Abstract
In response to DNA damage or replication fork stress, the Fanconi anemia (FA) pathway is activated, leading to monoubiquitination of FancD2 and FancI and their co-localization in foci. Here we show that, in the chicken DT40 cell system, multiple alanine-substitution mutations in 6 conserved and clustered S/TQ motifs of FancI largely abrogate monoubiquitination as well as focus formation of both FancI and FancD2, resulting in loss of DNA repair function. Conversely, FancI carrying phospho-mimic mutations on the same 6 residues induces constitutive monoubiquitination and focus formation of FancI and FancD2, and protects against cell killing and chromosome breakage by DNA interstrand crosslinking agents. We propose that the multiple phosphorylation of FancI serves as a molecular switch in activation of the FA pathway. Mutational analysis of putative phosphorylation sites in human FANCI indicates that this switch is evolutionarily conserved.
Introduction
Genome stability is crucial for maintaining integrity of the organism, and therefore all cells have elaborate systems to prevent, repair, or tolerate endogenous or exogenous DNA damage. In higher organisms, loss of these functions often leads to cancer predisposition1 as well as impaired stem cell proliferation2–4. A rare hereditary disorder Fanconi anemia (FA) is a prototype of such conditions. FA is clinically characterized by an increased occurrence of leukemias and solid tumors, progressive bone marrow failure, and developmental abnormalities5,6. Altogether 13 genes have been implicated in FA, and their products constitute a common pathway in DNA damage signaling termed the “FA pathway”. The FA pathway responds to stalled replication forks and interstrand crosslinks (ICLs) in addition to various types of DNA damage including double strand breaks and UV-induced damage. Upon treatment with ICL inducers such as mitomycin C (MMC) or cisplatin, FA cells display highly increased levels of cell death and chromosome breakages, reflecting a profoundly impaired ability to handle or repair ICLs. Although how the FA pathway participates in ICL repair is currently unknown, it is now presumed that it regulates molecular processes that stabilize and/or resume the arrested fork by affecting homologous recombination (HR) and/or translesion DNA synthesis5,6.
The newest member in the FA pathway, FancI, has been identified through a proteomic screen in an effort to identify ATM/ATR kinase substrates7, through a search for a FancD2 homolog in the database8, and by positional cloning9. FancI physically associates with the key factor FancD2, resulting in formation of the ID complex7,8. Upon DNA damage, FancD2 and FancI are monoubiquitinated in a manner dependent on each other7,8 as well as on the ATR kinase10, the E2 conjugating enzyme UBE2T11, and the FA core complex, which is a multi-subunit E3 ligase formed by eight FA proteins (FancA/B/C/E/F/G/L/M) and two associated proteins FAAP2412 and FAAP1006,13. In turn, FancD2 and FancI are both targeted to chromatin and form colocalizing foci together with the HR proteins BRCA1 and Rad517,8,14. Monoubiquitin on FancD2 serves as an attachable chromatin localization tag15, and is cleaved off by deubiquitinase USP116. Thus FancD2 monoubiquitination is crucial for DNA repair via the FA pathway with downstream or parallel effectors including BRCA2/FANCD117, PALB2/FANCN18, and BRIP1/FANCJ19. In addition, the core complex has been suggested to contribute to DNA repair besides having a role as an E3 ligase15.
Since FancI is an essential co-factor for FancD2 monoubiquitination, we set out to investigate how FancI contributes to triggering this key activation event in the FA pathway. We examined the functional role of monoubiquitination and phosphorylation of FancI in chicken DT40 cells, and found that multiple phosphorylation of FancI but not monoubiquitination is critical for FancD2 activation following DNA damage. Thus we propose that FancI phosphorylation serves as a molecular switch in the FA pathway.
Results
Generation of FANCI-deficient cells
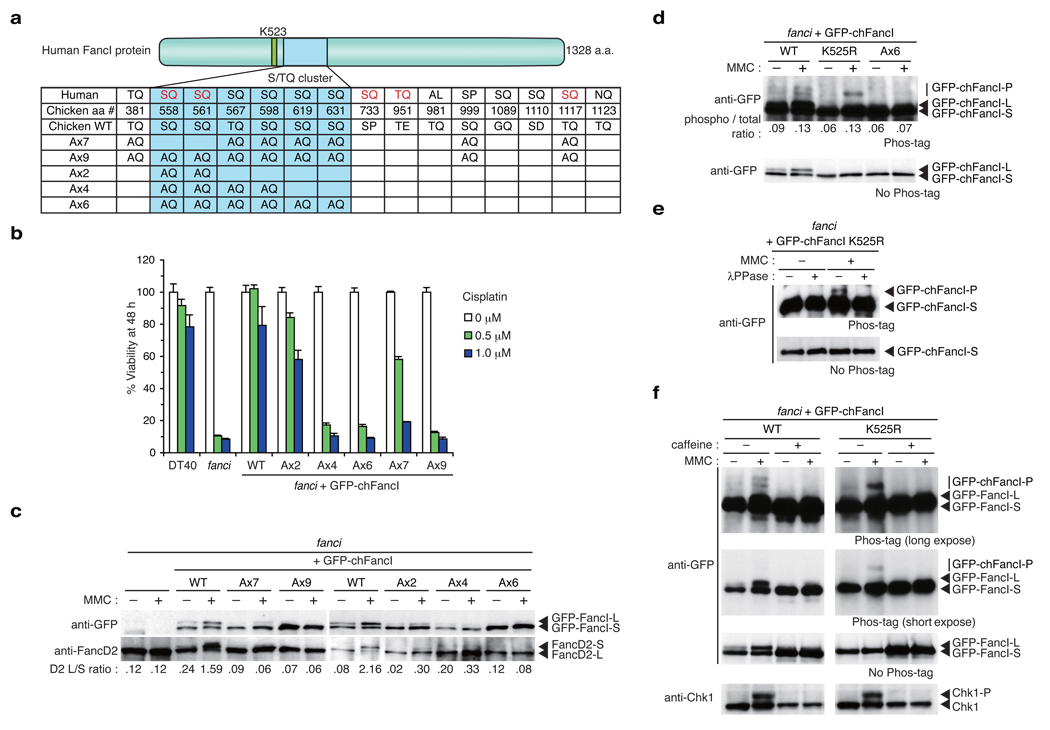
We have disrupted the FANCI gene in chicken DT40 cell line (Supplementary Fig. 1), and observed that fanci DT40 cells exhibited abrogated monoubiquitination as well as focus formation of FancD2 protein both before and after MMC treatment (Fig. 1), as expected from previous studies using human FANCI mutant cell lines20. Consistent with a critical role for FancD2 monoubiquitination in DNA repair21, fanci cells were extremely cisplatin-sensitive (Fig. 2a), and displayed increased levels of chromosome breakage induced by MMC (Fig. 2b). Expression of a GFP-tagged full-length chicken FANCI cDNA (GFP-chFancI WT) in fanci cells fully rescued ICL sensitivity in cell survival (Fig. 2a) and chromosome aberration assays (Fig. 2b), as well as monoubiquitination and focus formation of FancD2 (Fig. 2c,d). Similar to human FancI7,8, a slower mobility form of GFP-chFancI (designated GFP-chFancI-L) was detected by anti-GFP Western blotting (Fig. 2c). This L-form corresponds to monoubiquitinated FancI since the GFP-chFancI-L form accumulated following DNA damage, and when Lys525 - the equivalent residue to the monoubiquitination site in human FancI - was substituted with arginine (K525R), the L-form was undetectable.
