Abstract
Background
Atopy and plasma IgE concentration are genetically complex traits, and the specific genetic risk factors that lead to IgE dysregulation and clinical atopy are an area of active investigation.
Objective
To ascertain the genetic risk factors which lead to IgE dysregulation.
Methods
A genome wide association study (GWAS) was performed in 6,819 participants from the Framingham Heart Study (FHS). Seventy of the top SNPs were selected based on p-values and linkage disequilibrium among neighboring SNPs and evaluated in a meta-analysis with five independent populations from the KORA, B58C, and CAMP cohorts.
Results
Thirteen SNPs located in the region of three genes, FCER1A, STAT6, and IL-13, were found to have genome-wide significance in the FHS GWAS. The most significant SNPs from the three regions were rs2251746 (FCER1A, p-value 2.11×10-12), rs1059513 (STAT6, p-value 2.87×10-08), and rs1295686 (IL-13, p-value 3.55×10-08). Four additional gene regions - HLA-G, HLA-DQA2, HLA-A, and DARC - reached genome-wide statistical significance in meta-analysis combining FHS and replication cohorts, although the DARC association did not appear independent of SNPs in the nearby FCER1A gene.
Conclusion
This GWAS of the FHS has identified genetic loci in HLA genes that may have a role in the pathogenesis of IgE dysregulation and atopy. It also confirmed the association of known susceptibility loci, FCER1A, STAT6, and IL-13, for the dysregulation of total IgE.
Keywords: total IgE, atopy, asthma, GWAS
Introduction
Immunoglobulin E (IgE)-mediated allergy to environmental allergens plays a central role in the pathophysiology of asthma, allergic rhinoconjunctivitis, atopic dermatitis, and food allergy. Atopic persons have abnormally high concentrations of IgE reactive against one or more specific allergens and often have an elevated plasma total IgE concentration.1 Atopy and elevated IgE concentrations are genetically complex traits, and the specific genetic risk factors that lead to IgE dysregulation and clinical atopy are an area of active investigation.
Genetic linkage studies of total IgE levels have been conducted in numerous samples ascertained for asthma and atopy with associations reported for locations on multiple chromosomes. Fine mapping of genes at several of these locations as well as candidate gene studies have implicated a number of genetic variants as potentially important determinants of plasma IgE concentration, with IL-4, IL-13, and STAT6 among the most consistently replicated associations.2-3
To date, there have been two genome-wide association studies (GWAS) published investigating total IgE concentration. Weidinger et al.4, in 2008, reported findings from a population-based, German cohort of 1,530 individuals, with replication analyses performed in four independent population-based study samples that included a total n=9,769 subjects. The study found that functional variants of the alpha chain of the high affinity receptor for IgE (FCER1A) were strongly associated with total IgE levels. The study also confirmed STAT6 as a susceptibility locus as well as identifying RAD50, located adjacent to the IL-13 gene, as a potential determinant of IgE dysregulation. More recently, the GABRIEL asthma genetics consortium found, among both asthmatic cases and non-asthmatic controls, a SNP near HLA-DRB1 to be associated with total IgE with genome-wide statistical significance, as well as evidence of association for FCER1A, STAT6, and IL-13.5
In this study, we report the results of a GWAS of plasma total IgE concentration in the Framingham Heart Study. In addition to confirming FCER1A, STAT6, and IL-13 as susceptibility genes for IgE dysregulation, we identify genetic variants in HLA genes as potential determinants of atopy and IgE concentration.
Methods
Subjects
The Framingham Heart Study
In 1948, two thirds of the age-eligible men and women from the town of Framingham, Massachusetts, were recruited for the first round of physical examinations and lifestyle interviews to identify risk factors for cardiovascular disease. This Framingham Heart Study (FHS) Original Cohort included 5,209 men and women who were between the ages of 28 and 62 years. Beginning in 1971, the FHS Offspring Cohort was established, comprising 5,124 men and women who were either the offspring of the Original Cohort or spouses of those offspring. In 2002, 4,095 adult men and women who were the children of the Offspring Cohort were enrolled in the Third Generation Cohort.
During each examination cycle, the participants undergo a detailed examination including physical examination, medical history, laboratory testing, and electrocardiogram. Over the years, other tests have included pulmonary function, lifestyle questionnaires, cognitive function questionnaires, and noninvasive cardiovascular tests including echocardiograms.
