Medulloblastoma, the most common malignant pediatric brain tumour, arises in the cerebellum, and disseminates through the cerebrospinal fluid (CSF) in the leptomeningeal space to coat the brain and spinal cord1. Dissemination, a marker of poor prognosis, is found in up to 40% of children at diagnosis and most children at the time of recurrence. Therefore, affected children are treated with radiation to the entire developing brain and spinal cord followed by high dose chemotherapy with ensuing deleterious effects on the developing nervous system 2. The mechanisms of CSF dissemination are poorly studied and medulloblastoma metastases have been assumed to be biologically similar to the primary tumour 3,4. Here we show that in both mouse and human medulloblastoma, multiple metastases from a single patient are extremely similar to each other, but divergent from the matched primary tumour. Clonal genetic events in the metastases can be demonstrated in a restricted sub-clone of the primary tumour, suggesting that only rare cells within the primary tumour have the ability to metastasize. Failure to account for the bicompartmental nature of metastatic medulloblastoma could represent a major barrier to the development of effective targeted therapies.
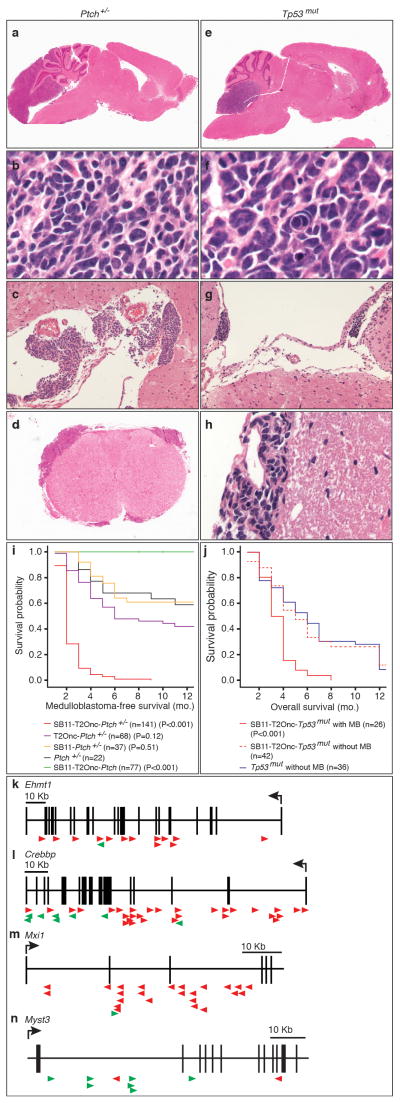
Thirty percent of Ptch+/− mice develop non-disseminated medulloblastoma by eight months of age5. Recently, the Sleeping Beauty (SB) transposon system was shown to be an effective tool for functional genomics studies of solid tumour initiation and progression 6,7. We expressed the SB11-transposase in cerebellar progenitor cells in transgenic mice under the Math1 enhancer/promoter, but did not observe any tumours when bred to transgenic mice with a concatemer of the T2/Onc transposon (Fig. 1, Supplemental Fig. S1, S2)8. However, Ptch +/−/Math1-SB11/T2Onc mice showed increased penetrance of medulloblastoma (~100% (271 out of 279) compared to ~40% (54 out of 139) of controls, and decreased latency (8 months to 2.5 months) (Fig. 1, Supplemental Fig. S2). While Ptch +/− medulloblastomas are usually localized, the addition of SB transposition results in metastatic dissemination through the CSF pathways, identical to the pattern seen in human children (Fisher’s exact test, p=1.8e-07, odds ratio=5.2, Supplemental Table S1) (Fig. 1c, d, g, h, Supplemental Figure S2). As neither transposon, nor transposase alone had an effect on tumour incidence, latency, or dissemination, we conclude that SB-induced insertional mutagenesis drives medulloblastoma progression on the Ptch +/− background (Fig. 1i, Supplemental Fig. S2).
Figure 1. Transposon mutagenesis models disseminated human medulloblastoma.