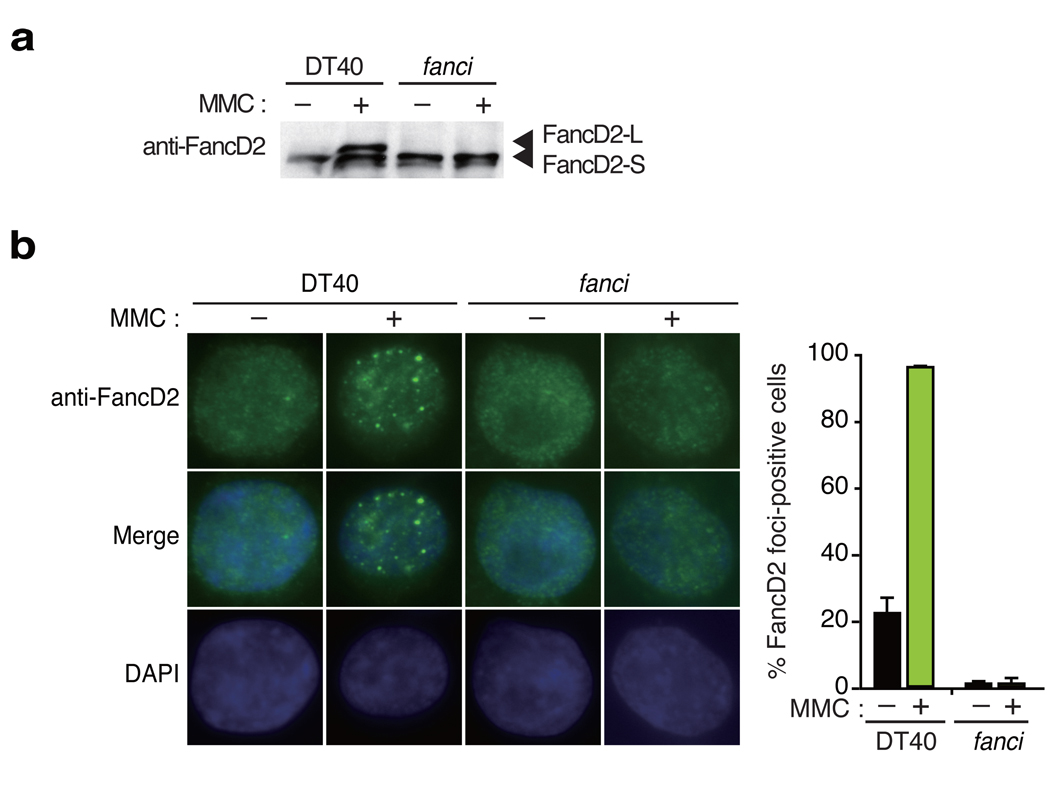
Figure 1. Gene targeting of FANCI resulted in loss of FancD2 monoubiquitination and focus formation.
(a) Western blot analysis of FancD2 monoubiquitination induced by MMC treatment. (b) FancD2 localization to nuclear foci by indirect fluorescence using anti-FancD2 antibodies and DAPI counter staining. Representative images of wild type (WT) or fanci cells with or without MMC treatment stained for endogenous FancD2 are shown. The mean and standard error (s.e.m.) of %FancD2 foci-positive cells in three independent experiments are shown in the bar graph. More than 100 cells were scored in each experiment, and cells containing more than 4 bright foci were defined as foci-positive.
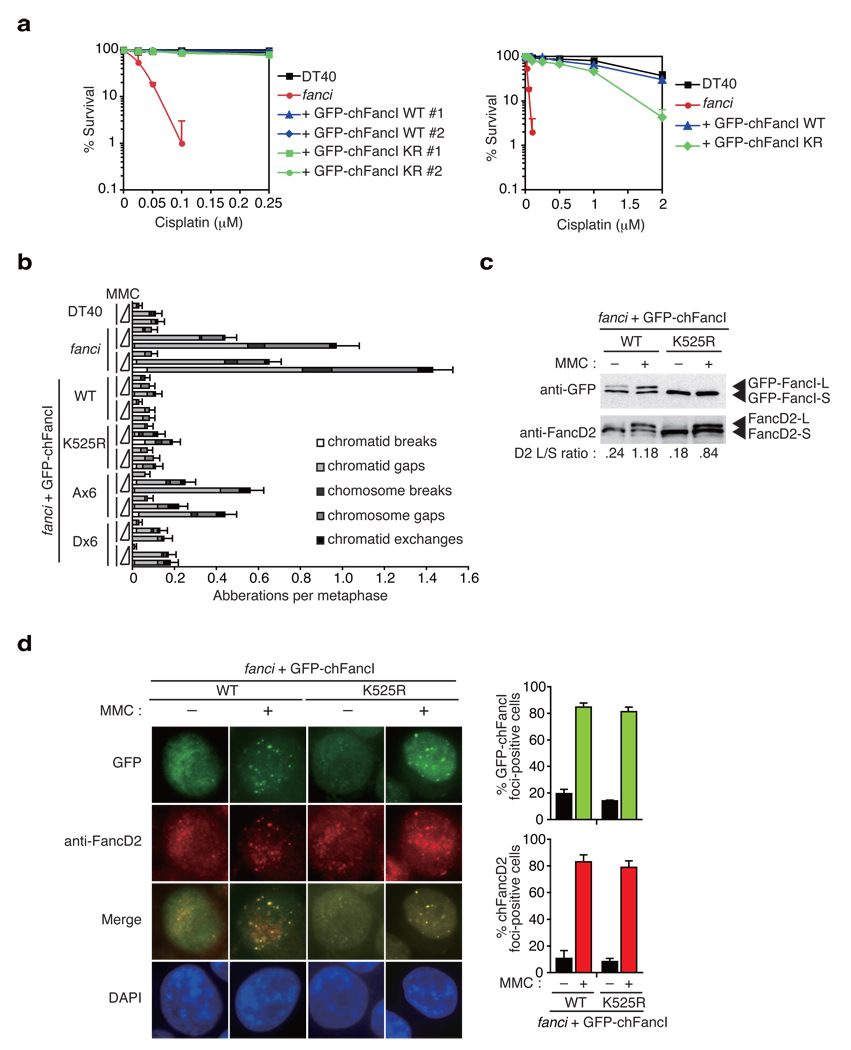
Figure 2. FancI monoubiquitination plays a minor role in inducing FA pathway activation.