Replication Cohorts
KORA studies
The Cooperative Health Research in the Region of Augsburg (KORA) cohorts KORA S3 and KORA S4 are independent population-based samples from the general population living in the region of Augsburg, Southern Germany, and were examined in 1994/95 (KORA S3) and 1999/2001 (KORA S4).6 The KORA S3 sample included 4,856 subjects (participation rate 75%), and the KORA S4 sample included 4,261 subjects (participation rate 67%). In the KORA S3 sample, 1,644 subjects were randomly selected for genotyping, including 1,530 individuals with total IgE level available. From KORA S4, 1,814 subjects were randomly selected for genotyping, including 1,764 individuals with measurements on total IgE. Total IgE concentration was measured using the FEIA CAP system (Pharmacia, Freiburg, Germany). Genotyping was performed using the Affymetrix 500K Gene Chip for KORA S3 and Affymetrix 6.0 for KORA S4. Imputation of SNP genotypes that were not directly measured was implemented using IMPUTE.7 For the selected SNPs, additive genetic models were fitted on logarithmically-transformed IgE levels adjusting for sex and age using SNPTEST (http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html).
British 1958 Birth Cohort
The British Birth Cohort is an ongoing follow-up of all persons born in Great Britain during one week in 1958, including a biomedical assessment during 2002 2004 8 at which blood samples and informed consent were obtained for creation of a genetic resource (http://www.b58cgene.sgul.ac.uk/). Through use of this resource as a nationally representative control sample, whole-genome typing was carried out on separate subsets of the cohort by the Wellcome Trust Case-Control Consortium (B58C-WTCCC) 9 and the Type 1 Diabetes Genetic Consortium (B58C-T1DGC).10 For the B58C-WTCCC subset, Affymetrix 500K genotypes were imputed to the HapMap 2 CEU template using IMPUTE, while for the B58C-T1DGC subset, genotyping was performed using an Illumina Infinium 550K array and imputed to the HapMap 2 CEU template using MACH. Total IgE was assayed by the HYTEC automated enzyme immunoassay (Hycor Biomedical, Edinburgh, United Kingdom) and results of its association with both Affymetrix and Illumina SNPs, and with HLA subtypes measured by Dynal11 are displayed online (http://www.b58cgene.sgul.ac.uk/phenosearch.php?pheno=1).
Childhood Asthma Management Program (CAMP)
CAMP was a multicenter, randomized, double-masked clinical trial initially designed to determine the effects of three inhaled treatments for mild to moderate childhood asthma.12 One thousand forty-one children aged 5 to 12 years at screening were enrolled, of which 568 Caucasian children with available genotype and IgE data were included in this analysis. Genome-wide SNP genotyping for CAMP subjects was performed on either Illumina’s HumanHap550 or Human610-Quad BeadChip and imputation was performed using MACH based on 1000 Genomes Project haplotypes. Serum total IgE was measured by radioimmunosorbent assay.
Measurement of Total IgE
The plasma samples used in the FHS study were collected and frozen at -80 degrees C at examination 24 for the Original Cohort, examination 7 for the Offspring Cohort, and at examination 1 for the Generation 3 Cohort. Total IgE measurements were performed using the Phadia Immunocap 100 system, in which an anti-IgE antibody is bound to a solid-phase carrier followed by flouroenzyme-based quantitative measurement of total IgE with high precision and reproducibility.13
Genome-wide genotyping and imputation procedures
Framingham participants were genotyped using the Affymetrix (Santa Clara, CA) GeneChip Human Mapping 500K Array Set, which was comprised of two arrays generating approximately 262,000 SNPs with Nsp arrays and 238,000 SNPs with Sty arrays. An additional Affymetrix 50K Array (HuGeneFocused50K) with gene-centric and coding SNPs was also genotyped for a total of approximately 550K SNPs. Imputation of SNP genotypes that were not directly measured was performed using the western European (CEU) reference panel in HapMap release 22, build 26 and the Markov Chain haplotyping (MACH) software (http://www.sph.umich.edu/csg/abecasis/MACH/) to increase genome coverage to 2,540,223 SNPs. This procedure also filled in missing data for the genotyped SNPs.
These genotype and comprehensive phenotype data have been made publicly available through the NHLBI’s SNP Health Association Resource (SHARe) initiative (http://public.nhlbi.nih.gov/GeneticsGenomics/home/share.aspx).
Statistical Analysis
Total IgE measures were log-transformed (base 10) to attain a normal distribution and, in the FHS family sample, were examined using linear-mixed effects (LME) models with fixed effects for SNP genotypes and random effects for individuals correlated within families due to polygenic/familial shared effects. Population stratification in the Framingham sample was assessed by principal component analysis using Eigenstrat14 (http://genepath.med.harvard.edu/~reich/Sortware.htm). Association of IgE with allele dose for each of the imputed genotypes was analyzed using an additive model. Age, sex, current smoking, pack-years of smoking, cohort membership, and principal component 1 were included as covariates in the LME model.
