singleton. GEOS: Mary J Sparks, Kathy Ryan, John Cole, Marcella Wozniak, Barney Stern, Robert Wityk, Constance Johnson, David Buchholz. Australian Stroke Genetics Collaborative membership: Jane Maguire, Simon Koblar, Jonathan Golledge, Jonathan Surm, Graeme Hankey, Jim Jannes, Martin Lewis, Rodney Scott, Lisa Lincz; Pablo Moscato; Ross Baker.
APPENDIX
Methods
Study subjects
All subjects were of self-reported European ancestry. Patients were classified into mutually exclusive etiologic subtypes according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST) (9). TOAST classification was performed in all stroke cases. The TOAST system has a category of “etiology unknown” which includes cases in which no cause has been found due to insufficient investigation, as well as cases where no cause is found despite full investigation. This “unknown” group was not analysed in subtype analyses described in this paper which focussed only in those patients where there were appropriate investigations to assign one of three subtypes; large vessel disease, cardioembolic and small vessel disease. The unknown cases were only included in the analyses of all ischaemic stroke which did not take into account subtype.
Our main analyses were of associations with all ischemic stroke and with the three main subtypes: large vessel, cardioembolic and small vessel stroke. We performed additional analyses in the discovery populations with young stroke (age <70 years at first stroke), and with the presence of large vessel stenosis and, separately, the presence of cardioembolic source, irrespective of assigned subtype. These last two analyses allowed inclusion of patients whose data was excluded from individual subtype analysis because they had more than one potential stroke subtype. Details of individual populations are given in Table 1 and in supplementary material.
DNA sample preparation
This was performed as described in the Supplementary Material.
GWA genotyping
Samples from the cases were genotyped at the WTSI on the Human660W-Quad (a custom chip designed by WTCCC2 comprising Human550 and circa 6000 common CNVs from the Structural Variation Consortium (24)). Samples from British control collections were genotyped on the Human1.2M-Duo (a WTCCC2 custom array comprising Human1M-Duo and the CNV content described above). Bead intensity data was processed and normalized in BeadStudio; data for successfully genotyped samples was extracted and genotypes called within collections using Illuminus(25). German controls were typed on Illumina Human 550k platform, and intensity data was processed and normalized for each sample in GenomeStudio using the Illumina cluster file HumanHap550v3.
GWA quality control
Samples
As previously described (26,27), we removed samples whose genome-wide patterns of diversity differed from those of the collection at large, interpreting them likely to be due to biases or artefacts. To do so we used a Bayesian clustering approach (28) to infer outlying individuals on the basis of call rate, heterozygosity, ancestry, and average probe intensity. To obtain a set of putatively unrelated individuals we used a hidden Markov model (HMM) to infer identity by descent and then iteratively removed individuals to obtain a set with pair-wise identity by descent <5%. To guard against sample mishandling we removed samples if their inferred gender was discordant with recorded gender, or if <90% of the SNPs typed by Sequenom on entry to sample handling (see above) agreed with the genome-wide data. Our final discovery dataset consisted of 3548 cases (2374 British, 1174 German) and 5972 controls (5175 British, 797 German) following sample quality control (Supplementary Table 4). A full breakdown of samples by cohort and subtype is in Table 1.
SNPs
A measure of (Fisher) information for allele frequency at each SNP was calculated using SNPTEST (see URLs). Autosomal SNPs were excluded if this information measure was below 0.98, if minor allele frequency was <0.01%, if the SNP had >5% missing data, or if Hardy Weinberg p-value was <1×10−20 in the case or control collections. In the 58C, UKBS and case data set, association between SNP and the plate on which samples were genotyped was calculated and SNPs with a plate effect p-value <1×10−6 were also excluded. An additional 45 SNPs were removed following visual inspection of cluster plots. A breakdown of the number of SNPs excluded is provided in Supplementary Table 5. Only SNPs genotyped on all the case and control collections were considered, leaving 495,851 autosomal SNPs after quality control. Hardy Weinberg p-values for the SNPs taken to replication are given in Supplementary Table 6.
