Abstract
BK polyomavirus (BKV) establishes persistent, low-level, and asymptomatic infections in most humans and causes polyomavirus-associated nephropathy (PVAN) and other pathologies in some individuals. The activation of BKV replication following kidney transplantation, leading to viruria, viremia, and, ultimately, PVAN, is associated with immune suppression as well as inflammation and stress from ischemia-reperfusion injury of the allograft, but the stimuli and molecular mechanisms leading to these pathologies are not well defined. The replication of BKV DNA in cell cultures is regulated by the viral noncoding control region (NCCR) comprising the core origin and flanking sequences, to which BKV T antigen (Tag), cellular proteins, and small regulatory RNAs bind. Six nuclear factor I (NFI) binding sites occur in sequences flanking the late side of the core origin (the enhancer) of the archetype virus, and their mutation, either individually or in toto, reduces BKV DNA replication when placed in competition with templates containing intact BKV NCCRs. NFI family members interacted with the helicase domain of BKV Tag in pulldown assays, suggesting that NFI helps recruit Tag to the viral core origin and may modulate its function. However, Tag may not be the sole target of the replication-modulatory activities of NFI: the NFIC/CTF1 isotype stimulates BKV template replication in vitro at low concentrations of DNA polymerase-α primase (Pol-primase), and the p58 subunit of Pol-primase associates with NFIC/CTF1, suggesting that NFI also recruits Pol-primase to the NCCR. These results suggest that NFI proteins (and the signaling pathways that target them) activate BKV replication and contribute to the consequent pathologies caused by acute infection.
INTRODUCTION
Human polyomavirus BK (BKV) persistently and asymptomatically infects approximately 80 to 90% of humans (25, 41). Kidneys are the major sites of replication, where BKV DNA is maintained at low levels (<0.01 copy/cell, on average) (20, 35) by the microRNA (miRNA)-mediated downregulation of the viral T antigen (Tag) (79) and the evasion of immune recognition (6). The activation of high levels of BKV replication in allografts occasionally occurs following kidney transplantation and can lead to viral titers exceeding 1,000 copies/cell (74), with concomitant viruria, viremia, and polyomavirus-associated nephropathy (PVAN), a major source of allograft loss. The causes of and mechanisms for the activation of viral DNA replication that occurs in the shift from persistent infection with low levels of replication to acute infection are not understood.
BKV replication in cell cultures is controlled by the viral noncoding control region (NCCR), within which the “core origin” (core-ori) serves as the initial binding site for the viral initiator-helicase protein, Tag, and small noncoding RNAs (21, 69, 84) (Fig. 1). Adjacent to the core-ori are the early flanking (EF) and the late flanking sequences (the “enhancer”), to which histones, cellular transcription factors, and perhaps also small noncoding RNAs bind and which control viral gene transcription and DNA replication (46, 52, 84, 85). The BKV archetype enhancer, comprised of four single-copy sequence blocks, termed P68, Q39, R63, and S63, rearranges by duplication, deletion, and insertion in late-stage PVAN or after passage in cell culture, providing a replication advantage and, perhaps, enhanced tropism (10, 30, 78).
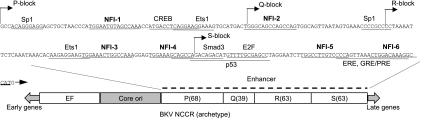
Fig 1.
BKV archetype NCCR. Shown is a schema of the BKV (Dik) NCCR with the sequence of the enhancer and predicted transcription factor binding sites; the six NFI sites are highlighted and numbered.
Binding sites for numerous cellular transcription factors, including nuclear factor I (NFI) (14–16, 22, 47), Sp1 (14, 22, 47), NFAT (40), AP1 (15, 22, 47), Smad3 (1), ERE and GRE/PRE (53), p53 (80), NF-κB (28), and C/EBP (28), have been identified in the archetype BKV enhancer and rearranged BKV variants, with experimental evidence supporting the importance of some of these sites for viral transcription and replication. Also, putative binding sites for Ets1, PEA3, AP-2, CREB, and granulocyte-macrophage colony-stimulating factor (GM-CSF) have been predicted by sequence homology (52, 75), but their functional importance is unproven. Notably, multiple NFI binding sites occur in the BKV archetype enhancer (Fig. 1) as well as in rearranged enhancers (14, 22, 47), suggesting that these sites may be functionally important. While some of these NFI sites regulate BKV early and late promoter activities (15, 16, 31, 42), the direct involvement of NFI sites in viral DNA replication has not been demonstrated.
NFI was originally identified as a cellular factor that stimulates adenovirus (Ad) DNA replication by recruiting the viral DNA polymerase to the viral origin of replication and distorting its structure (19, 62, 64). Subsequent studies indicated that NFI is a family of four isotypes, NFIA, NFIB, NFIC, and NFIX (or NFID), with almost identical N-terminal DNA binding/dimerization domains that bind to “TGGN5∼7GCCAA” sequences (32, 33). The expression pattern of NFI isotypes is cell type dependent and changes during differentiation and development (17, 43). NFI sites occur in many cellular promoters and enhancers as well as in viral genomes, including those of BKV (14–16, 22, 47), human JC virus (JCV) (55), variant murine polyomavirus (mPyV) (13, 86), human papillomavirus (HPV) (68), herpes simplex virus 1 (HSV-1) (44), and cytomegalovirus (CMV) (34). The functional importance of these NFI sites in regulating gene transcription is well established, but whether they also regulate DNA replication (other than adenoviral) has not been determined.
Here, we provide evidence for the functional importance of NFI for BKV archetype DNA replication: NFI sites placed proximal to the core-ori stimulate BKV DNA replication, and the mutation of NFI sites in the BKV enhancer diminishes replication in assays in which the mutant templates are in competition with the wild type (WT) for limiting factors. Furthermore, NFI interacts with two key replication proteins, BKV Tag and the p58 subunit of DNA polymerase-α primase (Pol-primase), as detected by in vitro pulldown assays and coimmunoprecipitation (co-IP) assays, and NFIC/CTF1 stimulates BKV DNA replication when Pol-primase is limiting in in vitro assays. We suggest that NFI and cognate signaling pathways help activate BKV replication and convert persistent infections into acute infections.
MATERIALS AND METHODS
Plasmids.
Test replication templates pUC-wt-BKV and pUC-Δen-BKV were generated by the insertion of PCR fragments of the intact BKV NCCR (positions 5031 to 282) and the NCCR without the enhancer (positions 5031 to 32) of the archetype Dik strain (GenBank accession number AB211369) into the XmaI/PstI sites of pUC18. pUC-6mtNFIs-BKV was generated by the ligation of a synthesized mutant BKV NCCR (GenScript) into the XmaI/PstI sites of pUC18. pUC-5mtNFIsW1-BKV, pUC-5mtNFIsW2-BKV, pUC-5mtNFIsW3-BKV, and pUC-5mtNFIsW6-BKV were derived from pUC-6mtNFIs-BKV by using the QuikChange site-directed mutagenesis kit (Stratagene). To construct test templates pUC-BKV-1fNFI, pUC-BKV-1rNFI, pUC-BKV-2rNFI, and pUC-BKV-4rNFI, the synthetic oligonucleotides 5′-CACATGGAATGTAGCCAAAACTGCA-3′ and 5′-GTTTTGGCTACATTCCATGTGTGCA-3′ (LNF1 consensus sites are underlined) were annealed, self-ligated, and inserted into the PstI site of pUC-Δen-BKV. Competition template pBC-wt-BKV was generated by the insertion of PCR fragments of the intact NCCR (positions 5031 to 282), enhancer (positions 33 to 282), and core origin (positions 5103 to 32) of BKV (Dik) into the XmaI/PstI, NotI/PstI, and NotI/XhoI sites of pBC-Sk(+). pBC-BKV-A89G, pBC-BKV-A143G, and pBC-BKV-A141T were mutated by using QuikChange site-directed mutagenesis.
The expression vector for BKV Tag, pCMV-BKTAg-Flag, lacking a simian virus 40 (SV40) origin, was described previously (46). NFI expression vectors pCH-NFIA, pCH-NFIB, pCH-NFIC, pCH-NFIX, and pCH-empty were kindly provided by Richard Gronostajski (18). pCH-hNFIC/CTF1 was generated by the insertion of the human NFIC/CTF1 cDNA (amplified by PCR from pCMV-CTF-1ΔUTR, kindly provided by Nicolas Mermod) (49), using the primers 5′-AGCTGGGCCCATGGATGAGTTCCAC-3′ and 5′-TTGCGCTAGCCTATCCCAGATACCAGGAC-3′, into the NheI/ApaI sites of the pCH-empty vector. The Pol-primase subunits were cloned into the expression vector pCMV with a T7 epitope tag at their N termini. The expression plasmids were kindly provided by E. Sock and M. Wegner (University of Erlangen—Nürnberg).