(a) Cisplatin sensitivity of cells with indicated genotypes were evaluated by colony survival. fanci cells were transfected with GFP-chFancI WT or K525R expression vector. The experiment was repeated at least three times, and a representative data set with mean and standard deviation of the triplicate cultures are shown. (b) Chromosome analysis of cells with the indicated genotypes. Along with wild type (DT40) and two clones of fanci cells, two independent clones of fanci cells expressing GFP-chFancI WT or mutant proteins were examined. Cells were treated with the increasing concentrations of MMC (0, 20, and 40 ng/ml) for 24 h. At least 50 metaphases were scored blindly for each preparation. Error bars indicate standard error (s.e.m.). (c) Monoubiquitination of FancD2 and FancI. Whole cell extracts were prepared from cells with the indicated genotypes and analyzed using antibodies against GFP or FancD2. (d) GFP-chFancI or FancD2 localization to DNA damage-induced nuclear foci. Representative images of fanci cells expressing GFP-chFancI wild type (WT) or the K525R mutant with or without MMC treatment are shown. The graph shows the mean and s.e.m. of %FancD2 or %GFP-chFancI foci-positive cells in fanci cells with GFP-FancI WT or K525R in three independent experiments. Cells with more than four bright foci were defined as foci-positive. More than 100 cells were scored for each data.
Genetic requirements for FancI monoubiquitination
Monoubiquitination of human FANCI is catalyzed by the core complex in a manner strictly dependent on the presence and monoubiquitination of FANCD27,8. We tested this in DT40 by expressing GFP-chFancI in wild type and various mutant cell lines followed by Western blotting. In wild type DT40, induction of the GFP-chFancI-L form was detected following MMC treatment, albeit the levels were quite low, perhaps due to the presence of endogenous FancI (Supplementary Fig. 2a). In contrast, the L-form was not induced in fancd222, fancd2-K563R-knock-in (the monoubiquitination site Lys563 was converted to Arg)21, fancc23, or fancl21 cells expressing GFP-chFancI (Fig. 3a, left panel, and Supplementary Fig. 2a). Consistently, we observed MMC-induced GFP-chFancI focus formation when expressed in wild type cells but not in fancd2, fancc, or fancl cells (Supplementary Fig. 2b). These results confirm the data obtained in human cells7,8, and further support the role of the core complex and FancD2 monoubiquitiation in inducing FancI monoubiquitination.
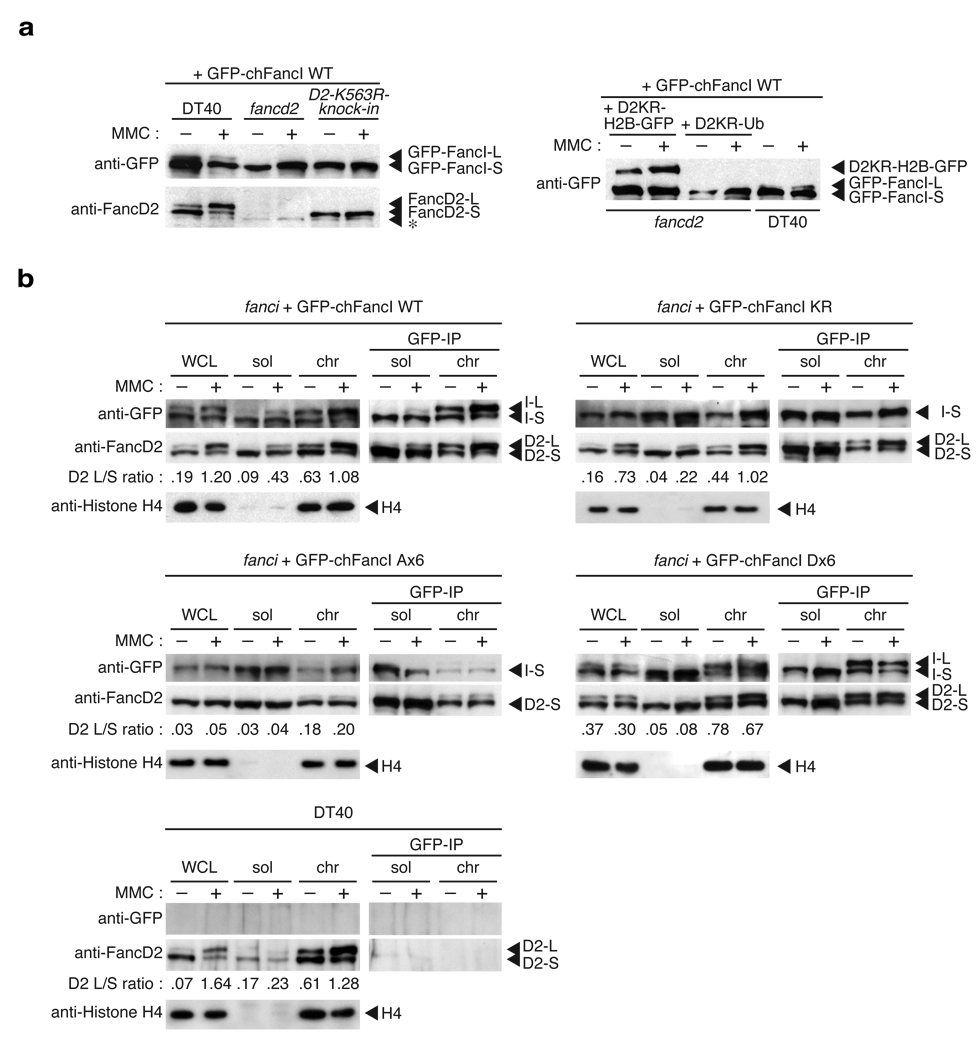
Figure 3. FancI monoubiquitination depends on FandD2 monoubiquitination but their physical interaction is constitutive.
(a) Genetic requirements for FancI monoubiquitination. Wild type DT40 cells, fancd2 cells, cells with the FANCD2 monoubiuqitination site K563R knockin mutation (D2-K563R-knock-in), and fancd2 cells carrying D2KR-H2B-GFP or D2KR-Ub fusions were stably transfected with GFP-chFancI wild type (WT), and treated with or without MMC. Whole cells lysates were blotted with anti-GFP or anti-FancD2 antibodies. Asterisk (*) indicates a non-specific band. (b) Co-immunoprecipitation between FancD2 and FancI. Indicated cells were stimulated with MMC, and fractionated into soluble and chromatin fractions. Immunoprecipitation was carried out using anti-GFP antibody beads. Whole cell lysates (WCL), fractions (5% of the input), and immunoprecipitates were separated by SDS-PAGE and blotted with antibodies as indicated. As a negative control for anti-GFP immunoprecipitation, wild type DT40 not expressing GFP-chFancI was similarly fractionated and analyzed. L or S indicates L-form or S-form, respectively.
