Summary
Background
Induction of plant immune responses involves significant transcription reprogramming that prioritizes defense over growth-related cellular functions. Despite intensive forward genetic screens and genome-wide expression-profiling studies, a limited number of transcription factors have been found that regulate this transition.
Results
Using the endoplasmic-reticulum-resident genes required for antimicrobial protein secretion as markers, we identified a heat-shock factor-like transcription factor that specifically binds to the TL1 (GAAGAAGAA) cis element required for the induction of these genes. Surprisingly, plants lacking this TL1-binding factor, TBF1, respond normally to heat stress but are compromised in immune responses induced by salicylic acid and by microbe-associated molecular pattern, elf18. Genome-wide expression profiling indicates that TBF1 plays a key role in the growth-to-defense transition. Moreover, the expression of TBF1 itself is tightly regulated at both the transcriptional and translational levels. Two upstream open reading frames encoding multiple aromatic amino acids were found 5′ of the translation initiation codon of TBF1 and shown to affect its translation.
Conclusions
Through this unique regulatory mechanism, TBF1 can sense the metabolic changes upon pathogen invasion and trigger the specific transcriptional reprogramming through its target genes expression.
Introduction
The sessile nature of plants subjects them to constant biotic and abiotic stresses. Even though plants do not have specialized immune cells, they can mount both local and systemic immune responses [1]. This requires extensive cross-talk between plant defense and other physiological processes. Induction of defense responses involves the recognition of microbe-associated molecular patterns (MAMPs) by membrane-associated receptors leading to MAMP-triggered immunity (MTI) and of pathogen-delivered effectors by cytosolic receptors resulting in effector-triggered immunity (ETI) [2]. Besides these local defense mechanisms, salicylic acid (SA) is produced during local infection that can lead to systemic acquired resistance (SAR). In Arabidopsis, SA signals through the key immune regulator NPR1 (nonexpressor of PR genes), which is required for subsequent transcriptional changes of as many as ~10% of all genes [3]. However, unlike the signal-specific MTI and ETI, SAR is broad spectrum and long lasting [4].
SAR-associated transcriptional reprogramming redirects cellular resources, normally dedicated to growth-related activities, toward de novo synthesis of antimicrobial proteins such as the pathogenesis-related (PR) proteins. Prior to PR protein accumulation, endoplasmic reticulum (ER)-resident genes encoding the secretory pathway machinery are coordinately upregulated to ensure efficient posttranslational modification and secretion of these antimicrobial peptides [3, 5]. The involvement of the ER function is not restricted to SAR. A connection between the ER-resident genes and MTI has been revealed in studies demonstrating that the biogenesis of EFR, a membrane-bound receptor for the MAMP signal elf18 (the N-terminal 18 amino acids of the bacterial translation elongation factor Tu, EF-Tu), requires the N-glycosylation pathway genes encoding the calreticulin 3 (CRT3) and the UDP-glucose:glycoprotein glycosyltransferase STT3A in the ER quality control mechanism (ERQC) [6, 7].
Previously, we showed that induction of both PR and ER-resident genes requires the transcription cofactor NPR1. Upon SA induction, NPR1 is nuclear translocated [8] and induces PR genes through its interaction with the TGA transcription factors (TFs) at PR genes promoters [9, 10]. However, it is not known how NPR1 regulates the ER-resident genes. TGA TFs are unlikely candidates, because in the tga mutants, expression of ER-resident genes is unaltered following induction [3]. Significant enrichment of a novel cis element TL1 (translocon 1; GAAGAAGAA) in the promoter regions of these NPR1-dependent ER-resident genes suggests the involvement of an unknown TF [3]. Point mutations in the TL1 elements of the BiP2 (Lumenal Binding Protein 2) promoter abolished the inducibility of this gene upon SA treatment [3]. Because the secretory pathway is required not only for defense but also for many other cellular functions, identification of this TL1-binding TF is vital to our understanding of the mechanism controlling the growth-to-defense transition.
Here, we report the identification of a heat-shock factor-like protein (HSF4/HsfB1) that binds to the TL1 cis element and regulates the expression of genes containing this element in their promoters. Because mutants of this TF have normal heat-shock responses but are compromised in the growth-to-defense transition upon pathogen challenge, we renamed it the TL1-binding transcription factor 1, or TBF1. Consistent with its key role in this general control, the translation of TBF1 is tightly regulated through two upstream open reading frames (uORFs) enriched in aromatic amino acids, which are precursors of a large array of plant secondary metabolites involved in defense.
Results
TBF1 Is a TF that Binds to the TL1 cis Element Enriched in Defense-Related Gene Promoters
The TL1 cis element (consensus sequence GAAGAAGAA) in the ER-resident genes is essential for their activation in response to SA induction [3]. We examined publically available microarray data (see Supplemental Information available online) and found that the TL1 cis element is enriched in promoters of genes regulated by elf26 (the first 26 amino acids of EF-Tu) (p < 0.001) and flg22 (p < 0.01), indicating that this novel element may play a role in MTI. Interestingly, we detected no significant enrichment of TL1 when all of the BTH (benzothiadiazole; an SA analog)-affected promoters were analyzed even though the element was first discovered in the SA-induced, NPR1-dependent ER-resident genes [3].
Next, we searched the TF that binds to TL1 using the TFSEARCH database and found HSFs of Saccharomyces cerevisiae and Drosophila melanogaster as potential candidates. The Arabidopsis genome contains 21 HSF-like genes. Several reports have indicated the involvement of the HSFs in immediate heat response, acquired thermotolerance, sensing of reactive oxygen species (ROS), and seed development [11, 12]. To identify a candidate gene for TBF1, we first examined publically available expression data for profiles of the Arabidopsis HSF family members (see Supplemental Information). Only one gene family member, HSF4 (also known as HsfB1; At4g36990), was strongly induced by BTH as well as virulent and avirulent Pseudomonas syringae pv. maculicola (Psm) ES4326 bacteria. Because Arabidopsis HSF4 and its tomato homolog do not functionally complement the yeast hsf1 mutant strain [13] (Daniel Neef and Dennis Thiele, personal communication) and its overexpression has little effect on heat-shock protein expression or thermotolerance [14, 15], we speculated that HSF4 does not encode a typical heat-shock factor. Its pathogen-inducible expression pattern suggests that it has a novel function related to plant immunity and is a candidate for TBF1.