Bacterial expression vectors for truncated BKV Tag (pGEX3X-BKTHD, pGEX3X-BKTHDHR, and pGEX3X-BKTHR) were generated by the insertion of PCR-amplified fragments into the EcoRI/BamHI sites of pGEX3X. Primers used for PCR were BKTHD (5′-ATAGGATCCCAGGCTTAAAGGAGCATGATTTTAAC-3′ and 5′-CGGCCAATTCTTAATCAAGAATACATTTCCCCATG-3′), BKTHDHR (5′-ATAGGATCCCAGGCTTAAAGGAGCATGATTTTAAC-3′ and 5′-ACGCGAATTCTTATTTTGGGGGTGGTGTTTTAG-3′), and BKTHR (5′-ATATGGATCCCAATTACAAGAGAAGAGGATTCAG-3′ and 5′-ACGCGAATTCTTATTTTGGGGGTGGTGTTTTAG-3′).
Cell cultures and antibodies.
HK-2 human proximal tubular kidney cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone), 4 mM l-glutamine, 100 μg/ml penicillin, and 100 μg/ml streptomycin (Lonza). HEK293 cells and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (low glucose) (Invitrogen) supplemented with 10% fetal bovine serum (HyClone), 4 mM l-glutamine, 100 μg/ml penicillin, and 100 μg/ml streptomycin (Lonza). RPTECs (renal primary tubular cells) were cultured in renal epithelial growth medium with supplements (Lonza). All cells were grown at 37°C with 5% CO2 in a humidified incubator.
Antibodies used for Western blotting included anti-NFI (sc-870; Santa Cruz), anti-p53 (Cell Signaling), anti-c-Jun (sc-1694; Santa Cruz), anti-Sp1 (sc-59; Santa Cruz), anti-Ets1 (sc-111; Santa Cruz), anti-NF-κB (sc-372; Santa Cruz), anti-CREB (sc-58; Santa Cruz), anti-Smad3 (sc-8332; Santa Cruz), M2 anti-Flag (Sigma), anti-β-actin (sc-47778; Santa Cruz), anti-hemagglutinin (HA) (Roche), and anti-T7 (Novagen) antibodies. Polyclonal rabbit antiserum against recombinant human primase p58-p48 expressed in Escherichia coli cells was prepared and purified as previously described (87).
DNA replication assays.
DNA replication assays with transfected cells were performed as previously described (46, 84), with transfection procedures optimized by luciferase/β-galactosidase reporter assays. The in vitro monopolymerase assays were performed as previously described (46, 85), with the following slight modifications: the assay mixture included 0.5 μg of BKV template DNA, 50 ng topoisomerase I, 1 μg replication protein A (RPA), and Pol-primase (as indicated in the figure legends) in a solution containing 30 mM HEPES (pH 7.8); 7 mM magnesium acetate; 0.1 mM EGTA; 0.5 mM dithiothreitol (DTT); 200 μM each UTP, GTP, and CTP; 4 mM ATP; 100 μM each dATP, dGTP, and dTTP; 10 μM dCTP; 40 mM creatine phosphate; 1 μg creatine kinase; 0.1 mg/ml heat-treated bovine serum albumin (BSA); and 5 μCi [α-32P]dCTP (3,000 Ci/mmol; Perkin-Elmer) in 40 μl. The amount of added NFI protein is indicated in the figure legends. Purified BKV Tag (0.4 μg) was added to start the reaction, and after incubation for 60 min at 37°C, the reaction products were precipitated with cold 10% (wt/vol) trichloroacetic acid (TCA) containing 2.5% (wt/vol) sodium pyrophosphate, spotted onto glass fiber filters (GF/C; Whatman), washed with 1 M HCl, and analyzed by scintillation counting.
Nuclear extracts.
HEK293 cells from 10 liters of a suspension culture were purchased from The National Cell Culture Center (NCCC) (Minneapolis, MN). Cell pellets were washed in 5 packed cell volumes (PCV) of a solution containing ice-cold hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM DTT) and centrifuged in a Beckman GH-3.7 rotor at 3,000 rpm for 5 min at 4°C. Collected cells were resuspended in 3 PCV of ice-cold hypotonic buffer and allowed to swell on ice for 10 min; swollen cells were transferred into a chilled glass Dounce tissue grinder, homogenized with 10 to 15 strokes using a type B pestle, and centrifuged in a Beckman GH-3.7 rotor at 3,500 rpm for 15 min at 4°C; the supernatants were removed; the pellets were resuspended in 1 packed nuclear volume (PNV) of low-salt buffer (20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT); and 0.183 PNV of 5 M NaCl was added dropwise to the resuspended nuclei while mixing gently by swirling, placed onto a rotating platform for 30 min at 4°C, and centrifuged at 16,000 rpm (37,000 × g) in a Sorvall SA-600 rotor for 1 h at 4°C. The supernatants were collected and dialyzed in dialysis buffer (20 mM HEPES [pH 7.9], 20% glycerol, 5 mM NaCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT). The protein concentration of the extracts was adjusted to 1.4 μg/μl, determined by a Bradford protein assay (9a). Small-scale preparations of HK-2 cell, HeLa cell, RPTEC, and HEK293 cell nuclear extracts for Western blotting were made by using nuclear extract kits (Active Motif).
Competitive EMSAs.
Competitive electrophoretic mobility shift assays (EMSAs) were performed according to previously described procedures (11), with the following modifications: 4 pmol competitor oligonucleotides, 2 μg of HeLa cell nuclear extracts (sc-2120; Santa Cruz) or 0.25 μg of purified human NFIC (hNFIC)/CTF1 (Abcam), and 50 ng of poly(dI · dC) were incubated in 1× HEPES binding buffer (25 mM HEPES [pH 7.5], 6 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50 mM NaCl, 0.5 mM PMSF, 5% glycerol) for 10 min at room temperature (RT); 20 fmol biotin-labeled probe for the NFI site was then added to the reaction mixture and incubated for 20 min at room temperature. For antibody supershift EMSAs, 1 μl of NFI antibody (sc-5567; Santa Cruz) was added after the binding reaction, and the mixture was incubated for another 10 min at room temperature. The reaction products were fractionated in 5% nondenaturing polyacrylamide gels (prerun in 0.5× Tris-borate-EDTA [TBE] at 100 V for 30 min) in 0.5× TBE at 100 V until the bromophenol blue dye reached the bottom of the gel. The products were transferred onto nylon membranes in 0.5× TBE at 65 V for 30 min, UV cross-linked for 15 min, and visualized by chemiluminescent nucleic acid detection (Pierce).
In vitro pulldown assays.
BKV Tag with an N-terminal Flag epitope was expressed by using the Bac-to-Bac baculovirus system (Invitrogen) as follows: 1.5 × 107 infected Hi-Five insect cells were harvested at 48 h postinfection (p.i.) and lysed in 1 ml of 0.5% NP-40 lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 5 mM KCl, 1.0 mM MgCl2, 0.5% NP-40, 10% glycerol, 1× PhosSTOP phosphatase inhibitors [Roche], 1× complete protease inhibitor cocktail [Roche]) by incubation on a rotating platform at 4°C for 30 min and homogenization with a glass Dounce tissue grinder. Lysates were cleared by centrifugation at 20,000 × g in a Sorvall SA-600 rotor for 30 min at 4°C. Supernatants were incubated with 60 μl of an anti-Flag M2 affinity gel (Sigma) by rotation at 4°C for 2 h and washed three times with 1 ml of ice-cold phosphate-buffered saline (PBS); each of the gel suspensions was then incubated with extracts of infected and uninfected Hi-Five cells and then incubated with 1 ml of HEK293 cell nuclear extracts (1.4 μg/μl) plus 1× complete protease inhibitor cocktail (Roche) by rotation at 4°C for 12 h. The gel was washed three times with 1 ml of ice-cold PBS and boiled for 5 min with 50 μl of 1× SDS sample buffer.