Functional role of FancI monoubiquitination
To ask whether FancI monoubiquitination is crucial for FancD2 monoubiquitination, we introduced GFP-chFancI carrying the K525R mutation (GFP-chFancI K525R) into fanci cells. We observed that it largely rescued cell survival (Fig. 2a) and chromosome breakage (Fig. 2b) in response to ICL treatment although less well than expression of GFP-chFancI wild type (WT), indicating that FancI without monoubiquitination is partially functional in DNA repair. Of note, the experiments in human fanci mutant cell lines have also provided similar data regarding MMC sensitivity and chromosome breakage (Fig. 6E and Fig. 6F in ref. 7, respectively). In support of this, MMC treatment of cells with GFP-chFancI K525R induced FancD2 monoubiquitination, albeit with slightly reduced efficiency (Fig. 2c,3b), leading to co-localizing focus formation of FancD2 and FancI (Fig. 2d). Consistently, the FancD2-L form was targeted to chromatin in these cells similarly to cells expressing GFP-chFancI WT (Fig. 3b). Furthermore, co-immunoprecipitation experiments showed that in the chromatin fraction of these cells, FancD2 interacted with both GFP-chFancI-S form (K525R expressing cells) and L form (WT expressing cells), respectively (Fig. 3b). Taken together, these data indicate that the role of FANCI monoubiquitination is largely dispensable for FancD2 monoubiquitination in the FA pathway and the interaction of the two proteins is monoubiquitination independent.
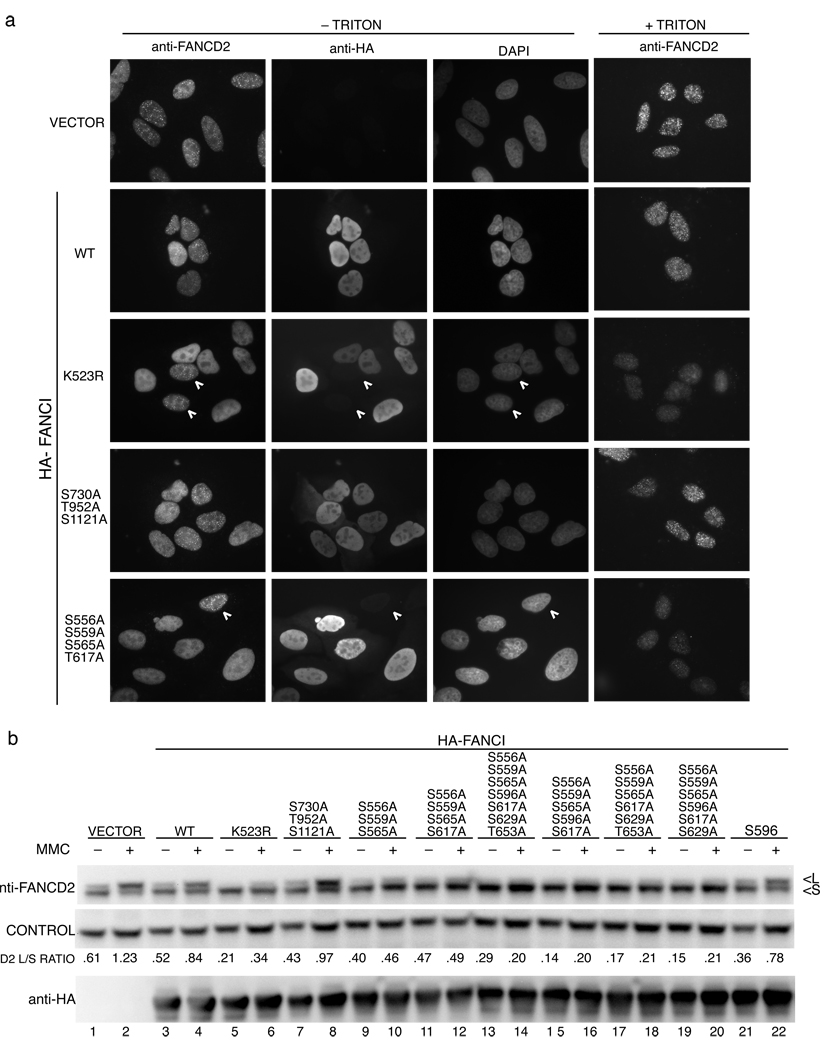
Figure 6. Analysis of phosphorylation mutants in human FANCI.
(a) Localization of FancD2 in U2OS cells expressing the indicated HA-tagged FANCI alleles. Cells were treated with 1 µM MMC and 24 h later were costained with an antibody against FANCD2 and the HA-tag. The white arrowheads indicate cells that show no expression of HA-tagged FancI serving as an internal control for FANCD2 staining. Triton treatment removes the majority of the nucleoplasmic FANCD2 allowing for a better visualization of FANCD2 foci. (b) Western analysis of FANCD2 in U2OS cells expressing HA-tagged FANCI alleles. Cells expressing the indicated alleles of FANCI were treated with 1 µM MMC and collected 24 h later. L indicates the long (monoubiquitinated) and S is the short form of FANCD2.
In a previous study, we have shown that cisplatin sensitivity in fancd2 cells is reversed to near wild type levels by transfection of FancD2 fusions with a single ubiquitin moiety or histone H2B (each termed D2KR-Ub or D2KR-H2B, respectively, since their monoubiuqitination site Lys563 is replaced with arginine)15. Both of the fusions of FancD2 are targeted to chromatin despite lacking the monoubiquitination site, so we asked if, under these conditions, the monoubiquitination of FancI is supported. To address this question, we transfected the GFP-chFancI expression vector into these cells, and examined monoubiquitination of FancI. Consistent with the above experiments, we found that GFP-chFancI was not converted to the L-form in these cells (Fig. 3a, right panel), despite near normal cisplatin tolerance of these cells15. Thus, these data further supported the conclusion that monoubiquitination of FancI is largely dispensable for DNA repair function of the FA pathway.
Functional analysis of FancI phosphorylation
The ATM/ATR kinase generally phosphorylates an S/TQ motif as a consensus phosphotylation site in substrates. Human FancI protein harbors a number of the S/TQ motifs, and at least five of these sites are phosphorylated in human or mouse FancI as detected by mass spectrometry7,24 (indicated by red letters in Fig. 4a, see also Supplementary Fig. 3). In comparison between human and chicken FancI amino acid sequences, we found eight conserved S/TQ sites, and six of these are found in a cluster spanning ~70 amino acids close to the monoubiquitination site Lys523 (human) or Lys525 (chicken) (Fig. 4a).
Figure 4. Phosphorylation sites in FancI are required for FA pathway activation and DNA repair.
(a) S/TQ sites in human and chicken FancI proteins were listed with corresponding sites in the other protein. Red lettering indicate sites whose IR-induced phosphorylation was detected by mass spectrometry7. Locations of multiple alanine substitutions in mutant chFancI proteins are indicated. (b) Cell survival in cisplatin containing medium was assayed using PI staining and FACSCalibur (Becton-Dickinson). (c) Monoubiquitination of GFP-chFancI and chFancD2 in response to MMC. Whole cell lysates were separated by SDS-PAGE and blotted using antibodies against GFP and chFancD2. (d) Phosphorylation of GFP-chFancI in response to MMC. Whole cell lysates were separated by SDS-PAGE using a gel containing Phos-tag-acrylamide, and blotted using anti-GFP antibodies. Shifted bands are indicated as GFP-chFancI-P. (e) GFP-chFancI was immunoprecipitated from the indicated cells, and treated with λ-phosphatase (λPPase), and detected as in d. (f) Cells were treated with caffeine (20 mM) and/or MMC (500 ng ml−1) or 6 h or left untreated, and analyzed by Phos-tag-containing gel and Western blotting as in d using antibodies against GFP or Chk1. For Phos-tag western, long and short exposures are shown.