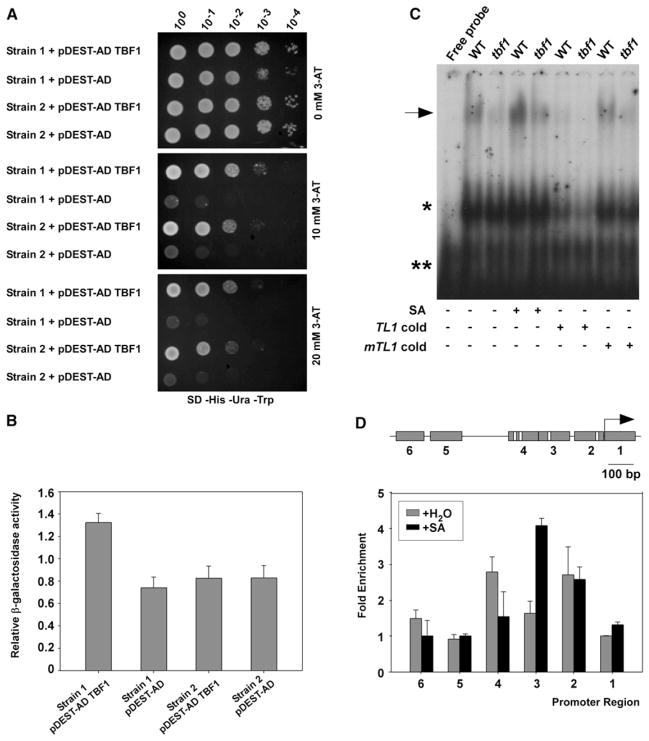
In the current study, we demonstrated HSF4 to be the TL1-binding TF (TBF1) through a yeast one-hybrid (Y1H) assay in which the fragment of BiP2 promoter containing multiple functional TL1 cis-elements [3] was used as bait. We constructed two yeast bait strains containing the wild-type (WT) and the mutant (mTL1) BiP2 promoters, respectively (Figures S1A and S1B). Expression of TBF1-AD (activation domain) in Strain 1 activated both HIS3 and LacZ reporters driven by the WT BiP2 promoter (Figures 1A and 1B). The binding specificity of TBF1 to TL1 was confirmed in Strain 2 where the two single nucleotide substitutions in the mTL1 core binding sequence blocked the induction of LacZ, whereas the control HIS3 reporter with the WT TL1 was induced normally (Figure 1B).
Figure 1. HSF4 Is the TL1-Binding TF, TBF1.
(A and B) TBF1 (HSF4) binding to the TL1 cis elements in the BiP2 promoter was detected in Y1H. Yeast growth on selective media (SD-His-Ura-Trp) supplemented with increasing concentrations of 3-AT was recorded at day 3 (A). β-galactosidase reporter activity was measured using ONPG as the substrate (B). Error bars represent standard deviation from three different technical replications. Both (A) and (B) were repeated three times with similar results. See also Figures S1A and S1B.
(C) Electrophoretic mobility shift assay was performed using plant extracts from WT and the tbf1 mutant upon 1 mM SA treatment. TL1 cold and mTL1 cold (mutant TL1; both at 5 pmol/μL) were used as unlabeled probes. The arrow marks the TBF-TL1 complex. Asterisks indicate nonspecific binding. (+) and (−) signify for the presence or absence of corresponding treatments. The experiment was repeated three times with similar results. See also Figures S1Cand S1D.
(D) TBF1-GFP binding to the TL1 elements in the BiP2 promoter was measured by ChIP after treatment with H2O or 1mMSA. The PCR amplicons, 1 to 6 (gray boxes) with TL1 elements highlighted in white are shown (upper panel). Arrow represents the BiP2 translational start site. Normalized fold enrichment for each amplicon was calculated (lower panel). Error bars represent standard deviation from three different replicates. Experiment was repeated five times with similar results. See also Figure S1E.
TBF1 binding to the TL1 cis element was further demonstrated by electrophoretic mobility shift assay using protein extracts from WT and an insertional knock-out TBF1 mutant, tbf1 (Figures S1C and S1D). WT displayed an upshifted band whose intensity was further enhanced in the extract made from plants treated with SA (Figure 1C). This band was diminished in the tbf1 mutant extract, indicating that TBF1 is required for the DNA-protein complex formation. Competition assay using nonradioactive TL1 and mutant mTL1 probes indicated that the observed TBF1 binding was specific to the TL1 consensus sequence.
To test TBF1 DNA-binding activity in vivo, we generated transgenic tbf1 plants expressing a translational fusion between TBF1 and GFP driven by the endogenous TBF1 promoter (TBF1p:TBF1-GFP). Because the fusion protein was proven to be biologically active through genetic complementation of the tbf1 mutant phenotype (Figure S1E), we used it for chromatin immunoprecipitation (ChIP). As shown in Figure 1D, using six pairs of primers spanning different regions of the BiP2 promoter, we detected sequence enrichment corresponding to the TL1-containing region 2 in both uninduced and SA-treated samples, region 3 in the SA-treated sample, and region 4 in the uninduced sample. No enrichment was found in regions 1, 5, and 6 that do not contain the TL1 element.
TBF1 Is a Major Molecular Switch Involved in Transcriptional Reprogramming Induced by SA and elf18
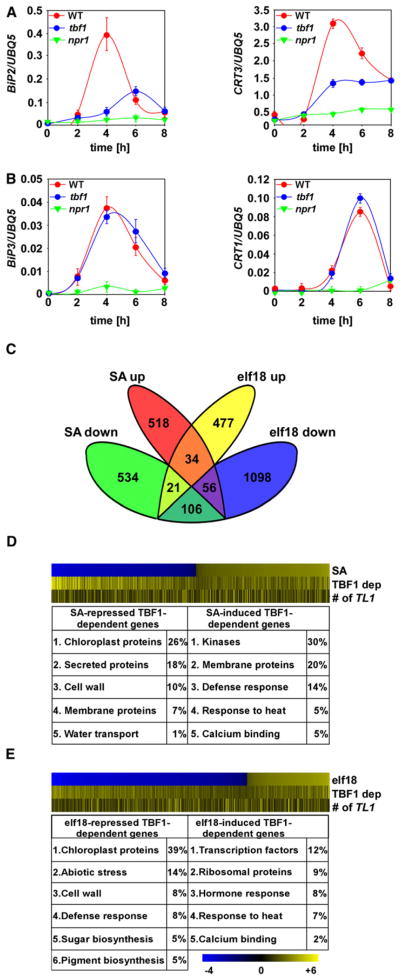
In the test for TBF1 dependency, we found that the induction of BiP2 and CRT3, whose promoters contain multiple copies of TL1 elements, was compromised in the tbf1 mutant and in npr1-1 (Figure 2A). Consequently, the BiP2 protein accumulation was inhibited in the SA-treated tbf1 mutant plants (Figure S2A). In contrast, induction of two other ER-resident genes, BiP3 and CRT1, which do not have TL1 in their promoters, was not affected in tbf1 (Figure 2B), confirming the specificity of TBF1 to TL1.