To identify SNPs for replication, a nominal p-value threshold of < 10-4 resulted in 527 eligible SNPs. For each gene within this group, the highest associated SNP based on p-value was chosen for replication (n=33). SNPs within +/- 500 kb of the original SNP were also selected if minor allele frequencies differed by > 0.05 (n=15). Of the remaining regions which lay outside of reference genes, the same strategy was used to select an additional 22 SNPs, with a total of 70 SNPs selected for replication.
Association of total IgE with SNPs in the Framingham dataset (discovery sample) was assessed in the replication datasets using meta-analysis based on log10 transformation of total IgE concentration. For cohorts in which the original analysis was performed using natural log transformation of IgE concentration, the coefficients were converted to log10 for the meta-analysis. Meta-analysis was performed using Metal software with weights based on the standard errors of the beta estimates (inverse variance method) (http://www.sph.umich.edu/csg/abecasis/metal/index.html). We performed a meta-analysis of the 70 SNPs selected for replication among the replication cohorts only and among the replication cohorts plus FHS. SNAP (version 2.0; http://broad.mit.edu/mpg/snap) was used to generate regional association plots.
Results
Characteristics of the FHS participants included in the IgE GWAS are provided in Table 1, and characteristics of replication cohorts along with FHS participants are provided in Table E1 in the Online Repository. A Manhattan plot of the GWAS findings is shown in Figure 1. The Q-Q plot (Figure E1) does not show strong departure from the distribution of expected p-values by chance, with a genomic control parameter of 1.029, suggesting that familial relationships and population stratification were appropriately controlled for in the analysis.
Table 1.
Descriptive characteristics of the Framingham Heart Study Participants
| Original Cohort N=860 | Offspring Cohort N=2377 | Generation 3 Cohort N=3681 | |
|---|---|---|---|
| Age | 73 (11) | 60 (9) | 40 (9) |
| Female | 56% | 55% | 53% |
| Current smoker | 34% | 11% | 16% |
| Pack years | 30 (22) | 27 (23) | 13 (13) |
| Asthma | n/a | 8% | 16% |
| Hay fever | n/a | 29%* | 47% |
| Log IgE | 1.59 (0.64) | 1.55 (0.59) | 1.51 (0.56) |
Prevalence of hay fever for the FHS Offspring Cohort was ascertained 5 years after plasma collection for IgE measurement.
Figure 1.
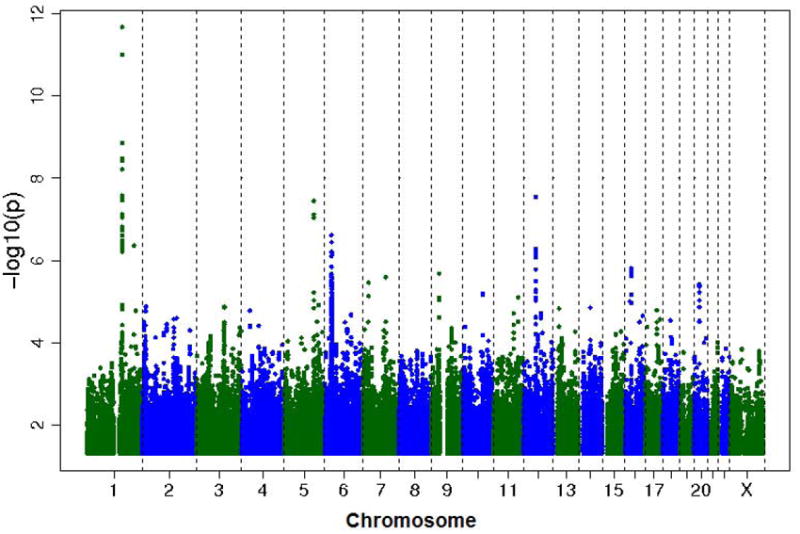
Genome wide association results for imputed SNPs and plasma total IgE concentration in the Framingham Heart Study. Coordinates on the X-axis represent chromosomes. Negative logarithm p-values are shown on the y-axis
In the FHS GWAS, 13 SNPs met genome-wide significance as determined by the commonly accepted significance threshold of 5 × 10-08.15 All 13 SNPs were within 60 kb of three genes, FCER1A, STAT6, and IL-13. Details of these 13 SNPs, as well as all 527 SNPs with a nominal p-value < 10 -4, can be seen in Figures E2-E4 and Table E2, respectively, in the Online Repository.