Initial replication genotyping and quality control
Genotyping of European replication samples was carried out at the WTSI using Sequenom iPLEX Gold assay and genotyping of the US samples at the Broad Institute, Boston, USA using the Sequenom platform, with the exception of the GEOS Study, in which genotyping was carried out using Illumina Human Omni1-Quad. Imputation to HapMap3 using BEAGLE software program (29) was performed. Individual samples were excluded from analysis if they had call rates <80% or if reported gender was discordant with gender specific markers. We removed pairs of samples showing concordance indicative of being duplicates.
The PoBI samples were genotyped on the custom Human1.2M-Duo array using Illumina’s Infinium platform and subjected to similar quality control as described above; for each SNP used in replication the cluster plot was visually inspected.
The PROCARDIS controls were genotyped with the Illumina HumanHap 610 Quad beadchip. PCA with HapMap2 reference population data allowed exclusion of individuals with non-European ancestry. Subsequent PCA with HapMap3 on German stroke samples with GWAS data and additional European reference population data, showed German PROCARDIS controls had similar ancestry to German stroke cases (data not shown).
Third stage replication of rs11984041 SNP
For the deCODE cases and controls genotyping was performed on 317K or 370K Illumina chips. The SNP rs11984041 was imputed using HapMap. ASGC cases and control samples were genotyped on the Illumina HumanHap610-Quad array, SNP rs11984041 was directly genotyped. Milan cases samples were genotyped using Illumina Human610-Quadv1_B or Human660W-Quad_v1_A beadchip; both include the rs11984041 SNP. Milan controls were genotyped with the Illumina HumanHap 610 Quad beadchip. PCA with HapMap3 on the Italian stroke samples showed that Italian PROCARDIS controls had similar ancestry to Italian stroke cases.
Genotype imputation
This was performed as described in the Supplementary Material.
Association analysis
We performed single SNP analysis separately in the British and German discovery data sets under an additive model (on the log-odds scale) using missing data likelihood score tests as implemented in SNPTEST. We conducted a fixed effect meta-analysis in R to combine the evidence of association, averaging the estimated effect size parameters associated with genotype risk across the two data sets, weighting the effect size estimates by the inverse of the square of corresponding standard errors. P-values were calculated assuming the combined data z-score to be normally distributed. The British and German cohorts had an inflation factor ranging from 1.014 to 1.058 and from 1.011 to 1.044 respectively, depending on the stroke subtype considered (Supplementary Figure 1). This analysis was also performed separately in males and females.
We also conducted a genome-wide scan analysis based on a Bayesian model which allows each stroke subtype to have its own effect and models relationships between these effects using a hierarchical prior specification. The same effects were assumed for the corresponding stroke subtype in both British and German populations (Supplementary Table 2).
Finally we performed a genome-wide scan using GENECLUSTER (31). This estimates genealogical tree of the case-control sample at a position of interest based on the genealogy of a reference panel (HapMap2 CEU in our case), by simultaneously phasing and clustering the case and control haplotypes to the tips of the reference genealogy. The method detects signals of association in the form of differential clustering of cases and controls underneath a branch, or a number of branches, in the estimated genealogy, which is equivalent to associations due to haplotypic effects or allelic heterogeneity (Supplementary Table 2).
Replication
Replication of potential associations found in the GWAS of the discovery cohorts was conducted in two stages in independent European and American samples. We investigated in the European replication cohorts 50 SNPs, that either were in loci reported in the literature from previous GWAS, or showed potential associations (P<1×10−5) with all stroke or one of the stroke subtypes in analysis of the discovery data set, and showed consistent direction of effect in both British and German cohorts (Supplementary Table 2). This threshold was chosen based on resources available for replication. After analysis of the combined results of the discovery and European replication populations 20 of these SNPs were taken forward to second stage replication in the American samples (Supplementary Table 3).
Association analysis was performed in each replication cohort separately via a logistic regression assuming an additive genetic model. Evidence of association across the replication data was combined using a fixed effect meta-analysis as previously described. Data on the presence or absence of a cardioembolic source or large vessel stenosis (irrespective of assigned TOAST subtype) were not available in all of replication cohorts. For replication of SNPs identified due to association with these phenotypes in the discovery cohorts, we assessed association in the replication cohorts with the cardioembolic or large vessel stroke subtypes respectively.
Bayesian model comparison
Details of this analysis are given in the Supplemental Material.