Figure 2. TBF1 Plays aMajor Role in Transcriptional Reprogramming during MTI and SAR.
(A and B) Relative transcript levels of TBF1-dependent and independent ER-resident genes were determined by qRT-PCR using cDNA generated from WT, tbf1, and npr1-1 plants treated with 1mMSA. Error bars represent standard deviation from nine technical replicates derived from three independent experiments. See also Figure S2A.
(C) Venn diagram shows the numbers of TBF1-dependent SA downregulated (SA down), SA upregulated (SA up), elf18 upregulated (elf18 up), and elf18 downregulated (elf18 down) genes (p < 0.05).
(D and E) Heat maps of TBF1-regulated genes in total numbers (top), degrees of TBF1 dependency (middle), and numbers of TL1 cis elements in the gene promoters (bottom) in response to SA (D) and elf18 (E) treatment. Top ranked functional groups were determined using DAVID Gene Ontology. Scale indicates the log-transformed p values of down-(blue) and up-(yellow) regulated genes (top), yellow lines indicate TBF1 dependency (middle), and yellow lines correspond to the numbers of TL1 cis elements in the gene promoters (bottom). See also Figures S2B–S2E.
We next performed a genome-wide transcriptional profiling on WT and tbf1 plants upon SA and elf18 treatments to determine the global effect of TBF1. We found 1,269 and 1,792 TBF1-dependent genes differentially regulated by SA and elf18, respectively (p < 0.05, fold change > 2). However, only ~8% of these genes were regulated by both signals (Figure 2C; Table S1), indicating that TBF1 controls distinct output genes in SAR and MTI. Moreover, the total numbers of significantly induced and repressed genes (the top heatmaps in Figures 2D and 2E), the degrees of TBF1 dependency (the middle heatmaps), and the numbers of TL1 cis elements present in the gene promoters (the bottom heatmaps) indicate that TBF1 plays a greater role in SA- and elf18-mediated transcription repression than in induction. This finding is in agreement with the previous work indicating that class B-Hsfs mainly act as repressors of target gene expression [16, 17].
Gene ontology (GO) analysis identified a significantly enriched cluster of SA-induced secretory pathway genes (p ≤ 0.001; Figure 2D “membrane proteins,” Table S1), recapitulating our earlier findings [3]. In addition, we identified several major SA- and elf18-mediated and TBF1-dependent functional categories including genes known to encode defense-related proteins, ribosomal proteins, chloroplast function-related proteins, and abiotic stress signal transducers (Figures 2D and 2E; Table S1; described in more detail in Supplemental Information) [18]. We postulate that these genes are likely directly controlled by TBF1 because their regulatory regions contain the TL1 element (Table S1).
Expression changes observed in the microarray experiment were further confirmed through qRT-PCR of independent biological samples on 26 selected genes representing various GO categories listed in Figures 2D and 2E (Figures S2B–S2E).
TBF1 Plays a Key Role in the Growth-to-Defense Transition
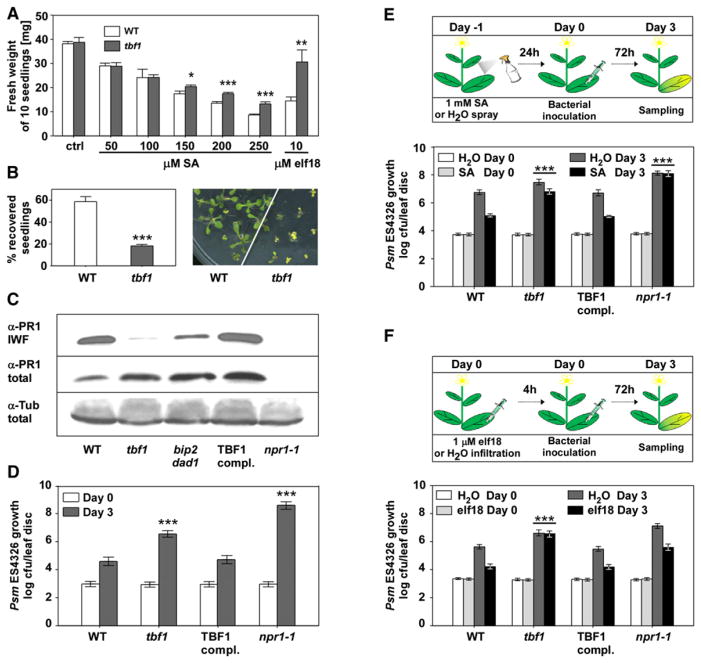
Upon SA treatment, TBF1 downregulates genes encoding chloroplast proteins (Figure 2D)—an effect that is known to be associated with SA [19]. Interestingly, chloroplast function-related genes were even more profoundly repressed by elf18 (Figure 2E), indicating that TBF1 might play a general role as a major molecular switch in the growth-to-defense transition. To test this, we first measured the growth of both WT and the tbf1 mutant plants. As shown in Figure 3A, in the absence of SA or elf18, the tbf1 mutant grew at a similar rate as WT. In the presence of elf18 or increasing SA concentrations, however, the growth of the WT plants was significantly inhibited. This effect was partially alleviated in the tbf1 mutant. In contrast, another MAMP signal, flg22, exerted a similar growth-suppressing effect on both WT and tbf1 seedlings (Figure S3A).
Figure 3. TBF1 Is a Major Molecular Switch for the Growth-to-Defense Transition.