To examine whether phosphorylation of these S/TQ sites plays an important role in the FA pathway, we mutated multiple S/TQ sites in chicken FancI simultaneously. We isolated chFancI mutant cDNAs each carrying seven or nine consecutive alanine-substitutions on the S/TQ sites (termed Ax7 or Ax9), and expressed them in fanci cells as GFP-fusion proteins (Fig. 4a). To facilitate rapid functional evaluation of the mutants, we developed a cell viability assay by PI staining and flow cytometry following 48 h cisplatin exposure in liquid culture (see methods). Interestingly, the FancI Ax7 mutant was able to provide an appreciable rescue of the cell death induced by cisplatin, albeit less efficiently than that of wild type FancI, whereas the Ax9 mutant could not (Fig. 4b). Colony survival assays confirmed these results (Supplementary Fig. 4a). These observations suggest that FancI S/TQ sites might be crucial for DNA repair, and also indicate that the two non-overlapping sites (Ser558 and Ser561) between the Ax7 and the Ax9 mutants are functionally important. However, another FancI mutant Ax2 in which these two sites were changed to alanine was still capable of reversing cisplatin sensitivity (Fig. 4a,b, and Supplementary Fig. 4a), suggesting that in addition to these two sites, other S/TQ motifs may function in a redundant manner.
To further investigate the importance of potential phosphorylation sites in FancI, we created Ax6 or Ax4 mutants in which all or four of the six S/TQ sites in the conserved S/TQ cluster were changed to AQ, respectively (Fig. 4a). fanci cells transfected with the Ax6 mutant displayed levels of cisplatin sensitivity close to those in fanci cells, similar to cells expressing the Ax9 protein (Fig. 4b and Supplementary Fig. 4a). MMC-induced chromosome aberrations in cells with the Ax6 mutant were increased compared to cells expressing GFP-chFancI WT (Fig. 2b). The Ax4 mutant showed partial sensitivity to cisplatin damage in the colony survival assay (Supplementary Fig. 4a) and a stronger defect in the liquid culture assay (Fig. 4b). We also looked at these cells expressing GFP-chFancI mutants by anti-GFP or anti-FancD2 Western blotting. Compared to cells expressing GFP-chFancI WT, cells with the Ax4, the Ax6, or the Ax9 mutants virtually abrogated FancI or FancD2 monoubiquitination both before and after MMC treatment (Fig. 4c). Conversely, weak levels of monoubiquitination were still observed in the Ax2 and the Ax7 mutants (Fig. 4c). These results suggest that multiple and redundant phosphorylation in the S/TQ cluster domain is a prerequisite for induction of monoubiquitination of FancD2 and FancI, and for their DNA repair function.
FancI is phosphorylated without prior ubiquitination
To support the hypothesis that chicken FancI protein is indeed phosphorylated following ICL damage, we utilized fanci cells expressing wild type or mutant FancI, and the Phos-tag reagent that selectively binds to phosphorylated amino acid residues25. In polyacrylamide gels containing Phos-tag-acrylamide, phosphorylated forms of protein could be detected by slower migration. We treated fanci cells expressing GFP-chFancI WT, K525R, or the Ax6 mutant with or without MMC, and whole cell lysates of these cells were separated by electrophoresis with or without Phos-tag, and blotted with anti-GFP antibodies. In the presence of Phos-tag, GFP-chFancI WT displayed two slower migrating bands in addition to the S- and L-forms of FancI, indicating that MMC treatment induced FancI phosphorylation. Consistent with this, phosphorylated FancI has been previously detected as two bands using an anti-phospho-SQ antibody7. The only minor fraction of FancI (~13% of total FancI as shown in Fig. 4d) displayed mobility shift in Phos-tag gel after MMC treatment. Interestingly, the GFP-chFancI K525R protein from MMC-treated cells displayed a single retarded band in a Phos-tag-containing gel (Fig. 4d), which was abrogated by λ-phosphatase treatment in vitro (Fig. 4e). The data suggest that FancI could be phosphorylated without prior monoubiquitination, and that the upper phosphorylated FancI might be monoubiquitinated. The phosphorylation occurs on residues that were mutated in the Ax6, since GFP-chFancI Ax6 mutant protein did not show a detectable band shift in the presence of Phos-tag (Fig. 4d).
FancI phosphorylation is caffeine-sensitive
Since replication fork arrests generate RPA-coated single strand gaps that activate ATR26, and FancI is a putative ATR/ATM substrate7, it seems likely that ATR kinase phosphorylates FancI following ICL treatment. Since caffeine is able to inhibit ATM and ATR kinases27, we examined its effects on FancI phosphorylation as well as monoubiquitination of FancI and FancD2 by Western blotting using gels with or without Phos-tag. Caffeine treatment inhibited the mobility shift of Chk1 following MMC treatment (Fig. 4f), indicating that ATR activity is suppressed. It also substantially inhibited MMC-induced monoubiquitination of GFP-chFancI and FancD2 (Fig. 4f and Supplementary Fig. 5a), as well as phosphorylation of GFP-chFancI WT or K525R (Fig. 4f). Collectively, these data establish that ICL treatment induces FancI phosphorylation in a caffeine-sensitive manner.