Histology of transposon driven medulloblastoma on the Ptch +/− background (a to d) resembles human medulloblastoma with leptomeningeal metastases on the surface of the brain (c) and spinal cord (d). Histology of transposon driven medulloblastoma on the Tp53 mut background (e to h) show histological features of large cell/anaplastic medulloblastoma including nuclear pleomorphism and nuclear wrapping (f). Dissemination to the leptomeningeal spaces of the brain (g) and spinal cord (h). (i) Ptch +/− mice with sleeping beauty transposition develop more frequent medulloblastomas with a shorter latency than Ptch +/− mice without transposition (P values are from t-tests of survival comparing individual genotypes to Ptch +/− mice; n= # of mice per genotype). (j) Medulloblastoma was not seen in Tp53mut mice without transposition, but was seen in 42% of Tp53mut mice with transposition. (P values are from t-tests comparing survival between Tp53mut mice and SB11-T2Onc-Tp53mut and Tp53mut mice; n = # of mice). (k–n). Insertion maps of notable gCISes. Insertions in the direction of transcription denoted by green arrows, those against the direction of transcription by red arrows.
Humans with germline mutations in TP53 have Li-Fraumeni syndrome and are at increased risk to develop medulloblastoma. While no medulloblastomas were found in Tp53mut (Tp53 +/− or Tp53−/−) mice, 40% of Tp53mut/Math1-SB11/T2Onc mice developed disseminated medulloblastoma (Fig. 1e–h, j, Supplemental Fig. S2)9. Human medulloblastomas with TP53 mutations frequently have large cell/anaplastic histology. Tp53mut/Math1-SB11/T2Onc medulloblastomas exhibit large cells, nuclear atypia, and nuclear molding typical of large cell/anaplastic histology (Fig. 1f). We conclude that SB transposition can drive the initiation and progression of metastatic medulloblastoma on a Tp53mut background.
We used linker-mediated PCR and Roche 454 sequencing to identify the site of T2/Onc insertions in Ptch+/−/Math1-SB11/T2Onc, and Tp53mut/Math1-SB11/T2Onc primary medulloblastomas and their matched metastases. Genes that contained insertions statistically more frequently than the background rate were identified as gene-centric commonly inserted sites (gCISes)10. We identified 359 gCISes from 139 primary tumours on the Ptch background and 26 gCISes from 36 primary medulloblastomas on the Tp53 background (Supplemental Tables S2–S7, Supplemental Figures S3–S5). A large number of gCISes targeted candidate medulloblastoma oncogenes/tumour suppressor genes (Supplemental Table S8)11. Insertions in candidate tumour suppressor genes including EHMT1, CBP, and MXI1 are predicted to be loss of function (Fig. 1k,l,m), while insertions in putative medulloblastoma oncogenes are largely gain of function, as exemplified by MYST3 (Fig. 1n).
Many gCISes mapped to regions of amplification, focal hemizygous deletion, and homozygous deletion, which we recently reported in the genome of a large cohort of human medulloblastomas (Supplemental Table S8) 11. There is a high level of overlap between gCISes and known cancer genes (COSMIC database) (Supplemental Table S9,10), suggesting that many gCISes are bona fide driver genes in medulloblastoma (Fisher’s exact test p=0.0012)12. Similarly, many mouse gCIS/ human amplified genes are over-expressed in human Shh medulloblastomas (Supplemental Fig. S6). Conversely, mouse gCISes deleted in human medulloblastomas were frequently expressed at a lower level in human medulloblastomas (Supplemental Fig. S6). Expression of 6/7 gCISes studied by immunohistochemistry on a human medulloblastoma tissue microarray were associated with a significantly worse overall and progression free survival in human medulloblastoma (Supplemental Table 11, Supplemental Figures S7, S8) 13. We conclude that our SB-driven leptomeningeal disseminated medulloblastoma model resembles the human disease anatomically, pathologically and genetically and thus represents an accurate model of the human disease that can be used to identify candidate driver events and understand the pathogenesis of human medulloblastoma.