(A) Fresh weight of ten seedlings grown for 10 days on plates with MS growth media (ctrl), or MS supplemented with increasing concentrations of SA or 10 μM elf18. Error bars represent standard deviation of three replicates. This experiment was repeated three times with similar results. Statistical analysis was performed using Student’s t test, *p < 0.05, **p < 0.01, ***p ≤ 0.001. See also Figure S3A.
(B) Seedling recovery after treatment with tunicamycin was measured by counting the percentage of surviving seedlings (left) and by phenotype observations (right). Error bars represent standard deviation of three replicates. This experiment was repeated five times with similar results. Statistical analysis was performed using Student’s t test, ***p ≤ 0.001. See also Figure S3B.
(C) PR1 accumulation in the intercellular wash fluid (IWF) and total protein extracts from leaves of 3-week-old WT, tbf1, tbf1 transformed with the WT TBF1 gene (TBF1 compl.), npr1-1, and bip2 dad2. For loading controls, an antibody against tubulin (α-Tub) was used to probe the total protein blot. See also Figure S3C.
(D) Enhanced disease susceptibility was measured in 3-week-old WT, tbf1, TBF1 complementation, and npr1-1 plants 3 days after infiltration with a bacterial suspension of Psm ES4326 (OD600nm = 0.0001). Error bars represent 95% confidence intervals of 24 replicates derived from three independent experiments. This experiment was repeated at least five times with similar results. Statistical analysis was performed using Bonferroni post hoc test, ***p < 0.0001.
(E) SA-induced resistance was determined according to the schematic representation (upper panel), and the growth of Psm ES4326 was plotted as in (D) but with a higher initial inoculum (OD600nm = 0.001) (lower panel). Error bars represent 95% confidence intervals of 24 replicates derived from three independent experiments. Statistical analysis was performed using two-way analysis of variance (ANOVA), ***p < 0.0001.
(F) elf18-induced resistance was measured according to the schematic representation (upper panel) and with the initial Psm ES4326 inoculum of OD600nm = 0.001 (lower panel). Error bars represent 95% confidence intervals of 24 replicates derived from three independent experiments. Statistical analysis was performed using two-way ANOVA, ***p < 0.0001. See also Figures S3D and S3E.
We performed an additional series of tests to determine the stress responses mediated by TBF1. Even though the tbf1 mutant has been shown to have normal heat-induced marker gene expression (Figure S3B) and plays no detectable role in the heat-shock response [20], its defect in the induction of multiple chaperone genes prompted us to examine the unfolded protein response (UPR). When the plants were treated with the UPR inducer tunicamycin, seedling survival rate for tbf1 was only ~20% compared to ~60% for WT (Figure 3B), indicating that TBF1 plays a role in UPR.
The capacity of TBF1 to efficiently secrete antimicrobial PR1 protein was evaluated next. Whereas the PR1 transcript induction and total PR1 protein levels were unchanged in tbf1 (Figure 3C; Figure S3C), secreted PR1 in the intercellular wash fluid was dramatically reduced in the tbf1 mutant, as in the control bip2 dad1 double mutant [3], compared to WT. This phenotype was complemented in the transgenic line carrying a genomic fragment encompassing upstream regulatory sequences (3,554 bp) and the coding region (1,047 bp) of TBF1 (TBF1 complementation). The defect in secretion of anti-microbial proteins in the tbf1 mutant correlated with more than 1 log higher growth of the bacterial pathogen Psm ES4326 compared to WT and the complementation line (Figure 3D). In response to SA induction, less than 1 log reduction in Psm ES4326 growth was observed in the tbf1 mutant as opposed to the ~2 log reduction detected in the WT plants (Figure 3E). SA-inducible defenses were restored in the TBF1 complementation line, whereas npr1-1 was completely deficient in establishing resistance.
We next examined whether the tbf1 mutant is capable of mounting effective MTI using elf18 and flg22. WT plants pre-treated with either signal showed a more than 1 log reduction in Psm ES4326 growth compared to mock-treated samples (Figure 3F; Figure S3D). The tbf1 mutant, on the other hand, completely failed to establish the resistance induced by elf18. Interestingly, this defect is specific to elf18 because flg22-induced resistance was intact in the tbf1 mutant (Figures S3D and S3E). Although previous studies showed that flg22 and elf18 induce largely overlapping sets of genes [21], our results, however, are consistent with the genetic data showing that the perception of elf18, but not flg22, specifically requires the ERQC mechanism [6, 7, 22, 23] and with the fact that TBF1 controls the induction of these ERQC genes (Figure 2A). The molecular mechanism underlying this TBF1 requirement has yet to be determined.
Another observation from the Psm ES4326 infection experiment was the near-normal response to elf18 observed in the SA-insensitive npr1-1 mutant (Figure 3F). Even though there was an overall increase in Psm ES4326 growth in npr1-1, elf18 pretreatment could still result in a more than 1 log reduction in pathogen growth similar to that observed in the WT. This result is in accordance with the expression profiling data showing that elf18 and SA induce distinct sets of genes (Figure 2C). Moreover, a recently published study demonstrated that MTI induced by flg22 and elf18 is largely intact in the sid2 mutant, which is deficient in SA synthesis [24]. MTI and SAR are two temporally and spatially separate immune responses. The former occurs locally and immediately upon pathogen challenge, whereas the latter is a systemic response induced after the local response.