FancI phospho-mimic mutants activate the FA pathway
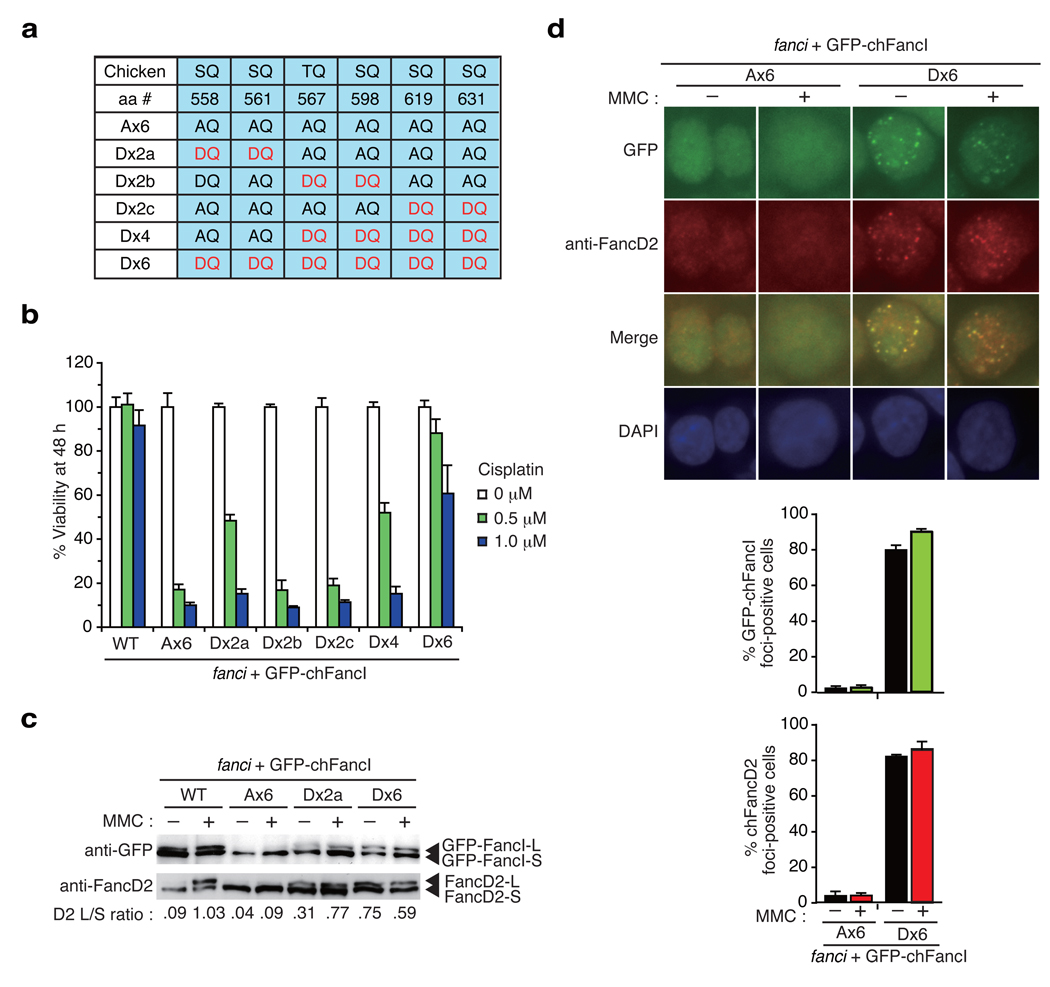
To gain more insight into the functional role of phosphorylation in the S/TQ cluster, we replaced all or some of the six alanine residues in the Ax6 mutant FancI with aspartic acid (denoted as D in single letter code) (Fig. 5a). Substitution to aspartic acid provides negative charge, thus mimicking phosphorylation. The phospho-mimic mutant (termed Dx6) in which all of the AQ sites in the Ax6 were changed to DQ quite effectively reversed cisplatin sensitivity (Fig. 5b and Supplementary Fig. 4b) as well as MMC-induced chromosome breakage (Fig. 2b). In contrast, among mutants in which two adjoining S/TQ sites were converted to DQ (Dx2a, Dx2b, and Dx2c), only the Dx2a mutant (Asp558 and Asp561) displayed protection against DNA damage with cisplatin. The Dx4 mutant, in which the Dx2b and the Dx2c mutations were combined, were as functional as the Dx2a mutant (Fig. 5b). In addition, we tried to dissect the Dx2a mutation to the two mutants each carrying single DQ motif (Asp558 or Asp561) and five AQ motifs, and found that neither mutant supported survival in the liquid culture assay (data not shown). We also found that the Dx2a and the Dx6 mutants were able to activate monoubiquitination of FancD2 as well as FancI even without ICL damage (Fig. 5c). The Dx6 mutant cell line displayed weak cisplatin sensitivity in liquid culture assay (Fig. 5b). This could be due to the fact that even though there is an increased level of ubiquitinated FancD2 without DNA damage, the further induction of FancD2 monoubiquitination upon damage seems to be blunted (Fig. 5c, 3b and Supplementary Fig. 5a). There is also a precedent in the literature in the setting of USP1 deficiency28, that persistent FANCD2 monoubiquitination results in low levels of DNA damage sensitivity. Additionally, there is an indication that FancI Dx6 protein may have suboptimal function since cells with Dx6/K525R combined mutations exhibited higher levels of ICL sensitivity than cells with either single mutant (data not shown). Collectively, results from the mutagenesis of the SQ/TQ sites suggest that, among the six sites in the S/TQ cluster, phosphorylation on the N-terminal two sites (Ser558 and Ser561) may have a higher impact on FancI function compared to other sites. However, if simultaneously phosphorylated, the other four sites may deliver sufficient signal to activate downstream events like monoubiquitination of FancD2.
Figure 5. Phospho-mimic mutants induce constitutive activation of the FA pathway.
(a) Phospho-mimic mutations in the S/TQ cluster in chFancI. (b) Phospho-mimic mutations protected fanci cells against DNA damage induced by cisplatin. Cell survival was assessed as in Fig. 4b. (c) Constitutive monoubiquitination of GFP-chFancI and chFancD2. Whole cell lysates were separated and blotted using antibodies against GFP or chFancD2. (d) GFP-chFancI Ax6 or Dx6 localization in cells with or without DNA damage. Fixed cells were stained with anti-FancD2 antibodies and detected using fluorescence microscopy. The mean and standard error (s.e.m.) of %FancD2 or %GFP-chFancI foci-positive cells in three independent experiments are shown in the bar graph. Cells with more than four bright foci were defined as foci-positive. More than 100 cells were scored for each sample.
Constitutive foci formation of the FancI Dx6 protein
Studying the localization of the phospho-mimic mutants of FancI, we found constitutive foci formation of the GFP-chFancI Dx6 without ICL treatment. These foci colocalized very well with FancD2 foci (Fig. 5d) but only partially localized with γH2AX or Rad51 foci29 (Supplementary Fig. 5b). Upon MMC treatment, the %GFP foci-positive cells were slightly increased with a modestly enhanced co-localization with γH2AX or Rad51 foci (Fig. 5d and Supplementary Fig. 5b). Furthermore, cell fractionation studies revealed that, in cells expressing the Dx6 protein, the FancD2 L-form constitutively localized in chromatin fraction together with L-form of the FancI Dx6, while the Ax6 mutant induced this relocalization much less efficiently even after MMC treatment (Fig. 3b). Collectively, these results suggest that FancI phosphorylation is sufficient to induce chromatin accumulation and foci formation of FancD2 and FancI. We have previously shown that the D2KR-Ub fusion translocates to chromatin without need for additional DNA damage15. Our current results are in keeping with this, and suggest that the machinery for chromatin loading of monoubiquitinated FancD2 and FancI is constitutively active and that the phosphorylation is the key activating event in the pathway.
Human FANCI serves an evolutionarily conserved function
To examine the role of FANCI phosphorylation in human cells, we studied mutations in ten S/TQ motifs in human FANCI protein. These included three sites known to be phosphorylated in the human protein (Ser730, Thr952, and Ser1121)7, three sites consisting of the analogous human sites to known mouse phosphorylation sites (Ser556 and Ser559) and a conserved site six residues downstream, Ser565, identified from alignments of vertebrate FancI proteins (Supplementary Fig. 3), and a cluster of four sites just downstream of these (Ser596, Ser617, Ser629, and Thr653) (summarized in Supplementary Fig. 6). Proteins carrying various combinations of mutations were expressed in U2OS cells, where we previously showed it out-competes endogenous FANCI and exerts a dominant effect7, and assayed for their effects on FANCD2 foci formation and mono-ubiquitination in response to MMC treatment.