Glutathione S-transferase (GST)-truncated BKV Tag proteins were expressed in E. coli Rosetta 2 cells (Novagen) cultured in LB medium on a shaking platform at 225 rpm at 25°C. Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the A600 reached 0.4 to 0.5. After induction, E. coli cells were cultured at 25°C with shaking at 225 rpm for 20 h and then sonicated (twice for 300 s each with a 20% duty cycle at maximum power) in L1 buffer (50 mM Tris-Cl [pH 8.0], 250 mM NaCl, 1 mM DTT, 10% glycerol, 1 mM PMSF, 1× complete protease inhibitor cocktail [Roche]). NP-40 (0.1%) was added to the lysates after sonication and incubated on ice for 10 min. After centrifugation at 20,000 × g in a Sorvall SA-600 rotor for 30 min at 4°C, supernatants were incubated with glutathione beads (GE Healthcare) at 4°C for 2 h, followed by washing 2 times with 20 bed volumes of L1 buffer. A small portion (1/20) of the beads for each fusion protein was boiled with SDS sample buffer, and the quantity of bound fusion proteins was determined by SDS-PAGE and Coomassie blue staining. Beads with approximately equal amounts of each fusion protein (∼50 μg) were incubated with 800 μl of HEK293 cell nuclear extract (1.4 μg/μl) supplemented with 1× complete protease inhibitor cocktail (Roche) by rotation at 4°C for 12 h. The beads were washed 5 times with 1 ml of L1 buffer and then boiled for 5 min with 100 μl 1× SDS sample buffer. All samples were fractionated by SDS-PAGE and analyzed by Coomassie blue staining, followed by Western blotting.
Coimmunoprecipitation assays.
HEK293 cells (approximately 6 × 106 cells) were transfected with expression vectors by using LipofectAMINE and Plus reagent (Invitrogen) in 100-mm-diameter plates as previously described (46). At 48 h, the cells were lysed with 750 μl of a solution containing 1% Triton lysis buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 30 μM ethidium bromide, 1 mM PMSF, complete protease inhibitor cocktail [Roche], 1× PhosSTOP phosphatase inhibitors [Roche]) by rotation for 1 h at 4°C, and lysates were cleared by centrifugation at 20,000 × g in a Sorvall SA-600 rotor for 30 min at 4°C. A sample (10 μl) was taken for the protein input control, and the remaining supernatants were incubated with 50 μl of an anti-Flag M2 affinity gel (Sigma) or 50 μl of an anti-HA affinity matrix (Roche) by rotation at 4°C for 2 h, followed by four washes with 1 ml of 1% Triton lysis buffer. Following the last wash, pelleted beads were suspended in 40 μl of 1× SDS sample buffer and boiled for 5 min. Samples of 20 μl of the supernatant were analyzed by SDS-PAGE and Western blotting.
RESULTS
Synthetic NFI sites proximal to the core origin stimulate BKV DNA replication in HK-2 cells.
Six putative NFI sites occur in the BKV archetype enhancer (Dik strain) (Fig. 1), and EMSAs were employed to evaluate NFI binding to these sites. As a positive control for NFI protein-oligonucleotide complexes, an EMSA was performed with a biotin-labeled oligonucleotide with the sequence of the NFI site in the adenovirus inverted terminal repeat (ITR) (45). As expected, a shift of the migration of the oligonucleotide caused by NFI binding was observed in the absence of a competitor (Fig. 2A, lanes 1 and 6), and the addition of a 200× excess of the unlabeled oligonucleotide fully competed for NFI binding (Fig. 2A, lanes 2 and 7); however, an oligonucleotide with a point mutation in the NFI site did not alter the band shift (Fig. 2A, lane 3). The addition of an anti-NFI antibody to the binding reaction mixture caused a supershift (Fig. 2A, lane 4, dashed arrow), confirming that the band shift was caused by NFI. An excess of BKV enhancer DNA also competed for NFI binding (Fig. 2A, lane 5), confirming that the BKV archetype enhancer contains functional NFI sites. Oligonucleotides with sequences of the six NFI binding sites in the BKV archetype enhancer (Fig. 1 and 3A) competed for NFI binding with different efficiencies (Fig. 2A, lanes 8 to 13), suggesting that these sites have different affinities for NFI proteins. Oligonucleotides with sequences of NFI sites closer to the BKV core-ori competed more efficiently than did those with distal sites.
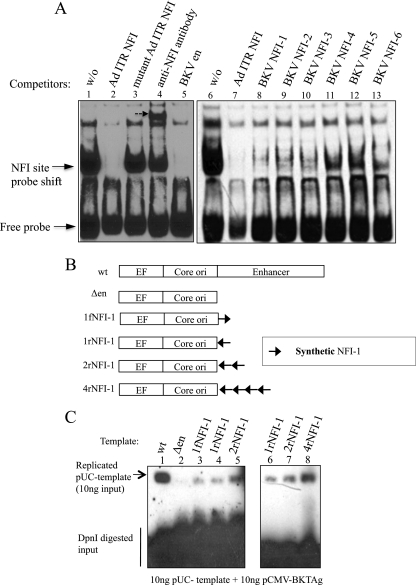
Fig 2.
EMSA of NFI sites and stimulation of BKV DNA replication by synthetic NFI sites. (A) EMSA with the NFI site in the Ad ITR as a probe. Lanes 1 and 6, absence of a competitor; lanes 2 and 7, 200× molar excess of oligonucleotide with the NFI site (Ad ITR) as a competitor; lane 3, 200× molar excess of oligonucleotide with the mutant NFI site as a competitor; lane 4, incubation with anti-NFI antibody for 10 min after the binding reaction; lane 5, 200× molar excess of PCR products of the BKV enhancer (en) as a competitor; lanes 8 to 13, 200× molar excess of oligonucleotides with NFI sites in the BKV enhancer as competitors, as indicated. Positions of the free probe and shifted probe are indicated by arrows. (B) Design of BKV templates with synthetic NFI sites in place of the enhancer. (C) Southern blotting of DNA replication in HK-2 cells of BKV templates containing synthetic NFI sites. A total of 10 ng BKV template and 10 ng pCMV-BKTAg-Flag were transfected into HK-2 cells with carrier DNA; at 48 h, cells were harvested, and DpnI-resistant, replicated DNA was analyzed by Southern blotting.
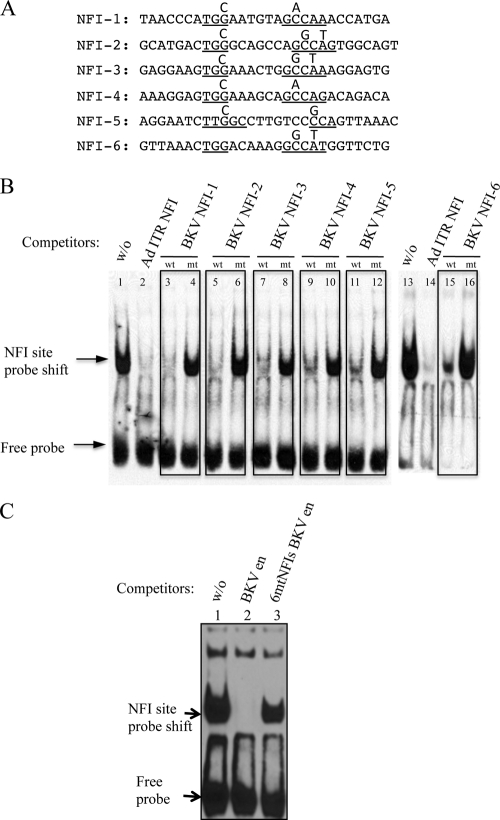
Fig 3.
Characterization of mutated NFI sites in the BKV enhancer. (A) Sequences of oligonucleotides with mutated NFI sites. NFI consensus motifs are underlined, with substitutions indicated above the mutated positions. (B) Competitive EMSA with wild-type (wt) NFI and mutant (mt) NFI sites in the BKV enhancer. (C) EMSA with the mutant BKV enhancer containing six mutated NFI sites (6mtNFIs). Lane 1, absence of a competitor; lane 2, wild-type BKV enhancer PCR fragment as a competitor; lane 3, 6mtNFIs mutant BKV enhancer PCR fragment as a competitor (200× molar excess of oligonucleotide, equivalent to 4 pmol).
Sequences corresponding to the first NFI site in the P block (Fig. 1) in different orientations and copy numbers were substituted for enhancer sequences (Fig. 2B) and assayed for Tag-dependent DNA replication in HK-2 cells (Fig. 2C, lanes 2 to 5). The addition of synthetic NFI sites to templates lacking the enhancer stimulated their replication, with the extent correlating with the number of sites (Fig. 2C, lanes 6 to 8) but not their orientations (Fig. 2C, lanes 3 and 4). The replication of templates containing multiple synthetic NFI sites was always weaker than the replication of templates with the archetype enhancer (Fig. 2C, lanes 1, 5, 7, and 8), suggesting that other elements also stimulate BKV DNA replication in these cells or that a particular spatial configuration of NFI sites is required.
Mutation of the BKV NCCR NFI sites reduces NFI binding and DNA replication.
Point mutations were introduced into the consensus sequence “TGGN5∼7GCCAA” of each of the six NFI sites in the BKV enhancer, with the design of the mutations confirmed with MatInspector (72) to ensure that no new transcription factor binding sites were created (Fig. 3A). Oligonucleotides with mutated NFI sites were demonstrably defective for NFI binding in competitive EMSAs (Fig. 3B). A 200-fold excess of the BKV enhancer DNA with mutations in all six NFI sites (6mtNFIs BK enhancer) only slightly reduced the NFI binding to the adenovirus ITR oligonucleotide (Fig. 3C, compare lanes 1 and 3), whereas the wild-type (WT) enhancer sequence completely abolished NFI binding (Fig. 3C, lane 2). These results indicate that the mutation of the six identified NFI sites greatly reduces NFI binding to the BKV archetype enhancer.