Translation of TBF1 Is Controlled by uORFs Sensitive to Cellular Metabolic Changes
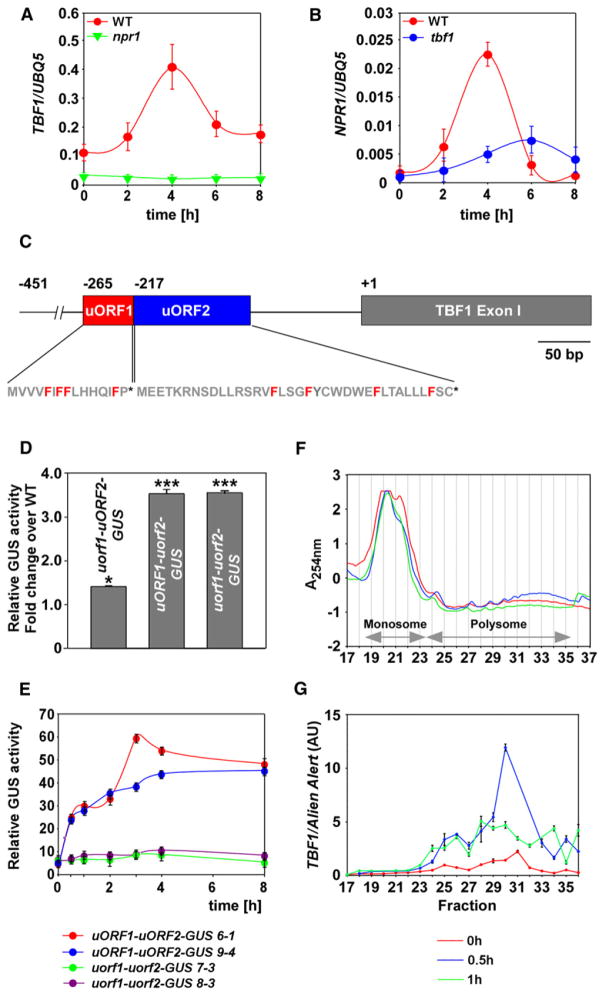
Our expression profiling and genetic data show that TBF1 is a major molecular switch that, upon pathogen challenge, turns on multiple defenses and inhibits primary growth and development (Figures 2 and 3). To understand how TBF1 itself is regulated, we analyzed its expression patterns upon SA treatment and detected the maximum accumulation of TBF1 transcript 4 hr after the treatment (Figure 4A). We further demonstrated that functional NPR1 is required for SA-mediated TBF1 transcription. Interestingly, TBF1 also plays a role in regulating NPR1 as the NPR1 transcript levels were diminished in tbf1 (Figure 4B), suggesting a feedback mechanism between these two key immune regulators.
Figure 4. TBF1 Expression Is Regulated at both Transcriptional and Translational Levels.
(A and B) Relative transcript levels of TBF1 (A) and NPR1 (B) genes in response to 1 mM SA treatment were determined by qRT-PCR. Error bars represent standard deviation from nine technical replicates derived from three independent experiments.
(C) Schematic representation of uORF1 and uORF2 and exon I of TBF1. The phenylalanines (F) in uORF1 and uORF2 are highlighted in red, and the stop codons are shown as asterisks. “+1” represents the translational start of TBF1 and −451, −265, and −217 represent the upstream positions of the 5′ end of the transcript, the start codon for uORF1, and the start codon for uORF2, respectively.
(D) Quantification of GUS activity in Nicotiana benthamiana leaves transiently expressing uORF1-uORF2-GUS (WT), uorf1-uORF2-GUS, uORF1-uorf2-GUS, and uorf1-uorf2-GUS. This experiment has been repeated three times with similar results.
(E) Quantification of translational inhibitory effect exerted by uORFs in transgenic T3 plants expressing uORF1-uORF2-GUS (two independent transformants 6-1 and 9-4) or uorf1-uorf2-GUS (two independent transformants 7-3 and 8-3) at various time points after inoculation with Psm ES4326/avrRpt2 (OD600nm = 0.02). Error bars represent standard deviation from three different replicates. Experiment was repeated at least three times with similar results.
(F and G) Polysome profiles (F) and TBF1 expression (G) in samples obtained from WT plants at 0, 0.5, and 1 hr after inoculation with Psm ES4326/avrRpt2 (OD600nm = 0.02). The fractions containing monosome and polysome were annotated based on the absorbance at 254 nm (A254nm). The TBF1 transcript abundance normalized against Alien Alert® control transcript is expressed in arbitrary units (AU). Error bars represent standard error. This experiment was repeated using two biological replicates (each with three technical replicates) with similar results. See also Figure S4.
In addition to transcriptional regulation, analysis of the TBF1 mRNA through the 5′ and 3′ rapid amplification of cDNA ends (RACE) detected two additional upstream open reading frames (uORFs) 5′ of the TBF1 start codon (Figure 4C). The second uORF (At4g36988) is well-conserved among TBF1 homologs in other plant species [25]. To test whether these two uORFs influence translation initiation of TBF1, we made a fusion between the 5′ UTR of TBF1 containing both uORFs, the first exon of TBF1 and the GUS reporter (abbreviated as uORF1-uORF2-GUS). We also created three additional constructs with the start codon mutated (ATG to CTG) for uORF1 (uorf1-uORF2-GUS), uORF2 (uORF1-uorf2-GUS), or both uORFs (uorf1-uorf2-GUS). These reporter constructs were driven by the 35S promoter to allow detection of only translational differences. We first transiently expressed these constructs in Nicotiana benthamiana leaves and quantified the GUS activities (Figure 4D). Using activity of the WT construct as a control, we detected 1.5- and 3.5-fold increases in GUS activities in uorf1-uORF2-GUS and uORF1-uorf2-GUS, respectively. Like the uorf2 mutant, mutating both uORFs in uorf1-uorf2-GUS resulted in a 3.5-fold elevation in GUS activity over WT. These observations suggest that both uORFs have inhibitory effects on TBF1 translation, with the effect of uORF2 epistatic to that of uORF1.
To further understand the regulatory mechanism of TBF1 translation during plant defense, we quantified GUS activities of these translational fusions in stable transgenic Arabidopsis lines in response to Psm ES4326 carrying the avirulent effector, avrRpt2. We found that perception of this avirulent bacterial strain, which can induce MTI, ETI, and SAR, caused a rapid increase in GUS activity in the uORF1-uORF2-GUS transgenic lines (Figure 4E). Interestingly, this increase was not observed in the uorf1-uorf2-GUS transgenic lines. These results suggest that the Psm ES4326/avrRpt2 challenge could rapidly alleviate the inhibitory effects of the uORFs on translation of the downstream gene. To determine whether the endogenous TBF1 was indeed translated in the plant cell upon pathogen challenge, we conducted a polysome profiling experiment (Figure 4F). We found a significant increase in TBF1 transcript in the polysomal fractions of the gradient within 30 min of Psm ES4326/avrRpt2 inoculation (Figure 4G), consistent with the results from the study using the GUS reporter (Figure 4E). Moreover, this polysomal association appeared to be transient as the TBF1 transcript decreased 1 hr post inoculation. In contrast, polysomal association of the housekeeping gene UBQ5 transcript was not significantly increased upon pathogen challenge (Figure S4).