Of the top 70 SNPs selected for the meta-analysis with replication cohorts (Table E3), 10 SNPs demonstrated replication based on a Bonferroni-corrected alpha criterion of p < 0.05 / 70 = 0.0007 in a meta-analysis of the replication cohorts. (Table 2). Two additional SNPs, near the gene regions, HLA-A and HLA-G, did not meet the p < 0.0007 criterion in the replication cohorts but had association p values meeting the genome-wide significance criterion of p < 5 × 10-08 in meta-analysis of the replication cohorts together with the FHS discovery cohort (Table 2). Cohort-specific association results for these 12 SNPs are shown in Table E4. The top 2 SNPs, rs2251746 and rs2494264, located in non-coding regions of FCER1A, had combined (FHS plus replication cohorts) meta-analysis p-values of 4.52 × 10-26 and 9.40 × 10-20, respectively. SNPs rs1059513 in STAT6 and rs20541 in IL-13 were also associated with total IgE level at genome-wide significance, each with a combined meta-analysis p-value < 10-11.
Table 2.
Association results in the Framingham Heart Study and in meta-analysis of replication cohorts for the 12 SNPs with the strongest evidence of association with IgE concentration*
| Nearest Gene | SNP | Chromo some | Position | Minor Allele | MAF | FHS Beta | FHS P-value | Replication† Beta | Replication† P-value | Combined‡ Beta | Combined‡ P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FCER1A | rs2251746 | 1 | 157538684 | C | 0.26 | -0.084 | 2.11E-12 | -0.089 | 3.07E-15 | -0.087 | 4.52E-26 |
| FCER1A | rs2494264 | 1 | 157533462 | A | 0.46 | -0.064 | 1.37E-09 | -0.067 | 1.15E-11 | -0.066 | 9.40E-20 |
| STAT-6 | rs1059513 | 12 | 55775976 | C | 0.09 | -0.164 | 2.87E-08 | -0.095 | 2.08E-06 | -0.116 | 1.99E-12 |
| IL-13 | rs20541 | 5 | 132023863 | A | 0.19 | 0.075 | 3.55E-08 | 0.086 | 1.41E-11 | 0.081 | 3.41E-18 |
| HLA-G | rs2523809 | 6 | 29957598 | T | 0.13 | 0.085 | 2.40E-07 | 0.041 | 8.19E-03 | 0.062 | 4.34E-08 |
| HLA-A | rs2517754 | 6 | 30004659 | A | 0.53 | 0.054 | 3.59E-07 | 0.060 | 2.73E-03 | 0.055 | 3.61E-09 |
| FCER1A | rs4656784 | 1 | 157593504 | G | 0.19 | -0.075 | 3.84E-07 | -0.084 | 7.43E-11 | -0.080 | 1.68E-16 |
| STAT6 | rs167769 | 12 | 55790042 | T | 0.35 | 0.056 | 5.18E-06 | 0.045 | 1.47E-05 | 0.049 | 4.02E-10 |
| IL-13 | rs2243297 | 5 | 132027070 | A | 0.04 | 0.128 | 7.19E-05 | 0.127 | 5.58E-05 | 0.128 | 1.54E-08 |
| HLA-A | rs2571391 | 6 | 30031817 | C | 0.32 | -0.054 | 2.13E-06 | -0.069 | 7.43E-11 | -0.062 | 1.23E-15 |
| DARC | rs13962 | 1 | 157442151 | A | 0.13 | -0.087 | 1.52E-05 | -0.087 | 3.29E-07 | -0.087 | 2.21E-11 |
| HLA-DQA2 | rs2858331 | 6 | 32789255 | G | 0.44 | 0.048 | 2.03E-05 | 0.039 | 1.51E-04 | 0.043 | 1.44E-08 |
The SNPs in this table demonstrate an association with either p < 0.0007 in meta-analysis of replication cohorts or p < 5 × 10-08 in combined meta-analysis of FHS discovery cohort and replication cohorts. For the FHS analysis and the combined meta-analysis, p-values which demonstrate genome-wide significance are shown in bold. For the replication analysis, p-values which demonstrate significance based on p < 0.0007 are shown in bold.
Meta-analysis of replication cohorts only
Meta-analysis of replication cohorts combined with FHS cohort
Among the genome wide-significant associations based on combined meta-analysis p-values were SNPs in or near the HLA-A, HLA-G, HLA-DQA2, and DARC genes (Table 2). Two SNPs within a single LD block near the HLA-A gene were found to have meta-analysis p-values < 5 × 10-8 (http://www.broad.mit.edu.mpg/haploview). rs2571391 is located within 60 kb of the HLA-A gene and was the most highly associated SNP in this region, with a combined p-value of 1.23 × 10-15. Association plots for the regions encompassing HLA-A and HLA-G (Figure E5) and HLA-DQA2 (Figure E6) depict the associations in these regions in FHS.