Simultaneous mutation of S730A, T952A, and S1121A did not affect FANCD2 foci formation or ubiquitination (Fig. 6). However, the triple mutant of S556A S559A S565A resulted in greatly diminished FANCD2 foci formation, although faint foci were still visible after pre-extraction, and a 50% reduction in FANCD2 monoubiquitination (Fig. 6a,b). Of the Ser596, Ser617, Ser629, and Thr653 sites mutated singly or simultaneously, only the S617A mutation resulted in a slight decrease in the intensity of DNA-damage induced FANCD2 foci and monoubiquitination (Fig. 6 and Supplementary Fig. 6). The greatest effect on FANCD2 foci formation was seen in FANCI S556A S559A S565A S617A mutant (Fig. 6a). Additional FANCI mutations did not visibly affect FANCD2 localization (Supplementary Fig. 6). Expression of FANCI S556A S559A S565A S596A S617A resulted in the lowest levels of FANCD2 monoubiquitination with or without DNA damage. The fact that the basal level of FANCD2 monoubiquitination was diminished when this mutant was expressed indicates that phosphorylation is important during normal S phase and not only after DNA damage. These data indicated that FANCI phosphorylation exerts an evolutionarily conserved function in inducing FANCD2 monoubiqutination in human cells as well.
Discussion
In this study, we have shown that multiple phosphorylation sites in the S/TQ cluster of FancI are functionally important for inducing monoubiquitination of FancD2, a critical event in the FA pathway and ICL repair. These sites are evolutionarily and functionally conserved even in human cells, and some of them overlap with the sites that were detectably phosphorylated following IR damage7,24. Using Phos-tag technology and the mutant GFP-chFancI proteins, we have shown that MMC-induced phosphorylation occurs on these sites in a caffeine-sensitive manner. Furthermore, introduction of phospho-mimic mutations into merely six sites in chFancI (Dx6 mutant) can activate constitutive monoubiquitination and focus formation of FancD2 and FancI.
On the basis of the data presented here, we propose that FancI phosphorylation functions as a molecular switch to turn on the FA pathway in response to DNA damage or replication fork stress. How is FancI phosphorylation triggered following DNA damage? Several lines of evidence support the notion that the ATR kinase is involved. First, we found that FancI phosphorylation is inhibited by caffeine treatment at concentration that also inhibited Chk1 mobility shift (20 mM, as shown in Fig. 4f). Second, FancI has been identified as a putative ATM/ATR substrate7, and we found that multiple phosphorylations on the S/TQ cluster domain, the signature ATM/ATR motif, is important for FancI function. Third, ATR has been previously implicated in inducing FancD2 monoubiquitination through phosphorylation of FancD210,30. However, we found that FancD2 carrying multiple alanine substitutions on the potential ATR sites (i.e. ten S/TQ motifs including six conserved residues that correspond to S222, S319, T691, S705, S1079 and S1401 in human FANCD2 protein) in chicken FancD2 still efficiently reversed cisplatin hypersensitivity of fancd2 cells (data not shown). Our data argue that the phosphorylation of FancI, not FancD2, plays the most crucial role for induction of FancD2 monoubiquitination. Of note, there remains a possibility that phosphorylation of FancD2 or the core complex components like FancE31 or FancM32 plays a minor role in the activation of the FA pathway. Indeed, caffeine treatment resulted in a mild decrease in FancI monoubiquitination in cells expessing Dx6 mutant (Supplementary Fig. 5a). This could be due to contribution by phosphorylation of FancD2 or the core complex components.
It is currently unclear how phosphorylated FancI promotes monoubiquitination of FancD2 and itself, a process that must be mediated by the ubiquitin E3 ligase activity of the core complex. In other systems, like the SCF E3 ligase complex, the E3 and the substrate often interact upon phosphorylation of the substrate, either by direct binding of phospho-epitope with the E3 in a manner analogous to the phosphoprotein-SH2 domain interaction, or by exposing cryptic binding sites on the substrate via conformational change33,34. Since FancI and FancD2 interact with each other (forming the ID complex)7,8, we tested whether stability of the ID complex is affected by FancI phosphorylation using co-immunoprecipitation (co-IP) or TAP-pull down. We could not detect any significant alteration in the interaction between endogenous FancD2 and GFP-chFancI (WT or mutant) in FANCI-deficient DT40 both with or without MMC treatment (Fig. 3b), or between FLAG- or TAP-tagged FancD2 and GFP-chFancI (WT or mutant) transiently expressed in 293T cells (Supplementary Fig. 7a,b). It is conceivable that phosphorylated FancI somehow recruits FancD2 to the core complex by interacting with a molecule that associates with the core complex or with a member of the complex itself. So far we could not detect interaction of the FancI Dx6 with any of the known core complex components or the E2 enzyme UBE2T using the mammalian two-hybrid assay (data not shown). Notably, our current observation is perhaps the first indication that phosphorylation on one protein is necessary for the in-trans monoubiquitination of its partner. It is important to elucidate how phosphorylated FancI is involved in the precise molecular mechanism of the E3 ligase activation.
Although the monoubiquitination site on FancI is exquisitely conserved through evolution, FancI monoubiquitination seems to be largely dispensable for the function of the FA pathway in DT40 cells. The chFancI K525R mutant can support monoubiquitination as well as focus formation/chromatin loading of FancD2. We have confirmed this by making DT40 cells carrying K525R “knock-in” mutation (MI and MT, unpublished results). The differences in the degree to which human and chicken cells need monoubiquitination of FancI may have to do with the absolute amount of mono-ubiquitinated FancD2 needed in their respective cells. The human FANCI K523R mutant produces less FancD2-L in response to MMC than the chFancI K525R mutant.
In conclusion, we propose that phosphorylation of FancI on multiple sites may serve as a molecular switch to turn on the FA pathway upon DNA damage. Our data provide a basis for further elucidation of the mechanistic detail of the FA pathway, which should aid development of more rational therapeutics for FA and related conditions.
Methods
Cells and plasmids
Wild type and various mutant chicken DT40 cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal calf serum, 1% (v/v) chicken serum, 2 mM L-glutamine, 50 µM 2-mercaptoethanol, penicillin, and streptomycin in a 5% (v/v) CO2 incubator at 39.5 °C. Generation of fancd2, fancc, fancl, and fancd2-K563R-knockin cell lines has been described previously21–23. Targeting vectors for FANCI gene disruption were constructed by subcloning PCR-amplified genomic fragments on both sides of the resistance gene cassettes. Transfections and selection of the clones were done as previously described35. Full-length chicken FANCD2 or FANCI cDNA was cloned into FLAG- or TAP-tag expression vectors or pEGFP-C1 (Clontech). The plasmid for TAP-tag, which consists of calmodulin binding protein and a part of Protien A, was provided by Tohru Natsume (National Institute of Advanced Industrial Science and Technology, Tokyo, Japan). Mutations were introduced using site-directed mutagenesis kits (Stratagene). 293T cells were maintained in Dulbecco Modified Eagle medium (DMEM) supplemented with 10% (v/v) FCS as described 36 and transfected using Lipofectamine2000 (Invitrogen).