Both uORFs of TBF1 are enriched in aromatic amino acids, particularly in phenylalanine (Phe). Intriguingly, this Phe enrichment is evolutionarily conserved for uORF2 among the TBF1 homologs in other plant species [25]. We hypothesize that pathogen challenge may result in a decrease in cellular Phe concentration and allow translation initiation of TBF1 downstream of uORFs.
Because repeated efforts in measuring Phe concentrations in plants infected with Psm ES4326/avrRpt2 have not produced consistent results, we employed a yeast-based reporter system (see Experimental Procedures). This system involves the use of the yeast strain aro7, which is auxotrophic for Phe and tyrosine, and the reporter containing the mouse DHFR (dihydrofolate reductase), which has been engineered to be less stable [26] and resistant to methotrexate (MTX) [27]. In the presence of MTX, yeast growth becomes dependent on the recombinant DHFR reporter expression. We inserted uORF1-uORF2-TBF11st exon upstream of the DHFR reporter to generate uORF1-uORF2-DHFR and transformed this construct into aro7. The resulting transformant was cultured in the presence of MTX and growth was recorded over the course of time.
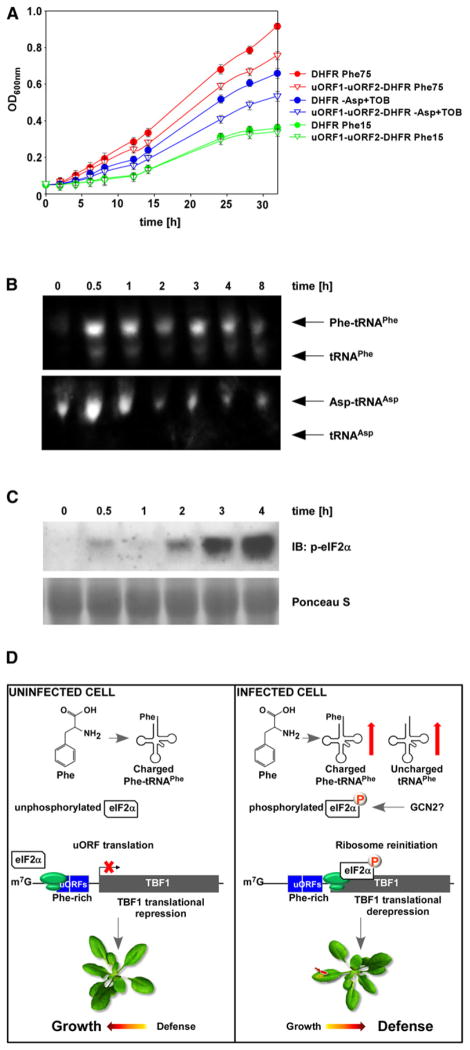
As shown in Figure 5A, under the Phe-rich conditions (75 mg/L; standard Phe concentration), the yeast strain carrying the DHFR displayed a much higher growth rate than the strain carrying the uORF1-uORF2-DHFR, showing the inhibitory effects of the uORFs on DHFR translation. However, under the Phe-restricting conditions (15 mg/L), both strains grew at similar rates, suggesting that low Phe level released the inhibitory effects of uORFs on DHFR translation (Figure 5A). To ensure that the TBF11st exon –DHFR fusion protein was not toxic to yeast cells, we grew the strains in the absence of MTX and noted no significant difference in their growth (Figure S5). The uORFs of TBF1 appear to be specifically sensitive to Phe starvation because aspartic acid (Asp) starvation caused by addition of tobramycin (TOB), a known inhibitor of yeast tRNAAsp aspartylation [28], did not eliminate the difference in growth rate between the strain carrying uORF1-uORF2-DHFR and the strain carrying DHFR.
Figure 5. TBF1 Translation Is Regulated in Response to Pathogen-Induced Changes in Phenylalanine Metabolism.
(A) The effects of phenylalanine and aspartate starvation on the translational inhibitory function of uORFs were measured by growth of the yeast strain aro7 (phe−, tyr−) transformed with the uORF1-uORF2-DHFR or DHFR reporter in medium containing methotrexate, an inhibitor of the endogenous DHFR. Optical densities for cultures containing two different concentrations of phenylalanine (Phe; 15 and 75 mg/L) as well as cultures lacking Asp and supplemented with tobramycin (TOB), an inhibitor of yeast tRNAAsp aspartylation, were recorded over the course of 32 hr. Error bars represent standard deviation from nine technical replicates derived from three independent experiments. See also Figure S5.
(B) tRNA analysis of WT plants at various time points after inoculation with Psm ES4326/avrRpt2 (OD600nm = 0.02). Northern blot using probes against tRNAPhe or tRNAAsp was performed to detect charged and uncharged tRNAPhe or tRNAAsp. This experiment was repeated using three biological replicates with similar results.
(C) Phosphorylated form of eIF2α was detected using a phospho-specific antibody in the total protein extracts from leaves of 3-week-old WT plants collected at various time points after inoculation with Psm ES4326/avrRpt2 (OD600nm = 0.02). Ponceau S stain was used to determine equal loading.
(D) A model illustrating the molecular mechanism by which the translation initiation of TBF1 is regulated through rapid increases in uncharged and charged tRNAPhe, phosphorylation of eIF2α, and ribosomal read-through of uORFs, leading to the growth-to-development transition.
We next sought to understand how Phe starvation alleviates uORFs-mediated repression of TBF1 translation. In yeast, amino acid starvation results in a series of events including uncharged tRNAs accumulation, activation, and autophosphorylation of GCN2, GCN2-mediated phosphorylation of eukaryotic initiation factor 2α (eIF2α), and translation initiation of GCN4 downstream of uORFs [29–32]. To determine whether a similar mechanism controls the translation of TBF1, we first performed northern blot analysis to measure charged and uncharged tRNA levels following inoculation with Psm ES4326/avrRpt2. We detected a dramatic increase in both charged and uncharged tRNAPhe within 30 min of the bacterial inoculation, which persisted for 8 hr (Figure 5B). In contrast, only a moderate increase in charged tRNAAsp level was observed under the same conditions. No uncharged tRNAAsp was detected. This is consistent with our hypothesis that pathogen challenge in plants can lead to specific changes in Phe metabolism.
Because a functional and stress-inducible GCN2-eIF2α pathway has been found in Arabidopsis [33], we investigated whether the pathogen-induced accumulation of uncharged tRNAPhe could lead to phosphorylation of eIF2α. In an immunoblot experiment, we detected a rapid accumulation of the phosphorylated eIF2α in Arabidopsis leaves infected with Psm ES4326/avrRpt2 (Figure 5C). The phosphorylated eIF2α may facilitate ribosome reattachment to the TBF1 translation start codon downstream of uORFs to initiate TBF1 translation.