The SNP rs13962 is a missense SNP in the DARC gene and was associated with total IgE with a combined p-value of 2.21 × 10-11. Given its close proximity to the FCER1A gene, a conditional model regression analysis was performed to assess the independence of this association. After adjusting for rs2251746, the top FCER1A SNP, the association of DARC was not found to be significant in FHS (p-value = 0.6). Similar findings were seen in all of the replication cohorts.
The GABRIEL consortium recently reported evidence of associations for both asthma and total IgE with SNPs in the HLA-DQB1 – HLA-DRB1 region.5 The total IgE peak was distinct from the asthma peak and best represented by rs9271300, which was not included in our meta-analysis because it is not part of the HapMap 2 panel. We were, however, able to impute rs9271300 with moderate accuracy (observed to expected variance ratio = 0.5) using the 1000 Genomes Project June 2010 haplotypes as a template.16 The association of this imputed SNP with log10-transformed total IgE in the FHS dataset was not significant (p=0.06); however, the 95% confidence interval (-0.001 to +0.061) around the regression coefficient of 0.03 in FHS included the GABRIEL regression coefficient (0.06) for this association, suggesting that the findings are not heterogeneous with those of the GABRIEL study.
Discussion
We found that SNPs in three gene regions, FCER1A, STAT6, and IL-13, were associated with total IgE concentration at a genome-wide significant level in the FHS cohort. The results confirm the findings from prior studies that have implicated these genes in the pathogenesis of IgE dysregulation. Additionally, we observed associations with SNPs near three other genes, HLA-A, HLA-G, and HLA-DQA2, for which relationships to IgE have not been clearly established.
Our analysis reveals associations, which replicate in independent cohorts and/or reach genome-wide significance in meta-analysis combining FHS with replication cohorts, in the region of the MHC superlocus on chromosome 6. While MHC II antigens, particularly HLA-DRB1 and HLA-DQB1, have been consistently associated with atopic phenotypes in the genetic literature,17-18 evidence for a genetic susceptibility via the classical MHC I antigens (HLA-A, -B, -C) is less clear. The stronger association of MHC II antigens versus MHC I antigens in the literature is consistent with their functional roles in the immune system, with MHC I responsible for initiating cell mediated immunity, targeting infected and cancer cells, and MHC II mediating humoral immunity, including the production of IgE antibodies to allergenic antigens.
Three SNPs in or near the HLA-A gene, an MHC I antigen, were found to have p-values < 2 × 10-7 in FHS, with the highest association seen for rs2571391 with a combined (FHS plus replication cohorts) meta-analysis p-value of 1.23 × 10-15. The first study to identify a linkage between HLA-A haplotypes and ragweed allergy was published in 1972 by Levine et al.,19 though a subsequent, larger study was unable to replicate the original findings.20 In 1979, Marsh et al published a regression analysis evaluating total IgE and found both positive (HLA-A2) and negative (HLA-A3) associations among the HLA-A antigens in patients with varying degrees of ragweed sensitivity.21 More recent studies have been unable to find significant associations between HLA-A and total IgE,22 intrinsic or extrinsic asthma,20 and mite sensitive allergy in Greek and Venezuelan populations.23-24 However, results published online from the British 1958 birth cohort show associations (at conventional levels of significance) between log-transformed total IgE and directly measured HLA-A subtypes HLA*02, *03, and *29 [see URLs below]. (http://www.b58cgene.sgul.ac.uk/phenotype0.php?snp=hlaa_02&pheno=1) (http://www.b58cgene.sgul.ac.uk/phenotype0.php?snp=hlaa_03&pheno=1) (http://www.b58cgene.sgul.ac.uk/phenotype0.php?snp=hlaa_29&pheno=1)
In the clinical literature, many studies in children have demonstrated a relationship between viral infection, particularly RSV, and the later development of asthma and increases in specific IgE.25-26 One hypothesis is that MHC I polymorphisms may influence IgE concentration through the processing of infectious pathogens, leading to immune dysregulation and, eventually, the clinical findings of atopy and asthma.
Our findings suggest that HLA-A may represent a susceptibility locus for IgE dysregulation. While both MHC I and MHC II loci were found to be associated with total IgE, the association of HLA-A was consistently stronger in both the original FHS GWAS and in the meta-analysis than the MHC II antigens, HLA-DQB1 and HLA–DRB1.