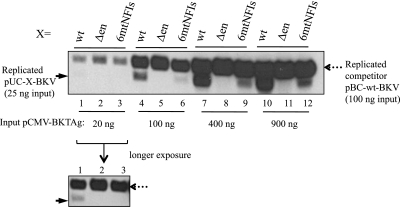
Assays of the replication of BKV templates containing mutations of all six NFI sites (pUC-6mtNFIs-BKV) compared with the replication of the wild-type template (pUC-wt-BKV) in HK-2 cells transfected with each template separately indicated that both templates replicated with similar efficiencies (data not shown). However, the use of a competitive assay in which a competitor (3.4-kb) template, pBC-wt-BKV, containing the archetype NCCR was cotransfected into HK-2 cells together with test mutant templates (2.6 kb) (Fig. 4A) and a Tag expression vector (pCMV-BKT-Flag), revealed that the level of replication of the test templates with the enhancer containing six mutant NFI sites (pUC-6mtNFIs-BKV) was greatly reduced (Fig. 4B, compare lane 3 with lane 1), as was also observed for a test template without the enhancer (Fig. 4B, lane 2). Test templates with mutations in Ets1 binding sites in the enhancer (pUC-mtEts1s-BKV) replicated with an efficiency similar to that of the wild-type template (Fig. 4B, compare lane 5 with lane 1), indicating that the effect of the mutations on DNA replication in the competitive assay is attributable to the NFI site.
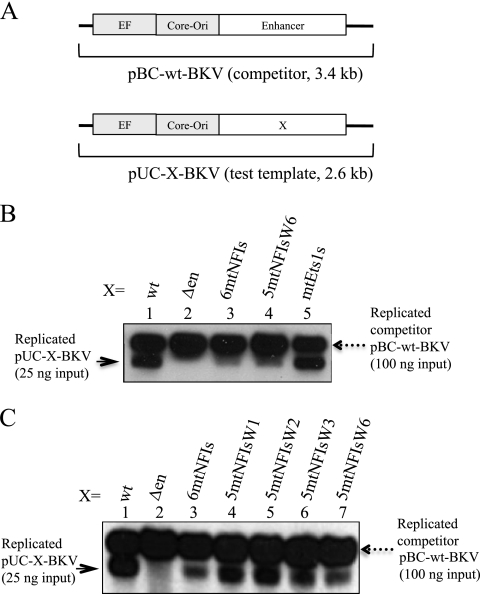
Fig 4.
Competitive DNA replication assays. The BKV test template (25 ng), 100 ng pBC-wt-BKV competitor, and 25 ng pCMV-BKTAg-Flag were transfected into HK-2 cells with carrier DNA; at 48 h posttransfection, cells were harvested, and DNA replication was analyzed by Southern blotting. (A) Schema of the BKV test template (X indicates variable sequences as specified) and the pBC-wt-BKV competitor. (B) Lane 1, wild-type BKV test template (pUC-wt-BKV); lane 2, enhancer-deleted BKV test template (pUC-Δen-BKV); lane 3, mutant BKV template with six mutant NFI sites (pUC-6mtNFIs-BKV); lane 4, mutant BKV template with five mutant NFI sites (5mtNFIs) and a sixth wild-type NFI site (W6) (pUC-5mtNFIsW6-BKV); lane 5, Ets1 site mutant BKV template (pUC-mtEts1s-BKV). (C) Lane 1, wild-type BKV test template (pUC-wt-BKV); lane 2, enhancer-deleted BKV test template (pUC-Δen-BKV); lane 3, 6mtNFIs mutant BKV template (pUC-6mtNFIs-BKV); lane 4, 5mtNFIs mutant BKV template with a wild-type NFI-1 site (W1) (pUC-5mtNFIsW1-BKV); lane 5, 5mtNFIs mutant BKV template with a wild-type NFI-2 site (W2) (pUC-5mtNFIsW2-BKV); lane 6, 5mtNFIs mutant BKV template with a wild-type NFI-3 site (W3) (pUC-5mtNFIsW3-BKV); lane 7, 5mtNFIs mutant BKV template with a wild-type NFI-6 site (W6) (pUC-5mtNFIsW6-BKV). The solid arrow at the left indicates the position of the replicated test template; the dashed arrow at the right indicates the position of the replicated competitor template.
To determine whether a specific NFI site is responsible for the stimulatory effect on DNA replication, pUC-6mtNFIs-BKV templates with single NFI sites reverted to the WT were assayed for replication. Wild-type NFI sites 1 and 2 (closer to core-ori) appeared to be more stimulatory for BKV DNA replication than NFI sites 3 and 6 (Fig. 4C, lanes 3 to 7).
The requirement of a competitor template to observe the stimulatory effect of NFI sites on BKV DNA replication might be explained by the recruitment of factors required for DNA replication that are made limiting by use of the competitor template. The observation that NFI sites closer to the core-ori are more stimulatory for replication than are sites distal to the core-ori suggests that NFI may target components of the initiation complex, such as Tag, Pol-primase, replication protein A (RPA), or topoisomerase I. Evidence favoring this notion is provided by the experiments described below.
NFI interacts with BKV Tag.
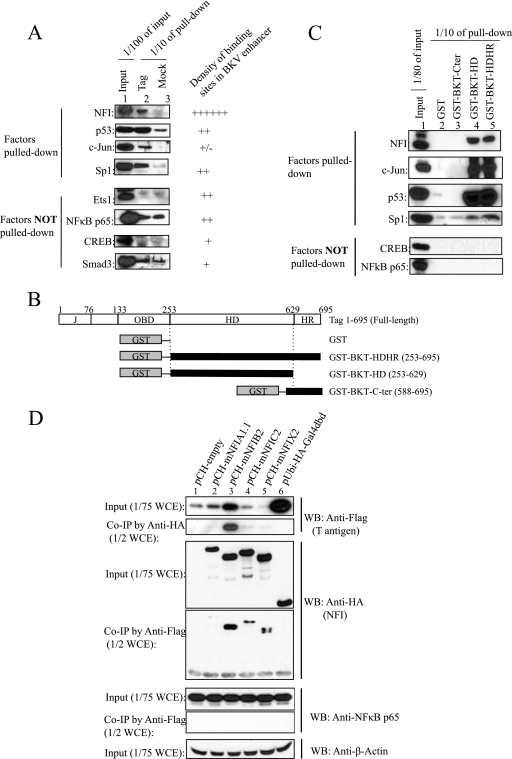
We tested whether NFI interacts with BKV Tag by antibody pulldown assays. Full-length Flag-tagged BKV Tag (BKT-Flag) was mixed with HEK293 nuclear extracts, protein complexes were collected by using Flag antibody resin, and associated candidate proteins were detected with specific antibodies to each one (Fig. 5A). Initially, a pan-NFI antibody recognizing all NFI isotypes was used in the pulldown assays; other antibodies also detected the association of BKV Tag with p53, Sp1, and c-Jun but not Ets1, CREB, NF-κB p65, or Smad3 (Fig. 5A). Domains of BKV Tag that interact with these transcription factors were determined with GST-tagged truncated BKV Tags (Fig. 5B): the BKV Tag helicase/ATPase domain (HD) pulled down NFI, p53, c-Jun strongly, and Sp1 weakly but not CREB or NF-κB (Fig. 5C, lanes 4 and 5), indicating that NFI, p53, and c-Jun interact with the helicase/ATPase domain, while Sp1 may also interact with other Tag domains in addition to the HD. The latter observations are consistent with previous reports that BKV Tag and SV40 Tag complex with p53 (80), the SV40 Tag origin binding domain (OBD) interacts with Sp1 (39), and SV40 interacts with c-Jun (7). None of the transcription factors tested appeared to interact with the BKV Tag C-terminal region (Fig. 5C, lane 3). Truncated BKV Tags did not pull down factors not observed to complex with full-length BKV Tag, including Ets1, NF-κB, CREB, and Smad3 (Fig. 5A and C and data not shown).
Fig 5.