Taken together, we concluded that TBF1 expression is tightly controlled not only at the transcriptional level by NPR1 but also at the translational level through uORFs. Pathogen challenge, which causes a temporary increase in uncharged tRNAPhe accumulation, triggers eIF2α phosphorylation and consequently derepression of TBF1 translation.
Discussion
The presence of TL1 in a wide array of defense-related gene promoters (Figures 2D and 2E) [3, 34, 35] suggests a critical role for its binding TF in plant immune responses. Consistent with these genomic data, we demonstrated that the tbf1 mutant plants are impaired in UPR, elf18-induced MTI and SA-mediated SAR, but not in the heat-shock response (Figure 3; Figure S3B). Evolution of such TFs with novel functions may explain the greater number of HSF-like genes in plants than in other organisms (one each in yeast and Drosophila, three in vertebrates) [36].
Activation of the immune system requires significant metabolic activities. It is well known that mutant plants with constitutively activated defense response often have stunted growth and retarded development [37]. Our study demonstrates that in response to MTI and SAR induction, TBF1 is a master molecular switch for the growth-to-defense reprogramming that involves activation and repression of nearly 3,000 genes, of which 46% contain at least one copy of the TL1 element in their promoters. TBF1 controls not only immune response genes but also genes involved in growth and development.
The pivotal role that TBF1 plays in the growth-to-defense transition underlines the importance of understanding its regulation. TBF1 expression is tightly controlled at both transcriptional and translational levels. Transcription of TBF1 and NPR1 appears to be interdependent because mutation of either gene affects transcription of the other (Figures 4A and 4B). TBF1 may directly regulate NPR1 expression through the TL1 elements in the NPR1 promoter. Because the NPR1 promoter also contains W-boxes [38], it is also possible that TBF1 regulates NPR1 indirectly through its transcriptional targets, WRKY TFs (Table S1). Reciprocally, NPR1 may regulate TBF1 expression through either WRKY or TGA TFs because the TBF1 promoter contains five W-boxes and three TGA binding sites known as the as-1 elements [39].
The two uORFs of TBF1 link its translation with cellular amino acid availability. Approximately 10% of eukaryotic mRNA contain uORFs, and a high percentage of them encode critical cellular regulators: protooncogenes, TFs, receptors, and proteins involved in immune responses [40]. Expression of such genes is highly regulated, because their protein products are essential for controlled cell growth and proliferation. Indeed, TBF1 is such a regulator as transgenic lines overexpressing the TBF1 cDNA under the constitutive 35S promoter were not viable (K.M.P.-M. and X.D., unpublished data).
Pathogen challenge and the subsequent increase in uncharged tRNAPhe and phosphorylation of eIF2α release the inhibitory effects of uORFs on TBF1 translation (Figure 5D). This is reminiscent of the regulatory mechanism described for the well-studied yeast GCN4 and mammalian ATF4 (reviewed in [32]). The yeast GCN4 transcript contains four uORFs in its 5′ region [41]. Under normal conditions, ribosomes bind to the 5′ cap of GCN4 mRNA and initiate translation of the first uORF. They are unable to reinitiate translation at the start codon of GCN4. During amino-acid starvation, uncharged tRNAs induce phosphorylation of eIF2α mediated by GCN2, which hinders reassembly of the 80S ribosome after translation of uORF1. This allows the 40S ribosomal subunit to continue scanning the mRNA and reinitiate translation at the start codon of GCN4 [41].
Whereas derepression of GCN4 translation can be triggered by starvation of any amino acid, uORF-mediated regulation of TBF1 in plants appears to be controlled by the metabolism of specific amino acids, such as Phe. It is uncertain whether pathogen infection causes a transient starvation of Phe. The rapid increase in the uncharged tRNAPhe after pathogen challenge coincided with an increase of the total tRNAPhe suggesting that the accumulation of uncharged tRNAPhe may result from decreased availability of Phe as well as increased tRNAPhe synthesis (Figure 5B). However, it is tempting to hypothesize that pathogen infection affects Phe availability for translation because aromatic amino acids are known precursors for a large array of plant metabolites such as the growth hormone auxin, the SAR signal SA, cell wall components, and pigments such as anthocyanins [4, 42, 43]. Similar to GCN4, the accumulation of uncharged tRNAPhe triggers phosphorylation of eIF2α, ribosomal movement through uORFs, and translation of TBF1. The translation regulatory mechanism of TBF1 allows the cell to quickly detect pathogen-triggered metabolic changes and produce the TBF1 protein to activate immune responses. Our data suggest that plants employ TBF1 in response to infection to rapidly reprogram cellular transcription, which diverts energy resources to cope with pathogens at the expense of growth and development (Figure 5D).
Experimental Procedures
Translational Analysis of uORF1-uORF2-GUS
The DNA fragment containing the 5′UTR and the first exon of TBF1 (designated uORF1-uORF2) was PCR amplified using primers TBF1 5′UTR-GW-F and TBF1 5′UTR-GW-R (Table S3) and cloned into the Gateway vector pDONR207 (Invitrogen). Two A-to-C point mutations were introduced, either separately or together, into the start codons (ATG) of uORF1 and uORF2. The WT and mutant uORF1-uORF2 sequences were inserted downstream of the constitutive 35S promoter and upstream of the coding region of the GUS reporter in pMDC140 through recombination [44]. The resulting translational reporters were transformed into Col-0 WT plants or transiently expressed in Nicotiana benthamiana using Agrobacterium tumefaciens [45]. For Arabidopsis stable transgenic lines, two independent T3 lines homozygous for each construct were chosen for quantitative GUS assay [3] at 0, 0.5, 1, 2, 3, 4, and 8 hr after Psm ES4326/avrRpt2 infiltration (OD600nm = 0.02). Further detailed experimental procedures are described in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank N. Spivey for sharing microarray data, D. Thiele and D. Neef for performing TBF1 complementation assay in yeast hsf1 mutant, C. Nicchitta for assistance in the polysome profiling experiment, T. Pan for input in the tRNA study, M. Froneberger for helping with the experiments, and J. Siedow and S. Mukhtar for critical reading of the manuscript and helpful discussions. This project was supported by a grant from the National Science Foundation (MCB-0519898) to X.D., Grants-in-Aid for Scientific Research (No. 23120520) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to Y.T., and the Hargitt Fellowship to K.M.P.-M. X.D. is a Howard Hughes Medical Institute – Gordon and Betty Moore Foundation investigator.