Analysis of cell sensitivity toward ICL treatments
Cell viability in liquid culture containing cisplatin (Nippon Kayaku, Tokyo, Japan) was assessed after 48 h using FACSCalibur (BD) and propidium iodide (PI) staining. Percentage of viable cells was calculated on the basis of forward scatter profile and exclusion of PI fluorescence among the acquired ten thousand events. Colony formation assay was carried out in medium containing 1.4% (w/v) methylcellulose and indicated dosages of cisplatin. Chromosomal analysis following MMC (Kyowa Hakkou, Tokyo, Japan) treatment was done as described35.
Subnuclear focus formation assay
After MMC exposure (500 ng per ml for 6 h), cytospin slides were fixed and stained with antibodies against chicken FancD2 (provided by Kenshi Komatsu, Kyoto University) or human RAD51 (provided by Hitoshi Kurumizaka, Waseda University). To detect subnuclear γH2AX foci, fixed cells were further treated with 70% (v/v) ethanol and stained with anti-γH2AX monoclonal antibody (UBI) as described37. Then cells were stained with FITC- or Alexa Fluor 594-conjugated secondary antibody (Invitrogen) with DAPI counterstaining. Images were captured by fluorescent microscopy (Axioplan2 equipped with AxioCam MRm and Axiovision, Zeiss) or confocal laser scanning microscopy (LSM510META, Zeiss).
Fractionation of cells, immunoprecipication, and Western blotting
Cells were stimulated with MMC 500 ng per ml for 6h or left untreated unless stated otherwise. Samples were separated by polyacrylamide gel electrophoreses, transferred to a nitrocellulose membrane, and detected with antibodies against GFP (MBL), chicken FancD2, or Chk1 (SantaCruz), and ECL plus reagents (GE Healthcare). For immunoprecipitation, cells were lysed in lysis buffer23 containing phosphatase inhibitor cocktail (Nacalai Tesque, Kyoto, Japan), or were fractionated into soluble and chromatin fractions as described previously15, and precipitated using anti-GFP (MBL) or anti-FLAG (Sigma) beads. TAP-tagged chFancD2 was pulled down using rabbit IgG-beads (Sigma). After extensive washing, the immunoprecipitates were treated with λ-phosphatase (NEB) and analyzed by Western blotting. Phos-tag-containing polyacrylamide gels were made with Phos-tag-acrylamide (3.5 µM) (Phos-tag Consortium, Hiroshima, Japan) and 7 µM MnCl2 to the separation gel. Densitometric analysis of the band intensity was carried out using ImageJ software (National Institute of Health, USA).
Methods for human U2OS cell experiments
The U2OS cells were grown in DMEM supplemented with 100 units of penicillin per ml, 0.1 mg streptomycin per ml, L-glutamine (2 mM), non-essential amino acids (0.1 mM), and 10% (v/v) FBS (Invitrogen). QuikChange multisite-directed mutagenesis kit (Stratagene) was used to make mutation in human FANCI. The FANCI mutant cDNAs were introduced into U2OS cells using a lentiviral vector pHAGE_CMV_IP_HAFLAG (kindly provided by J. Lin and J.W. Harper, Harvard Medical School). Antibodies were rabbit anit-FANCD2 (Novus) and anti-HA (Covance). For immunofluorescence, cells grown on autoclaved cover slips were rinsed in PBS and were fixed in 3.7% (w/v) formaldehyde (Sigma) diluted in PBS for 10 min at room temperature. Cells were washed once with PBS, permeabilized in 0.5% (v/v) NP40 in PBS for 10 min, washed again in PBS, and blocked with PBG (0.2% (w/v) cold fish gelatin, 0.5% (w/v) BSA in PBS) for 20 min. Coverslips were incubated for 2 h at room temperature or at 4° C overnight in a humidified chamber with a primary antibody and after washing 3 times for 5 min in PBG, were incubated with the appropriate secondary antibody. After three additional washes in PBG, the coverslips were embedded in Vectashield (Vector Laboratories) supplemented with DAPI. Triton X-100 pre-extraction was performed by incubating cells for 5 min at room temperature with 0.5% (v/v) Triton X-100 in PBS. Cells were fixed and processed as above. Images were captured with Axioplan2 Zeiss microscope with a AxioCam MRm Zeiss digital camera supported by Axiovision 4.6 software.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Drs. Tohru Natsume (National Institute of Advanced Industrial Science and Technology, Japan), Kenshi Komatsu (Kyoto University), Hiroshi Kurumizaka (Waseda University), J. Lin and J.W. Harper (Harvard Medical School) for reagents; Mr. Ryo Sasaki (Digital Microsystems, Inc., Kyoto) for advice and help in microscopy; Ms Keiko Namikoshi for expert technical help; Ms. Hiroko Shimamoto and Seiko Arai for secretarial assistance. This work was supported in part by Grants-in aid from the Ministry of Education, Science, Sports, and Culture of Japan (MT), by grants from Ministry of Health, Labour, and Welfare (MT), and by NIAID 1U19A1067751 and NIH grants (SJE). A. Smogorzewska is supported by T32CA09216 to the MGH Pathology Department. Financial support was also provided by The Novartis Foundation (Japan) for the Promotion of Science (MT) and The Uehara Memorial Foundation (MT). SJE is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Accession number for chicken FANCI cDNA: AB378696
Note: Supplementary information is available on the website.
AUTHOR CONTRIBUTIONS
MT, A. Smogorzewska, and SJE conceived and designed the experiments; MI, HK, A. Saberi, EU performed most of the DT40 cell experiments; MI, AK and ST performed microscopic analysis; EK, EK-K, TK and JT designed and performed phos-tag experiments; A. Smogorzewska performed human cell experiments; MI and HK analyzed the DT40 cell data; MT, A. Smogorzewska, and SJE wrote the paper.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 3.Ito K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 4.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 5.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 6.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 7.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 9.Dorsman JC, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell. Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 12.Ciccia A, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Ling C, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi T, et al. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 15.Matsushita N, et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol. Cell. 2005;19:841–847. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Nijman SM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 18.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Cantor SB, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 20.Levitus M, et al. Heterogeneity in Fanconi anemia: evidence for two new genetic subtypes. Blood. 2004;103:2498–2503. doi: 10.1182/blood-2003-08-2915. [DOI] [PubMed] [Google Scholar]
- 21.Seki S, et al. A requirement of FancL and FancD2 monoubiquitination in DNA repair. Genes Cells. 2007;12:299–310. doi: 10.1111/j.1365-2443.2007.01054.x. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto K, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell. Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano S, et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 27.Sarkaria JN, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 28.Oestergaard VH, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol. Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 30.Ho GP, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol. Cell. Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 2007;27:3098–3108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meetei AR, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 34.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto K, et al. Fanconi anemia FANCG protein in mitigating radiation-and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell. Biol. 2003;23:5421–5430. doi: 10.1128/MCB.23.15.5421-5430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishiai M, et al. DNA cross-link repair protein SNM1A interacts with PIAS1 in nuclear focus formation. Mol. Cell. Biol. 2004;24:10733–10741. doi: 10.1128/MCB.24.24.10733-10741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitao H, et al. Functional interplay between BRCA2/FANCD1 and FANCC in DNA repair. J. Biol. Chem. 2006;281:21312–21320. doi: 10.1074/jbc.M603290200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.