Interaction of BKV Tag with NFI. (A) Flag antibody pulldown assays to identify transcription factors interacting with BKV Tag. Lane 1, input of HEK293 cell nuclear extracts (1/100 of the total input); lane 2, BKV full-length Tag pulled-down fraction (1/10 of the total pulldown fraction); lane 3, mock pulldown fraction in the absence of Tag (1/10 of the total pulldown fraction). The number of specific transcription factor binding sites in the BKV archetype enhancer is indicated by the number of pluses; “+/−” indicates a partially conserved site. NA, not available. (B) Schema of truncation mutants of BKV Tag. (C) GST affinity pulldown assays to determine the interaction of specific Tag truncation mutants with transcription factors. Lane 1, HEK293 nuclear extract input (1/80 of the total input); lane 2, GST affinity pulldown fraction control; lane 3, GST-tagged BKT C-terminal region (Cter) pulldown fraction; lane 4, GST-tagged BKT helicase/ATPase domain (HD) pulldown fraction; lane 5, GST-tagged BKV Tag helicase and host range domain (HDHR) pulldown fraction. The loading of lanes 2 to 5 is 1/10 of the total pulled-down fraction. (D) Coimmunoprecipitation (co-IP) assays to detect the interaction of BKV Tag with different NFI isotypes. A BKV Tag expression vector (pCMV-BKTAg-Flag) was cotransfected with expression vectors for NFI isotypes (pCH-mNFIA, -mNFIB, -mNFIC, -mNFIX, or -empty) or a control vector expressing the Gal4 DNA binding domain (pUbi-HA-Gal4dbd). HA-tagged NFI isotypes and Flag-tagged Tag were detected with anti-HA and anti-Flag antibodies, respectively. NF-κB p65 was the internal negative control; β-actin was the loading control.
Isotype-specific antibodies directed against NFIA, NFIB, and NFIC were used to attempt to distinguish isotype-specific interactions with BKV Tag, but only NFIA was detected at a high level in the HEK293 cell extracts and was pulled down by Tag (data not shown). To determine other isotypes of NFI that interact with BKV Tag, HA-tagged NFI isotypes and Flag-tagged BKV Tag were overexpressed in HEK293 cells, and their interactions with Tag were assessed by co-IP assays using either anti-HA or anti-Flag antibodies (Fig. 5D). Four HA-tagged NFI isotypes were expressed at similar levels (Fig. 5D); however, NFIB strongly upregulated the expression of Tag driven by the CMV promoter (Fig. 5D, lane 3), leading to a higher level of coprecipitation of Tag with NFIB than with other NFI isotypes. In contrast, HA-NFIA only weakly coprecipitated with Tag (Fig. 5D, lane 2), which might be due to a high level of endogenous NFIA competing against the transiently expressed NFIA in an interaction with Tag (Fig. 5D, lane 2, and data not shown). In control assays, no interaction between Tag and the HA-tagged Gal4 DNA binding domain (Gal4dbd) was detected, even though Tag was expressed at an extremely high level (Fig. 5D, lane 6), and no interaction of Tag with endogenous NF-κB p65 was detected (Fig. 5D). These results are in agreement with data from in vitro pulldown assays (Fig. 5A and C), indicating a specific interaction of BKV Tag with HA-tagged NFI isotypes.
Increased Tag expression partially rescues the replication deficiency of BKV templates with mutant NFI sites.
The interaction of BKV Tag with NFI provides a possible basis for the stimulatory effect upon replication by NFI sites in the enhancer. Initially, BKV Tag was expressed at a low level in the competitive DNA replication assays (Fig. 4B); to investigate if BKV Tag is the only limiting factor in these assays, increasing amounts of a pCMV-BKV Tag expression vector were introduced into HK-2 cells, and the levels of DNA replication of wild-type and mutant (NFI site) templates were compared (Fig. 6). BKV Tag expressed from 400 ng of pCMV-BKTAg saturated replication (Fig. 6, lane 7 to 9), but this amount, and even the addition of 900 ng pCMV-BKTAg, only partially rescued the replication of the template with the mutant NFI sites (pUC-6mtNFIs-BKV), compared with the robust replication of the wild-type template (pUC-wt-BKV) (Fig. 6, compare lane 9 with land 7 and compare lane 12 with lane 10). Furthermore, templates with a deleted enhancer (pUC-Δen-BKV) replicated poorly, even when BKV Tag was highly expressed (Fig. 6, lanes 8 and 11). These observations indicate that high levels of BKV Tag alone cannot correct the replication deficiency of templates with mutant NFI sites and suggest that BKV Tag is not the sole limiting factor in these replication assays.
Fig 6.
DNA replication assays with wild-type BKV (pBC-wt-BKV) as a competitor and variable levels of BKV Tag. Twenty-five nanograms of pUC templates, 100 ng of the pBC-wt-BKV competitor, and increasing amounts of pCMV-BKT-Flag were transfected into HK-2 cells with carrier DNA. At 48 h posttransfection, cells were harvested, and DNA replication was analyzed by Southern blotting. The amount of the transfected BKV Tag expression vector pCMV-BKTAg-Flag was increased from 20 ng to 900 ng. Three test templates (wild-type BKV template [pUC-wt-BKV], enhancer deletion BKV template [pUC-Δen-BKV], and 6mtNFIs BKV template [pUC-6mtNFIs-BKV]) are indicated. Pictures of replication assays are presented in the first three lanes, with a picture of the longer exposure shown below the main panel. The solid arrow indicates the replicated test template; the dashed arrow indicates the replicated competitor template.
NFIC/CTF1 stimulates BKV DNA replication in vitro when DNA polymerase-α primase is limiting.
BKV Tag interacts with different NFI isotypes in co-IP assays (Fig. 5D), but their importance for BKV replication is difficult to test in vivo due to the concurrent expression of multiple endogenous NFI isotypes. The NFIC/CTF1 isotype stimulates the initiation of adenovirus DNA replication by recruiting adenovirus DNA polymerase to the origin of replication (9, 19), and the proline-rich transactivation domain of the NFIC/CTF1 isotype stimulates SV40 DNA replication when tethered to the SV40 origin (61). These observations prompted us to assess whether NFIC/CTF1 can stimulate BKV DNA replication in the monopolymerase replication system that contains Pol-primase, RPA, and topoisomerase I (46, 85) in the absence of other cellular factors.
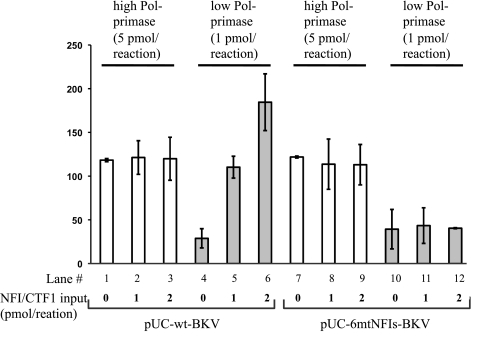
When Pol-primase was made limiting in the monopolymerase system, the DNA replication of the wild-type BKV template was stimulated strongly by NFIC/CTF1 in a concentration-dependent manner (Fig. 7, lanes 4 to 6). In contrast, with high levels of Pol-primase, the addition of NFIC/CTF1 had no effect on the replication of the wild-type BKV template (Fig. 7, lanes 1 to 3). Furthermore, no stimulation was observed with the NFI binding-site mutant BKV template regardless of the Pol-primase levels (Fig. 7, lanes 7 to 12). These findings indicate that NFIC/CTF1 stimulates the initiation of BKV DNA replication in vitro only when Pol-primase is limiting and suggest that Pol-primase is a limiting factor targeted by NFI to stimulate BKV DNA replication in vivo in the competitive replication assays.
Fig 7.
Monopolymerase DNA replication assays. DNA replication assays of the wild-type BKV template (pUC-wt-BKV) and mutant NFI site BKV template (pUC-6mtNFIs-BKV) were carried out with high levels (white columns) (5 pmol) and low levels (gray columns) (1 pmol) of purified Pol-primase and variable amounts of purified NFIC/CTF1 (0, 1, and 2 pmol), as indicated.
NFIC/CTF1 interacts with the cellular DNA polymerase-α primase p58 subunit.