Additional references
The authors of this paper are:
Céline Bellenguez, Steve Bevan, Andreas Gschwendtner, Chris C A Spencer, Annette I. Burgess, Matti Pirinen, Caroline A Jackson, Matthew Traylor, Amy Strange, Zhan Su, Gavin Band, Paul D Syme, Rainer Malik, Joanna Pera, Bo Norrving, Robin Lemmens, Colin Freeman, Renata Schanz, Tom James, Deborah Poole, Lee Murphy, Helen Segal, Lynelle Cortellini, Yu-Ching Cheng, Daniel Woo, Michael A. Nalls, Bertram Müller-Myhsok, Christa Meisinger, Udo Seedorf, Helen Ross-Adams, Steven Boonen, Dorota Wloch-Kopec, Valerie Valant, Julia Slark, Karen Furie, Hossein Delavaran, Cordelia Langford, Panos Deloukas, Sarah Edkins, Sarah Hunt, Emma Gray, Serge Dronov, Leena Peltonen, Solveig Gretarsdottir, Gudmar Thorleifsson, Unnur Thorsteinsdottir, Kari Stefansson, Giorgio B. Boncoraglio, Eugenio A. Parati, John Attia, Elizabeth Holliday, Chris Levi, Maria-Grazia Franzosi, Anuj Goel, Anna Helgadottir, Jenefer M Blackwell, Elvira Bramon, Matthew A Brown, Juan P Casas, Aiden Corvin, Audrey Duncanson, Janusz Jankowski, Christopher G Mathew, Colin NA Palmer, Robert Plomin, Anna Rautanen, Stephen J Sawcer, Richard C Trembath, Ananth C Viswanathan, Nicholas W Wood, Bradford B. Worrall, Steven J Kittner, Braxton D Mitchell, Brett Kissela, James F. Meschia, Vincent Thijs, Arne Lindgren, Mary Joan Macleod, Agnieszka Slowik, Matthew Walters, Jonathan Rosand, Pankaj Sharma, Martin Farrall, Cathie LM Sudlow, Peter M Rothwell, Martin Dichgans, Peter Donnelly, Hugh S Markus
These authors contributed equally.
These authors jointly directed the study.
Footnotes
Author Contributions
SBe, CCAS, PS, MF, CLMS, PMR, MD, PDo and HSM designed the experiment. SBe, AG, AIB, CAJ, TJ, DP, LM, HS, CLMS, PMR, MD and HSM were responsible for collecting and phenotyping discovery samples.
Replication sample or replication data were provided PDS, JP, BN, RL, RS, LC, Y-CC, DW, MAN, US, HRA, SBo, DW-K, VV, JS, KF, HD, SG, GT, UT, KS, GBB, EAP, JA, EH, CL, M-GF, AH, BBW, SJK, BJM, BK, JFM, VT, AL, MJM, AS, MW, JR, PS
Genotyping, quality control and informatics were conducted by CB, SBe, CCAS, MP, MT, AS, ZS, GB, CF, RM, BM-M, CM, CL, SE, SH, EG, SD, AG, MF, PD, HSM.
Genetic and statistical analysis was performed by CB, SB, CCAS, MP, AS, ZS, GB, CF, MT, RM, AH, MF, and PDo.