Footnotes
Accession Numbers
The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE34047.
Supplemental Information includes five figures, three tables, and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.cub.2011.12.015.
References
- 1.Nishimura MT, Dangl JL. Arabidopsis and the plant immune system. Plant J. 2010;61:1053–1066. doi: 10.1111/j.1365-313X.2010.04131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 4.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 5.Kwon C, Bednarek P, Schulze-Lefert P. Secretory pathways in plant immune responses. Plant Physiol. 2008;147:1575–1583. doi: 10.1104/pp.108.121566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, Dong X, Robatzek S, Schulze-Lefert P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339–2350. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J, Durrant WE, Wang S, Yan S, Tan EH, Dong X. DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe. 2011;9:115–124. doi: 10.1016/j.chom.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Curr Opin Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci. 2004;29:471–487. doi: 10.1007/BF02712120. [DOI] [PubMed] [Google Scholar]
- 13.Boscheinen O, Lyck R, Queitsch C, Treuter E, Zimarino V, Scharf KD. Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1997;255:322–331. doi: 10.1007/s004380050503. [DOI] [PubMed] [Google Scholar]
- 14.Busch W, Wunderlich M, Schöffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 2005;41:1–14. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- 15.Prändl R, Hinderhofer K, Eggers-Schumacher G, Schöffl F. HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen Genet. 1998;258:269–278. doi: 10.1007/s004380050731. [DOI] [PubMed] [Google Scholar]
- 16.Czarnecka-Verner E, Pan S, Salem T, Gurley WB. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol. 2004;56:57–75. doi: 10.1007/s11103-004-2307-3. [DOI] [PubMed] [Google Scholar]
- 17.Czarnecka-Verner E, Yuan CX, Scharf KD, Englich G, Gurley WB. Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol Biol. 2000;43:459–471. doi: 10.1023/a:1006448607740. [DOI] [PubMed] [Google Scholar]
- 18.Panstruga R, Parker JE, Schulze-Lefert P. SnapShot: Plant immune response pathways. Cell. 2009;136:978. doi: 10.1016/j.cell.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Sugano S, Jiang CJ, Miyazawa S, Masumoto C, Yazawa K, Hayashi N, Shimono M, Nakayama A, Miyao M, Takatsuji H. Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol Biol. 2010;74:549–562. doi: 10.1007/s11103-010-9695-3. [DOI] [PubMed] [Google Scholar]
- 20.Kumar M, Busch W, Birke H, Kemmerling B, Nürnberger T, Schöffl F. Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant. 2009;2:152–165. doi: 10.1093/mp/ssn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, Zipfel C, Jones JD. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Tintor N, Mentzel T, Kombrink E, Boller T, Robatzek S, Schulze-Lefert P, Saijo Y. Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc Natl Acad Sci USA. 2009;106:22522–22527. doi: 10.1073/pnas.0907711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden CA, Jorgensen RA. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 2007;5:32. doi: 10.1186/1741-7007-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker CL, Fields S. A yeast sensor of ligand binding. Nat Biotechnol. 2001;19:1042–1046. doi: 10.1038/nbt1101-1042. [DOI] [PubMed] [Google Scholar]
- 27.Ercikan-Abali EA, Waltham MC, Dicker AP, Schweitzer BI, Gritsman H, Banerjee D, Bertino JR. Variants of human dihydrofolate reductase with substitutions at leucine-22: effect on catalytic and inhibitor binding properties. Mol Pharmacol. 1996;49:430–437. [PubMed] [Google Scholar]
- 28.Walter F, Pütz J, Giegé R, Westhof E. Binding of tobramycin leads to conformational changes in yeast tRNA(Asp) and inhibition of aminoacylation. EMBO J. 2002;21:760–768. doi: 10.1093/emboj/21.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey M, Cao C, Sicheri F, Dever TE. Conserved intermolecular salt bridge required for activation of protein kinases PKR, GCN2, and PERK. J Biol Chem. 2007;282:6653–6660. doi: 10.1074/jbc.M607897200. [DOI] [PubMed] [Google Scholar]
- 30.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 31.Padyana AK, Qiu H, Roll-Mecak A, Hinnebusch AG, Burley SK. Structural basis for autoinhibition and mutational activation of eukaryotic initiation factor 2alpha protein kinase GCN2. J Biol Chem. 2005;280:29289–29299. doi: 10.1074/jbc.M504096200. [DOI] [PubMed] [Google Scholar]
- 32.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 33.Lageix S, Lanet E, Pouch-Pélissier MN, Espagnol MC, Robaglia C, Deragon JM, Pélissier T. Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 2008;8:134. doi: 10.1186/1471-2229-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabro G, Di Rienzo JA, Voigt CA, Savchenko T, Dehesh K, Somerville S, Alvarez ME. Genome-wide expression profiling Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant Physiol. 2008;146:1421–1439. doi: 10.1104/pp.107.111286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphry M, Bednarek P, Kemmerling B, Koh S, Stein M, Göbel U, Stüber K, Pislewska-Bednarek M, Loraine A, Schulze-Lefert P, et al. A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc Natl Acad Sci USA. 2010;107:21896–21901. doi: 10.1073/pnas.1003619107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heidel AJ, Clarke JD, Antonovics J, Dong X. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics. 2004;168:2197–2206. doi: 10.1534/genetics.104.032193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 39.Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller PF, Hinnebusch AG. cis-acting sequences involved in the translational control of GCN4 expression. Biochim Biophys Acta. 1990;1050:151–154. doi: 10.1016/0167-4781(90)90157-w. [DOI] [PubMed] [Google Scholar]
- 42.Holton TA, Cornish EC. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pajerowska-Mukhtar KM, Mukhtar MS, Guex N, Halim VA, Rosahl S, Somssich IE, Gebhardt C. Natural variation of potato allene oxide synthase 2 causes differential levels of jasmonates and pathogen resistance in Arabidopsis. Planta. 2008;228:293–306. doi: 10.1007/s00425-008-0737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.