Additionally, a SNP near the HLA-G gene, a non-classical MHC I antigen that has been associated with asthma and bronchial hyper-responsiveness in a positional candidate gene study27 was also found to reach genome wide significance in the combined meta-analysis. HLA-G has not previously been associated with total IgE dysregulation but is thought to have immunomodulatory effects via actions on NK cells, T lymphocytes, and antigen presenting cells.28
The HLA-DQA2 region had the strongest association among MHC II loci, with rs2858331, a SNP within 27 kb from the HLA-DQA2 gene, observed at a genome-wide significant level in the combined meta-analysis (p-value = 1.44 × 10-08). HLA-DQA2 is thought to play a central role in peptide loading of MHC II molecules. Though it has never been associated with total IgE in the literature, a genome-wide association study by Li et al. found a strong association of asthma with a nearby SNP, rs3916765, in the same region.29
In the GABRIEL Consortium study,5 the rs9271300 SNP near HLA-DRB1 was significantly associated with total IgE and was the strongest association observed for this phenotype. As discussed previously, this SNP was not found to be strongly associated with total IgE in FHS. It is not clear why the association with HLA-DRB1 was considerably weaker in the FHS population compared to the GABRIEL population. This finding may be partly explained by the difference in study populations as well as the difficulty in discriminating separate signals from the MHC superlocus. This discordance could also reflect the imperfect imputation of rs9271300 in our study.
The DARC gene on chromosome 1, in close proximity to FCER1A, was also identified as a susceptibility locus in the meta-analysis. The DARC gene was of particular interest given the biologic plausibility of DARC as a modulator of the body’s inflammatory response and literature suggesting a role for DARC in the development of asthma and IgE dysregulation. A recent study by Vergara et al investigated the relationship between DARC and asthma phenotypes in subjects of African descent. A SNP in the DARC promoter region, rs2814778, located in the same LD block as the top DARC SNP in FHS (http://www.broad.mit.edu.mpg/haploview), was found to be significantly associated with total IgE in Colombian and African Caribbean populations and with asthma in Colombian, Brazilian, and African Caribbean populations.30
Based on the HapMap CEPH sample, there is low to moderate LD between the highest associated SNP in DARC, rs13962, and the nearby top SNP in FCER1A (rs2251746) with r2 = 0.27. (Figure E1) (http://broad.mit.edu/mpg/snap) Conditional regression analysis, performed in FHS and the 5 independent replication cohorts, revealed that the association of DARC with total IgE concentration was not significant once rs2251746 was included in the model. While this suggests that the DARC association in the Vergara study may be confounded by the effect of the neighboring FCER1A gene, differences in the populations studied in this meta-analysis (predominantly Caucasian of European descent) and the Vergara study (African descent) likely play a role as the minor allele frequencies of the DARC SNP are significantly higher in populations of African descent compared to populations of European descent. Given the known function of the DARC receptor to act as a chemokine sink and modulate the inflammatory cascade,31 the DARC associations with asthma and total IgE remain plausible, particularly in African populations. Further studies to better delineate the roles of DARC and FCER1A in IgE dysregulation are warranted.
Our study also confirmed, at a genome-wide significant level, the findings from prior genetic literature implicating FCER1A, STAT6, and IL13 as susceptibility loci for IgE dysregulation. In the first published GWAS of total IgE concentration, Weidinger and colleagues identified FCER1A as a susceptibility gene in a population based German cohort.4 The top FCER1A SNP in the Weidinger study, rs2427837, was also highly associated with total IgE in FHS and had the second lowest p-value among all top SNPs (p=9.82 × 10-12). STAT6 and IL-13 are known to play significant roles in the pathogenesis of IgE dysregulation. IL-13, in conjunction with IL-4, is a critical mediator of the TH2 allergic response and STAT6 is considered the master regulator of IgE class switch recombination.32-33 Our observation of a strong association of STAT6 SNPs with IgE concentration confirms previous linkage and candidate gene studies which have shown STAT6 genetic variants to be associated with food allergy,34 atopic dermatitis,35 asthma,36 and total IgE.37 In a recent review of asthma and atopy genetics, Ober and Hoffman identified IL-13 as one of 8 “elite” genes which had been associated with an asthma or atopy phenotype in greater than 10 studies.3
The RAD50 gene is located within 20 kb of IL-13 on chromosome 5. In the Weidinger study,4 as well as the subsequent GWAS investigating asthma by Li and colleagues,29 the RAD50 gene was found to be more highly associated with total IgE and asthma, respectively, than the neighboring IL-13 gene. This is in contrast to the FHS cohort in which SNPs located in IL-13 were found to have the strongest associations in the region, with no SNPs within the RAD50 gene found to have a p-value < 10-4 (see Figure E3). The highest associated SNPs in each (rs1295686 for IL-13, rs2240032 for RAD50) were in moderate LD with an r2=0.58 in the HapMap CEPH sample. Given the findings from three separate GWAS, it is difficult to separate the two signals on a genetic level. With mouse models showing a TH2 locus control region in the 3’ end of RAD50, 38 both Il-13 and RAD50 are plausible candidate genes in the pathogenesis of IgE dysregulation and asthma.