The WTCCC2 management committee (PDo (Chair), LP (Deputy Chair), JMB, EB, MAB, JPC, AC, PDe, AD, JJ, HSM, CGM, CNAP, RP, AR, SJS, RCT, ACV, NWW) monitored the execution of the study. CB, SBe, CCAS, MP, MF, PD, HSM contributed to writing the first draft of the manuscript. All authors reviewed and commented on the final manuscript
No financial competing interests
References
- 1.Reducing brain damage: faster access to better stroke care. National Audit Office, Department of Health; London, UK: 2005. [Google Scholar]
- 2.Sacco RL, Ellenberg JH, Mohr JP, Tatemichi TK, Hier DB, Price TR, Wolf PA. Infarcts of undetermined cause: the NINCDS Stroke Data Bank. Ann Neurol. 1989;25:382–90. doi: 10.1002/ana.410250410. [DOI] [PubMed] [Google Scholar]
- 3.Dichgans M. Genetics of ischaemic stroke. Lancet Neurol. 2007;6:149–161. doi: 10.1016/S1474-4422(07)70028-5. [DOI] [PubMed] [Google Scholar]
- 4.Jerrard-Dunne P, Cloud G, Hassan A, Markus HS. Evaluating the genetic component of ischemic stroke subtypes: a family history study. Stroke. 2003;34:1364–1369. doi: 10.1161/01.STR.0000069723.17984.FD. [DOI] [PubMed] [Google Scholar]
- 5.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 6.Lemmens L, Buysschaert I, Geelen V, Fernandez-Cadenas I, Montaner J, et al. The Association of the 4q25 Susceptibility Variant for Atrial Fibrillation With Stroke Is Limited to Stroke of Cardioembolic Etiology. Stroke. 2010;41:1850–1857. doi: 10.1161/STROKEAHA.110.587980. [DOI] [PubMed] [Google Scholar]
- 7.Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, et al. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol. 2009;65:531–539. doi: 10.1002/ana.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markus HS. Genetics Studies in Ischaemic Stroke. Translational Stroke Research. 2010;1:238–245. doi: 10.1007/s12975-010-0041-5. [DOI] [PubMed] [Google Scholar]
- 9.Adams HP, Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 10.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Stroke Genetics Consortium. Wellcome Trust Case-Control Consortium 2 Failure to validate association between 12p13 variants and ischemic stroke. N Engl J Med. 2010;22:362–1547. doi: 10.1056/NEJMc0910050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsson S, Melander O, Jood K, Smith JG, Lövkvist H, et al. the International Stroke Genetics Consortium (ISGC) Genetic Variant on Chromosome 12p13 Does Not Show Association to Ischemic Stroke in 3 Swedish Case-Control Studies. Stroke. 2011;42:214–216. doi: 10.1161/STROKEAHA.110.594010. [DOI] [PubMed] [Google Scholar]
- 13.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, et al. Genomewide association studies of stroke. N Engl J Med. 2009;23:360–1718. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, Xiao ZS, Hou SQ, Zhao RT, Liu YF, et al. Strong association between the NINJ2 gene polymorphism and the susceptibility of stroke in Chinese Han population in Fangshan district. Beijing Da Xue Xue Bao. 2010;42:498–502. [PubMed] [Google Scholar]
- 15.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;5:466–707. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–76. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, et al. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2:151–63. [PubMed] [Google Scholar]
- 22.Langley B, Brochier C, Rivieccio MA. Targeting histone deacetylases as a multifaceted approach to treat the diverse outcomes of stroke. Stroke. 2009;40:2899–905. doi: 10.1161/STROKEAHA.108.540229. [DOI] [PubMed] [Google Scholar]
- 23.Kubo M, Hata J, Ninomiya T, Matsuda K, Yonemoto K, Nakano T, et al. A nonsynonymous SNP in PRKCH (protein kinase C η) increases the risk of cerebral infarction. Nat Genet. 2007;39:212–217. doi: 10.1038/ng1945. [DOI] [PubMed] [Google Scholar]
- 24.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo Y, Inouye M, Small K, Gwilliam R, Deloukas P, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–2746. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genetic Analysis of Psoriasis Consortium. the Wellcome Trust Case Control Consortium 2. Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The UK Parkinson’s Disease Consortium. The Wellcome Trust Case Control Consortium 2 Dissection of the genetics of Parkinson’s disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20:345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellenguez C, Strange A, Freeman C; Wellcome Trust Case Control Consortium,
- 29.Donnelly P, Spencer CC. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics. 2011 Nov 3; doi: 10.1093/bioinformatics/btr599. [Epub ahead of print] PubMed PMID: 22057162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browning B, Browning S. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Z, Cardin N, Wellcome Trust Case Control Consortium. Donnelly P, Marchini J. A Bayesian Method for Detecting and Characterizing Allelic Heterogeneity and Boosting Signals in Genome-Wide Association Studies. Statist. Sci. 2009;24:430–450. [Google Scholar]
URL section
- PROCARDIS www.procardis.org.
- SNPTEST http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html.
- NINDS Human Genetics Resource Center DNA and Cell Line Repository http://ccr.coriell.org/ninds.
- Biowulf Linux cluster at the National Institutes of Health Bethesda, Md. ( http://biowulf.nih.gov)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.