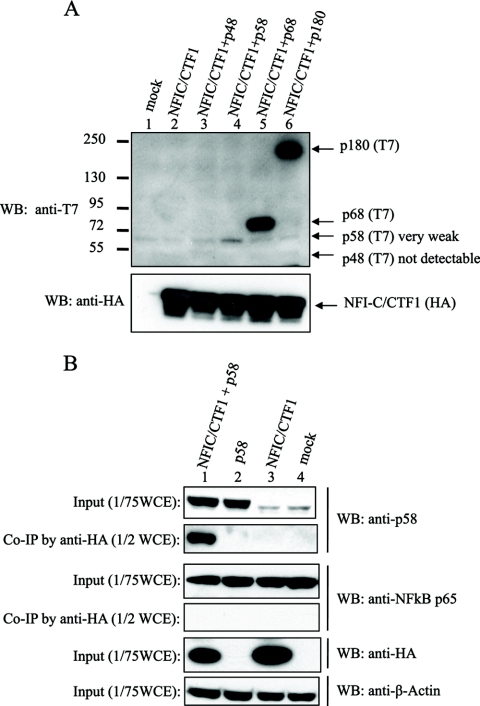
To further investigate the stimulation of replication by NFI, we studied whether it and Pol-primase proteins interact. Pol-primase consists of four subunits: two smaller subunits, p48 and p58, constitute the primase, and two larger subunits, p180 and p68, constitute the DNA polymerase-α catalytic and regulatory subunit p68, respectively (50, 51, 65, 82). HA-tagged human NFIC/CTF1 was ectopically expressed with each of four T7-tagged human Pol-primase subunits in HEK293 cells (Fig. 8A), and their interaction was examined by co-IP assays. The expression of p180 and p68 subunits was much more efficient than the expression of the primase subunits (as detected with the anti-T7 antibody) (Fig. 8A, lanes 3 to 6), but no interaction between NFIC/CTF1 and either the p180 or p68 subunits was detected (data not shown). Because the p58 expression level was low (Fig. 8A, lane 4), a more sensitive antibody against p58 was used, instead of the anti-T7 antibody, for the detection of p58 (Fig. 8B). In this assay, p58 was found to coprecipitate with NFIC/CTF1 (Fig. 8B, lane 1), and p58 co-IP was not detected in three control experiments, in which only p58 (Fig. 8B, lane 2), only NFIC/CTF1 (Fig. 8B, lane 3), or neither (Fig. 8B, lane 4) was expressed. No other significant distinct band was detected in the coprecipitated fraction. Unfortunately, despite several attempts, the level of expression of p48 was extremely low. An interaction between p48 and NFIC/CTF1 was not detected (Fig. 8A, lane 3, and data not shown), but we cannot exclude that p48 might bind to NFI.
Fig 8.
Interaction of NFI with Pol-primase. (A) NFIC/CTF1 and each subunit of Pol-primase were coexpressed in HEK293 cells as indicated. Lane 1, mock transfection; lane 2, NFIC/CTF1 alone; lane 3, NFIC and p48; lane 4, NFIC/CTF1 and p58; lane 5, NFIC/CTF1 and p68; lane 6, NFIC/CTF1 and p180. The expressions of NFIC/CTF1 and subunits of Pol-primase were detected by Western blotting using anti-T7 tag and anti-HA tag antibodies, respectively. (B) NFIC/CTF1 and p58 subunits of Pol-primase were coexpressed in HEK293 cells as indicated. Lane 1, NFIC/CTF1 and the p58 subunit; lane 2, the p58 subunit alone; lane 3, NFIC/CTF1 alone; lane 4, blank control; specific antibodies for the Western blots (WB) are indicated beside each panel. The p58 subunit was detected with a rabbit polyclonal anti-p58 antibody, the expression of NFIC/CTF1 was detected with an anti-HA antibody, NF-κB p65 was the internal negative control, and β-actin was the loading control.
DISCUSSION
Almost 2 decades after the etiology of PVAN was linked to acute BKV replication, the cause of the activation of BKV replication in kidney allografts still remains elusive. Immune suppression is associated with the activation of BKV replication (2, 26, 89), and stress-related injury, repair, regeneration, and differentiation are also associated (27, 37), but the responsible mechanisms have not been defined.
We have determined that NFI binds to six NFI sites in the BKV archetype NCCR and stimulates DNA replication. NFI sites in P24–37 (NFI-1) and at the P68-Q13 junction (NFI-2), proximal to the core-ori, appear to have a higher affinity for NFI and also stronger stimulatory effects on BKV DNA replication than distal sites. Almost all rearranged viruses contain the P block and P-Q junction region spanning these two NFI sites (30, 54, 70, 71, 75), supporting the notion that they might be particularly important for efficient viral replication in vivo. Other NFI sites have been implicated in the early-late transcription switch (NFI-3) (42), the regulation of viral gene transcription in response to the induction of transforming growth factor β (TGF-β) in kidney allografts (NFI-4) (1), and the modulation of the hormone-mediated stimulation of BKV replication (NFI-5 and -6) (53).
We attempted to distinguish the binding of different NFI isotypes to NFI sites using isotype-specific antibodies. Unfortunately, none of the available isotype-specific antibodies work in EMSAs. As NFI isotypes have an identical DNA binding domain that determines the specificity of binding to consensus sites, we speculate that the different NFI isotypes have similar binding affinities in vitro. However, the binding activity of NFI isotypes in vivo is likely affected by their interaction and/or competition with other transcription factors (66, 73, 77) and is challenging to demonstrate with in vitro assays using purified proteins.
The topography of NFI sites in the archetype BKV enhancer resembles that of NFI sites in the archetype JCV enhancer, except that JCV lacks the NFI-4 site overlapping the Smad3 site (48). The NFI site closest to the core origin of JCV also stimulates JCV DNA replication in vivo (81). JCV also persistently infects the kidney, and the reactivation of JCV in immunocompromised individuals causes progressive multifocal leukoencephalopathy (PML) (88). Previous analyses of rearranged JCV enhancers in PML patients also revealed a trend similar to that for rearranged BKV enhancers in PVAN: sequences close to the core origin (A to C for JCV and P and the P-Q junction for BKV), which contain the first two NFI sites (NFI-1 and NFI-2), are usually preserved and duplicated (29, 30). NFI isotype-specific expression determines the tropism of JCV (55, 76), but functions for different NFI isotypes in replication have not been identified.
A characterization of the NFI isotype-specific function for BKV DNA replication has been attempted with an in vivo replication system, but the results are complicated by the endogenous expression of different NFI isotypes/splicing variants (data not shown). Using the in vitro monopolymerase assay, we have defined the stimulatory activity of the NFIC/CTF1 isotype, the prototype of the NFI family of transcription factors, on the initiation of BKV DNA replication (Fig. 7). The role of other NFI isotypes in DNA replication will be tested with similar systems as purified NFI isotype proteins and antibodies become available.
As NFI stimulates adenovirus (Ad2/5) DNA replication through the recruitment of the Ad pol-pTP (adenovirus DNA polymerase-preterminal protein) complex to the replication origin (9, 19, 60) and/or the stabilization of the preinitiation complex (59), we suggest that similar mechanisms promote BKV DNA replication via the recruitment of BKV Tag and Pol-primase to the replication origin. In support of this, we observed that NFI stimulated the initiation of BKV DNA replication in vitro only at low concentrations of Pol-primase and NFI, whereas at high concentrations of Pol-primase, no stimulation was observed (Fig. 7), which is reminiscent of the stimulation of adenovirus DNA replication in vitro by NFI (60) and which is also consistent with the results of our in vivo competitive replication assays. Furthermore, NFI forms complexes with BKV Tag and Pol-primase; swine NFI also binds to calf primase and stimulates primase activity in a concentration-dependent manner in biochemical assays (24). All four NFI isotypes were found to interact with BKV Tag in co-IP assays (Fig. 5D).
These results suggest that the formation of NFI-primase and NFI-Tag complexes is important for the initiation of DNA replication, but elucidating the functions of individual complexes is challenging due to the coexpression of different NFI isotypes/splicing variants. Further studies are required to characterize the nature and functions of these interactions.
Although the NFI sites in the BKV NCCR are not required for BKV DNA replication in the absence of a competitor template, NFI sites in the enhancer stimulate BKV DNA replication when Tag or Pol-primase is limiting. This stimulatory activity might be essential in persistent infections, where BKV replicates at very low levels in kidney tubular epithelial cells when low levels of Tag and Pol-primase are expressed (20, 35, 74), and for the reactivation of BKV replication. The tubular epithelial cells in normal kidneys are terminally differentiated quiescent cells that divide at a low rate (8, 63) and express small amounts of Pol-primase (36, 83). Ubiquitously expressed NFI may facilitate the low-level replication of persistent BKV in kidney epithelial cells by increasing the level (and activity) of Pol-primase at the core-ori. Also, signaling mediated through TGF-β (3, 4), tumor necrosis factor alpha (TNF-α) (4), and oxidative stress (5, 56–58) induced by kidney ischemia/reperfusion injury and/or inflammatory responses (12, 23, 38) during kidney transplantation or by the administration of immunosuppressive drugs such as tacrolimus and cyclosporine (67) might alter NFI isotype expression or activity and thereby promote the NFI-mediated recruitment of Tag and/or Pol-primase to the viral core-ori. These notions can be tested experimentally.
ACKNOWLEDGMENTS
We thank members of the Folk laboratory, especially Sarah Scanlon and Olga Kenzior for their advice and assistance and David Pintel, Mark Hannink, David Setzer, and Michael Imperiale for their constructive criticisms.
This work was supported by funds from the University of Missouri—Columbia (W.R.F.), grants from the Science Foundation Ireland (H.P.N.), an NUIG student fellowship, and a Thomas Crawfort Hayes fellowship (both I.T.).