In summary, this GWAS of plasma total IgE in the Framingham Heart Study has identified potential susceptibility loci in the HLA-A, HLA-G, HLA-DQA2 gene regions for which an association with total IgE has not been clearly established in the previous literature. We have also confirmed the associations of previously described genetic variants in FCER1A, STAT6, and IL-13 as risk factors for IgE dysregulation. Based on these findings, further genetic investigation of the relationship between HLA loci and total IgE and other atopy phenotypes is warranted. These findings may help target molecular pathways for future interventions to prevent or treat IgE-mediated allergy.
Clinical Summary.
This genome-wide association study identified genetic loci near HLA genes that are associated with plasma total IgE concentration. It also confirmed the association of susceptibility loci in the FCER1A, STAT6, and IL-13 genes.
Acknowledgments
Sources of funding: G.O’Connor, J Wilk: N01 HC 25195 and P01 AI 050516; J Wilk: the Flight Attendant Medical Research Institute; D.Strachan: the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02; S.Weidinger: DFG (grant WE 2678/6-1) and the German Ministry of Education and Research (BMBF) as part of the National Genome Research Network (NGFN grant 01GS 0818); E. Albrecht.:German National Genome Research Network (NGFN-2 and NGFNPlus: 01GS0823); G.M.Hunninghake: K08 HL092222. B.E.Himes.: 2T15LM007092-16 from the National Library of Medicine; The CAMP Genetics Ancillary Study is supported by U01 HL075419, U01 HL65899, P01 HL083069, N01 HR16049, and T32 HL07427 from the NIH/NHLBI.
Abbreviations
- IgE
Immunoglobulin E
- GWAS
genome wide association study
- FHS
Framingham Heart Study
- KORA
Cooperative Health Research in the Region of Augsburg
- CAMP
Childhood Asthma Management Program
- LME
linear mixed effects
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, et al. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Postma DJ, Postma DS, Howard TD, Koppelman GH, Zheng SL, et al. Major genes regulating total serum immunoglobulin E levels in families with asthma. Am J Hum Genet. 2000;67(5):1163–73. doi: 10.1086/321190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7(2):95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 4.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4(8):e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 363(13):1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wichmann HE, Gieger C, Illig T. KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 7.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 8.Strachan DP, Rudnicka AR, Power C, Shepherd P, Fuller E, Davis A, et al. Lifecourse influences on health among British adults: effects of region of residence in childhood and adulthood. Int J Epidemiol. 2007;36:522–531. doi: 10.1093/ije/dyl309. [DOI] [PubMed] [Google Scholar]
- 9.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. The Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, et al. The Wellcome Trust Case Control Consortium. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–92. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childhood asthma management program research group. The childhood asthma management program (camp): Design, rationale, and methods. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 13.Yunginger JW, Ahlstedt S, Eggleston PA, Homburger HA, Nelson HS, Ownby DR, et al. Quantitative IgE antibody assays in allergic diseases. J Allergy Clin Immunol. 2000 Jun;105(6 Pt 1):1077–78. doi: 10.1067/mai.2000.107041. [DOI] [PubMed] [Google Scholar]
- 14.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 2006;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 16.Durbin R, Altshuler D. 1000 Genomes. Retrieved September 30, 2010, from http://www.1000genomes.org.
- 17.Woszczek G, Kowalski ML, Borowiec M. Association of asthma and total IgE levels with human leucocyte antigen-DR in patients with grass allergy. Eur Respir J. 2002;20(1):79–85. doi: 10.1183/09031936.02.01002001. [DOI] [PubMed] [Google Scholar]
- 18.Aron Y, Desmazes-Dufeu N, Matran R, Polla BS, Dusser D, Lockhart AN, et al. Evidence of a strong, positive association between atopy and the HLA class II alleles DR4 and DR7. Clin Exp Allergy. 1996;26(7):821–8. [PubMed] [Google Scholar]
- 19.Levine BB, Stember RH, Fotino M. Ragweed hay fever: genetic control and linkage to HL-A haplotypes. Science. 172;178(66):1201–3. doi: 10.1126/science.178.4066.1201. [DOI] [PubMed] [Google Scholar]
- 20.Turton CW, Morris L, Buckingham JA, Lawler SD, Turner-Warwick M. Histocompatibility antigens in asthma: population and family studies. Thorax. 1979;34(5):670–6. doi: 10.1136/thx.34.5.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh DG, Chase GA, Freidhoff LR, Meyers DA, Bias WB. Association of HLA antigens and total serum immunoglobulin E level with allergic response and failure to respond to ragweed allergen Ra3. PNAS. 1979;76(6):2903–2907. doi: 10.1073/pnas.76.6.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyers DA, Hasstedt S, Marsh DG, Skolnick M, King MC, Bias WB, et al. The inheritance of immunoglobulin E: genetic linkage analysis. Am J Med Genet. 1983;16(4):575–81. doi: 10.1002/ajmg.1320160414. [DOI] [PubMed] [Google Scholar]
- 23.Parapanissiou E, Papastavrou T, Deligiannidis A, Adam K, Kanakoudi F, Daniilidis M. HLA antigens in Greek children with allergic bronchial asthma. Tissue Antigens. 2005;65(5):481–4. doi: 10.1111/j.1399-0039.2005.00392.x. [DOI] [PubMed] [Google Scholar]
- 24.Lara-Marquez ML, Yunis JJ, Layrisse Z, Ortega F, Carvallo-Gil E, Montagnani S, et al. Immunogenetics of atopic asthma: association of DRB1*1101 DQA1*0501 DQB1*0301 haplotype with Dermatophagoides spp.-sensitive asthma in a sample of the Venezuelan population. Clin Exp Allergy. 1999;29(1):60–71. doi: 10.1046/j.1365-2222.1999.00461.x. [DOI] [PubMed] [Google Scholar]
- 25.Schauer U, Hoffjan S, Bittscheidt J, Kochling A, Hemmis S, Bongartz S, et al. RSV bronchiolitis and risk of wheeze and allergic sensitisation in the first year of life. Eur Respir J. 2002;20(5):1277–83. doi: 10.1183/09031936.02.00019902. [DOI] [PubMed] [Google Scholar]
- 26.Sigurs N, Gustafsson PM, Bjarnason R, Lundberg F, Schmidt S, Sigurbergsson F, et al. Severe respiratory syncytial virus bronchiolitis in infancy and asthma and allergy at age 13. Am J Respir Crit Care Med. 2005;171(2):137–41. doi: 10.1164/rccm.200406-730OC. [DOI] [PubMed] [Google Scholar]
- 27.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Nicolae D, Cox NJ, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76(2):349–57. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N, Carosella E. HLA-G: from biology to clinical benefits. Trends Immunol. 2008;29(3):125–32. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 125(2):328–335 e11. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vergara C, Tsai YJ, Grant AV, Rafaels N, Gao L, Hand T, et al. Gene encoding Duffy antigen/receptor for chemokines is associated with asthma and IgE in three populations. Am J Respir Crit Care Med. 2008;178(10):1017–22. doi: 10.1164/rccm.200801-182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luster AD. Chemokines--chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338(7):436–45. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 32.Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300(5625):1527–8. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 33.Linehan LA, Warren WD, Thompson PA, Grusby MJ, Berton M. STAT6 is required for IL-4-induced germline Ig gene transcription and switch recombination. J Immunol. 1998;161(1):302–10. [PubMed] [Google Scholar]
- 34.Amoli MM, Hand S, Hajeer AH, Jones KP, Rolf S, Sting C, et al. Polymorphism in the STAT6 gene encodes risk for nut allergy. Genes Immun. 2002;3(4):220–4. doi: 10.1038/sj.gene.6363872. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Suzuki M, Arakawa H, Tokuyama K, Morikawa A. Linkage and association studies of STAT6 gene polymorphisms and allergic diseases. Int Arch Allergy Immunol. 2003;131(1):33–8. doi: 10.1159/000070432. [DOI] [PubMed] [Google Scholar]
- 36.Shao C, Suzuki Y, Kamada F, Kanno K, Tamari M, Hasegawa K. Linkage and association of childhood asthma with the chromosome 12 genes. J Hum Genet. 2004;49(3):115–22. doi: 10.1007/s10038-003-0118-z. [DOI] [PubMed] [Google Scholar]
- 37.Weidinger S, Klopp N, Wagenpfeil S, Rummler L, Schedel M, Kabesch M. Association of a STAT 6 haplotype with elevated serum IgE levels in a population based cohort of white adults. J Med Genet. 2004;41(9):658–63. doi: 10.1136/jmg.2004.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–53. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]