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Abend JR, Imperiale MJ. 2008. Transforming growth factor-beta-mediated regulation of BK virus gene expression. Virology 378:6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abend JR, Low JA, Imperiale MJ. 2007. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J. Virol. 81:272–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alevizopoulos A, et al. 1995. A proline-rich TGF-beta-responsive transcriptional activator interacts with histone H3. Genes Dev. 9:3051–3066 [DOI] [PubMed] [Google Scholar]
- 4. Alevizopoulos A, Mermod N. 1996. Antagonistic regulation of a proline-rich transcription factor by transforming growth factor-beta and tumor necrosis factor-alpha. J. Biol. Chem. 271:29672–29681 [DOI] [PubMed] [Google Scholar]
- 5. Bandyopadhyay S, Starke DW, Mieyal JJ, Gronostajski RM. 1998. Thioltransferase (glutaredoxin) reactivates the DNA-binding activity of oxidation-inactivated nuclear factor I. J. Biol. Chem. 273:392–397 [DOI] [PubMed] [Google Scholar]
- 6. Bauman Y, et al. 2011. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 9:93–102 [DOI] [PubMed] [Google Scholar]
- 7. Bharucha VA, Peden KW, Tennekoon GI. 1994. SV40 large T antigen with c-Jun down-regulates myelin P0 gene expression: a mechanism for papovaviral T antigen-mediated demyelination. Neuron 12:627–637 [DOI] [PubMed] [Google Scholar]
- 8. Bonventre JV. 2003. Dedifferentiation and proliferation of surviving epithelial cells in acute renal failure. J. Am. Soc. Nephrol. 14(Suppl 1):S55–S61 [DOI] [PubMed] [Google Scholar]
- 9. Bosher J, Robinson EC, Hay RT. 1990. Interactions between the adenovirus type 2 DNA polymerase and the DNA binding domain of nuclear factor I. New Biol. 2:1083–1090 [PubMed] [Google Scholar]
- 9a. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 10. Broekema NM, et al. 2010. A system for the analysis of BKV non-coding control regions: application to clinical isolates from an HIV/AIDS patient. Virology 407:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buratowski S, Chodosh LA. 2001. Mobility shift DNA-binding assay using gel electrophoresis. John Wiley & Sons, Inc, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 12. Campistol JM, Iñigo P, Larios S, Bescos M, Oppenheimer F. 2001. Role of transforming growth factor β-1 in the progression of chronic allograft nephropathy. Nephrol. Dial. Transplant. 16:114–116 [DOI] [PubMed] [Google Scholar]
- 13. Caruso M, Iacobini C, Passananti C, Felsani A, Amati P. 1990. Protein recognition sites in polyomavirus enhancer: formation of a novel site for NF-1 factor in an enhancer mutant and characterization of a site in the enhancer D domain. EMBO J. 9:947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cassill JA, Deyerle KL, Subramani S. 1989. Unidirectional deletion and linker scan analysis of the late promoter of the human papovavirus BK. Virology 169:172–181 [DOI] [PubMed] [Google Scholar]
- 15. Chakraborty T, Das GC. 1989. Identification of HeLa cell nuclear factors that bind to and activate the early promoter of human polyomavirus BK in vitro. Mol. Cell. Biol. 9:3821–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chakraborty T, Das GC. 1991. Proteins of the nuclear factor-1 family act as an activator of the late promoter in human polyomavirus BK in vitro. J. Gen. Virol. 72(Pt 8):1935–1942 [DOI] [PubMed] [Google Scholar]
- 17. Chaudhry AZ, Lyons GE, Gronostajski RM. 1997. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 208:313–325 [DOI] [PubMed] [Google Scholar]
- 18. Chaudhry AZ, Vitullo AD, Gronostajski RM. 1998. Nuclear factor I (NFI) isoforms differentially activate simple versus complex NFI-responsive promoters. J. Biol. Chem. 273:18538–18546 [DOI] [PubMed] [Google Scholar]
- 19. Chen M, Mermod N, Horwitz MS. 1990. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J. Biol. Chem. 265:18634–18642 [PubMed] [Google Scholar]
- 20. Chesters PM, Heritage J, McCance DJ. 1983. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J. Infect. Dis. 147:676–684 [DOI] [PubMed] [Google Scholar]
- 21. Deyerle KL, Sajjadi FG, Subramani S. 1989. Analysis of origin of DNA replication of human papovavirus BK. J. Virol. 63:356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deyerle KL, Subramani S. 1988. Linker scan analysis of the early regulatory region of human papovavirus BK. J. Virol. 62:3378–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Djamali A. 2007. Oxidative stress as a common pathway to chronic tubulointerstitial injury in kidney allografts. Am. J. Physiol. Renal Physiol. 293:F445–F455 [DOI] [PubMed] [Google Scholar]
- 24. Dornreiter I. 1991. Herstellung monoklonaler Antikörper gegen DNA-Polymerase-α-Primase und ihre Anwendung zur Untersuchung von Protein-Protein-Wechselwirkungen in der Replikation viraler DNA. Ph.D. thesis, LMU München, München, Germany [Google Scholar]
- 25. Egli A, et al. 2009. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J. Infect. Dis. 199:837–846 [DOI] [PubMed] [Google Scholar]
- 26. Egli A, Köhli S, Dickenmann M, Hirsch HH. 2009. Inhibition of polyomavirus BK-specific T-cell responses by immunosuppressive drugs. Transplantation 88:1161–1168 [DOI] [PubMed] [Google Scholar]
- 27. Fishman JA. 2002. BK virus nephropathy—polyomavirus adding insult to injury. N. Engl. J. Med. 347:527–530 [DOI] [PubMed] [Google Scholar]
- 28. Gorrill TS, Khalili K. 2005. Cooperative interaction of p65 and C/EBPbeta modulates transcription of BKV early promoter. Virology 335:1–9 [DOI] [PubMed] [Google Scholar]
- 29. Gosert R, Kardas P, Major EO, Hirsch HH. 2010. Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate. J. Virol. 84:10448–10456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gosert R, et al. 2008. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J. Exp. Med. 205:841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grinnell BW, Berg DT, Walls JD. 1988. Negative regulation of the human polyomavirus BK enhancer involves cell-specific interaction with a nuclear repressor. Mol. Cell. Biol. 8:3448–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gronostajski RM. 2000. Roles of the NFI/CTF gene family in transcription and development. Gene 249:31–45 [DOI] [PubMed] [Google Scholar]
- 33. Gronostajski RM, Adhya S, Nagata K. 1985. Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol. Cell. Biol. 5:964–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hennighausen L, Fleckenstein B. 1986. Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J. 5:1367–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heritage J, Chesters PM, McCance DJ. 1981. The persistence of papovavirus BK DNA sequences in normal human renal tissue. J. Med. Virol. 8:143–150 [DOI] [PubMed] [Google Scholar]
- 36. Hirose F, Yamamoto S, Yamaguchi M, Matsukage A. 1988. Identification and subcellular localization of the polypeptide for chick DNA primase with a specific monoclonal antibody. J. Biol. Chem. 263:2925–2933 [PubMed] [Google Scholar]
- 37. Hirsch HH, Steiger J. 2003. Polyomavirus BK. Lancet Infect. Dis. 3:611–623 [DOI] [PubMed] [Google Scholar]
- 38. Hribova P, et al. 2005. Cytokines and chemokine gene expression in human kidney transplantation. Transplant. Proc. 37:760–763 [DOI] [PubMed] [Google Scholar]
- 39. Johnston SD, Yu XM, Mertz JE. 1996. The major transcriptional transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J. Virol. 70:1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jordan JA, Manley K, Dugan AS, O'Hara BA, Atwood WJ. 2010. Transcriptional regulation of BK virus by nuclear factor of activated T cells. J. Virol. 84:1722–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Knowles WA. 2006. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV), p 19–45 In Ahsan N. (ed), Polyomaviruses and human diseases, vol 577 Springer, New York, NY: [DOI] [PubMed] [Google Scholar]
- 42. Kraus RJ, Shadley L, Mertz JE. 2001. Nuclear factor 1 family members mediate repression of the BK virus late promoter. Virology 287:89–104 [DOI] [PubMed] [Google Scholar]
- 43. Kulkarni S, Gronostajski RM. 1996. Altered expression of the developmentally regulated NFI gene family during phorbol ester-induced differentiation of human leukemic cells. Cell Growth Differ. 7:501–510 [PubMed] [Google Scholar]
- 44. Kwun HJ, Yim SW, Lee DH, Jang KL. 1999. Activation of the thymidine kinase promoter by herpes simplex virus type 1 immediate early proteins. Mol. Cells 9:277–280 [PubMed] [Google Scholar]
- 45. Leegwater PA, van Driel W, van der Vliet PC. 1985. Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J. 4:1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahon C, et al. 2009. Restriction of human polyomavirus BK virus DNA replication in murine cells and extracts. J. Virol. 83:5708–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Markowitz RB, Dynan WS. 1988. Binding of cellular proteins to the regulatory region of BK virus DNA. J. Virol. 62:3388–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Markowitz RB, et al. 1991. BK virus and JC virus shed during pregnancy have predominantly archetypal regulatory regions. J. Virol. 65:4515–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mermod N, O'Neill EA, Kelly TJ, Tjian R. 1989. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell 58:741–753 [DOI] [PubMed] [Google Scholar]
- 50. Mizuno T, et al. 1998. The second-largest subunit of the mouse DNA polymerase alpha-primase complex facilitates both production and nuclear translocation of the catalytic subunit of DNA polymerase alpha. Mol. Cell. Biol. 18:3552–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mizuno T, et al. 1996. Identification of the nuclear localization signal of mouse DNA primase: nuclear transport of p46 subunit is facilitated by interaction with p54 subunit. J. Cell Sci. 109(Pt 11):2627–2636 [DOI] [PubMed] [Google Scholar]
- 52. Moens U, Johansen T, Johnsen JI, Seternes OM, Traavik T. 1995. Noncoding control region of naturally occurring BK virus variants: sequence comparison and functional analysis. Virus Genes 10:261–275 [DOI] [PubMed] [Google Scholar]
- 53. Moens U, Subramaniam N, Johansen B, Johansen T, Traavik T. 1994. A steroid hormone response unit in the late leader of the noncoding control region of the human polyomavirus BK confers enhanced host cell permissivity. J. Virol. 68:2398–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moens U, Van Ghelue M. 2005. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology 331:209–231 [DOI] [PubMed] [Google Scholar]
- 55. Monaco MCG, Sabath BF, Durham LC, Major EO. 2001. JC virus multiplication in human hematopoietic progenitor cells requires the NF-1 class D transcription factor. J. Virol. 75:9687–9695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morel Y, Barouki R. 1998. Down-regulation of cytochrome P450 1A1 gene promoter by oxidative stress. J. Biol. Chem. 273:26969–26976 [DOI] [PubMed] [Google Scholar]
- 57. Morel Y, Barouki R. 2000. The repression of nuclear factor I/CCAAT transcription factor (NFI/CTF) transactivating domain by oxidative stress is mediated by a critical cysteine (Cys-427). Biochem. J. 348:235–240 [PMC free article] [PubMed] [Google Scholar]
- 58. Morel Y, Mermod N, Barouki R. 1999. An autoregulatory loop controlling CYP1A1 gene expression: role of H2O2 and NFI. Mol. Cell. Biol. 19:6825–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mul YM, Van der Vliet PC. 1992. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 11:751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mul YM, Verrijzer CP, van der Vliet PC. 1990. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol. 64:5510–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Muller K, Mermod N. 2000. The histone-interacting domain of nuclear factor I activates simian virus 40 DNA replication in vivo. J. Biol. Chem. 275:1645–1650 [DOI] [PubMed] [Google Scholar]
- 62. Mysiak ME, Wyman C, Holthuizen PE, van der Vliet PC. 2004. NFI and Oct-1 bend the Ad5 origin in the same direction leading to optimal DNA replication. Nucleic Acids Res. 32:6218–6225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nadasdy T, Laszik Z, Blick KE, Johnson LD, Silva FG. 1994. Proliferative activity of intrinsic cell populations in the normal human kidney. J. Am. Soc. Nephrol. 4:2032–2039 [DOI] [PubMed] [Google Scholar]
- 64. Nagata K, Guggenheimer RA, Enomoto T, Lichy JH, Hurwitz J. 1982. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc. Natl. Acad. Sci. U. S. A. 79:6438–6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nasheuer HP, Smith R, Bauerschmidt C, Grosse F, Weisshart K. 2002. Initiation of eukaryotic DNA replication: regulation and mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 72:41–94 [DOI] [PubMed] [Google Scholar]
- 66. Nehls MC, Grapilon ML, Brenner DA. 1992. NF-I/Sp1 switch elements regulate collagen alpha 1(I) gene expression. DNA Cell Biol. 11:443–452 [DOI] [PubMed] [Google Scholar]
- 67. Nowak C, et al. 2007. Consequences of inbreeding and reduced genetic variation on tolerance to cadmium stress in the midge Chironomus riparius. Aquat. Toxicol. 85:278–284 [DOI] [PubMed] [Google Scholar]
- 68. O'Connor M, Bernard H-U. 1995. Oct-1 activates the epithelial-specific enhancer of human papillomavirus type 16 via a synergistic interaction with NFI at a conserved composite regulatory element. Virology 207:77–88 [DOI] [PubMed] [Google Scholar]
- 69. Oehlmann M, Mahon C, Nasheuer HP. 2007. Comparison of DNA replication in Xenopus laevis and simian virus 40. Adv. Exp. Med. Biol. 604:3–16 [DOI] [PubMed] [Google Scholar]
- 70. Olsen GH, Hirsch HH, Rinaldo CH. 2009. Functional analysis of polyomavirus BK non-coding control region quasispecies from kidney transplant recipients. J. Med. Virol. 81:1959–1967 [DOI] [PubMed] [Google Scholar]
- 71. Perets TT, et al. 2009. High frequency and diversity of rearrangements in polyomavirus BK noncoding regulatory regions cloned from urine and plasma of Israeli renal transplant patients and evidence for a new genetic subtype. J. Clin. Microbiol. 47:1402–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Quandt K, Frech K, Karas H, Wingender E, Werner T. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rafty LA, Santiago FS, Khachigian LM. 2002. NF1/X represses PDGF A-chain transcription by interacting with Sp1 and antagonizing Sp1 occupancy of the promoter. EMBO J. 21:334–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Randhawa P, Shapiro R, Vats A. 2005. Quantitation of DNA of polyomaviruses BK and JC in human kidneys. J. Infect. Dis. 192:504–509 [DOI] [PubMed] [Google Scholar]
- 75. Randhawa P, et al. 2003. Viral regulatory region sequence variations in kidney tissue obtained from patients with BK virus nephropathy. Kidney Int. 64:743–747 [DOI] [PubMed] [Google Scholar]
- 76. Ravichandran V, Major EO. 2008. DNA-binding transcription factor NF-1A negatively regulates JC virus multiplication. J. Gen. Virol. 89:1396–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ravichandran V, Sabath BF, Jensen PN, Houff SA, Major EO. 2006. Interactions between c-Jun, nuclear factor 1, and JC virus promoter sequences: implications for viral tropism. J. Virol. 80:10506–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rubinstein R, Schoonakker BC, Harley EH. 1991. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J. Virol. 65:1600–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS. 2008. Evolutionarily conserved function of a viral microRNA. J. Virol. 82:9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shivakumar CV, Das GC. 1996. Interaction of human polyomavirus BK with the tumor-suppressor protein p53. Oncogene 13:323–332 [PubMed] [Google Scholar]
- 81. Sock E, Wegner M, Grummt F. 1991. DNA replication of human polyomavirus JC is stimulated by NF-I in vivo. Virology 182:298–308 [DOI] [PubMed] [Google Scholar]
- 82. Stadlbauer F, et al. 1994. DNA replication in vitro by recombinant DNA-polymerase-alpha-primase. Eur. J. Biochem. 222:781–793 [DOI] [PubMed] [Google Scholar]
- 83. Sugawara I, et al. 1989. Intracellular localisation of a subunit of human DNA polymerase alpha affecting primase activity recognised by monoclonal antibody (HDR-854-E4) and its application to distinction between proliferative and non-proliferative lesions. Br. J. Cancer 60:176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tikhanovich I, Liang B, Seoighe C, Folk WR, Nasheuer HP. 2011. Inhibition of human BK polyomavirus replication by small noncoding RNAs. J. Virol. 85:6930–6940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tikhanovich I, Nasheuer HP. 2010. Host-specific replication of BK virus DNA in mouse cell extracts is independently controlled by DNA polymerase alpha-primase and inhibitory activities. J. Virol. 84:6636–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tseng RW, Williams T, Fujimura FK. 1988. Unique requirement for the PyF441 mutation for polyomavirus infection of F9 embryonal carcinoma cells. J. Virol. 62:2896–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Weisshart K, et al. 2000. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 275:17328–17337 [DOI] [PubMed] [Google Scholar]
- 88. White MK, Khalili K. 2011. Pathogenesis of progressive multifocal leukoencephalopathy—revisited. J. Infect. Dis. 203:578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhou W, et al. 2007. Functional characterization of BK virus-specific CD4+ T cells with cytotoxic potential in seropositive adults. Viral Immunol. 20:379–388 [DOI] [PubMed] [Google Scholar]