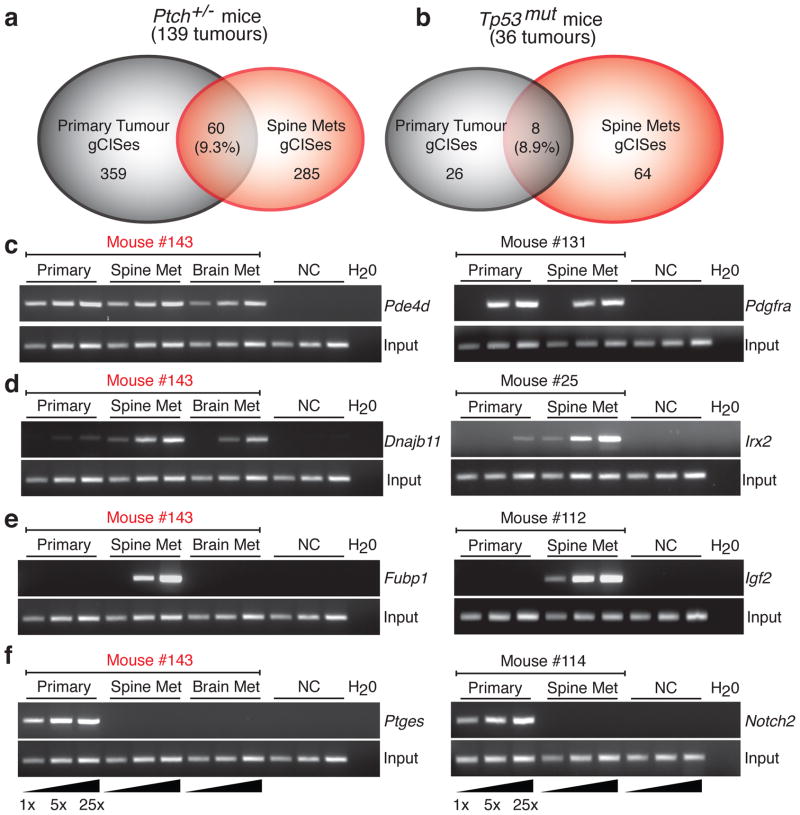
We compared the gCISes identified from Ptch+/−/Math1-SB11/T2Onc, and Tp53mut/Math1-SB11/T2Onc primary medulloblastomas and matched metastases (Supplemental Table S2). Strikingly, the overlap between primary tumour gCISes (pri-gCISes) from Ptch+/−/Math1-SB11/T2Onc tumours and those from the metastases (met-gCISes) from the same animals was only 9.3% of gCISes (Figure 2a). Similarly, the overlap in pri-gCISes from primary Tp53mut/Math1-SB11/T2Onc gCISes and the matching met-gCISes was only 8.9% (Figure 2b). Leptomeningeal metastases and the matched primary tumour share identical, highly clonal insertion sites (Fig. 2c). The chances of two (or three) unrelated tumours having SB insertions in exactly the same TA dinucleotide are extremely low. We conclude that leptomeningeal metastases and matched primary tumour arise from a common transformed progenitor cell, and have subsequently undergone genetic divergence. Sequencing also identified insertions that are highly clonal in the metastases, but not seen in the matched primary tumour (not shown). Endpoint PCR for these insertions in the matched primary/metastatic tumours show that the insertion is highly clonal in the metastase(s), and present in a very small subclone of the primary tumour (Fig. 2d, Supplemental Figure S9). These data are consistent with a model in which metastatic disease arises from a minor restricted subclone of the primary tumour. Dissemination could occur repeatedly from the same subclone of the primary tumour, which seeds the rest of the CNS, or could occur once followed by reseeding of the rest of the leptomeningeal space by the initial metastasis. Insertions that are restricted to a minor subclone of the primary tumour, but which are clonal in the metastases, could correspond to the ‘metastasis virulence’ genes, described previously, that offer a genetic advantage during dissemination, but not to the primary tumour 14. Another explanation of our data could be reseeding of the primary tumour by a metastatic clone that had acquired additional genetic events in the periphery. This latter hypothesis is mitigated by the presence in the same animal of highly clonal insertions in the metastasis that are completely absent from the primary tumour 15. As reseeding should be accompanied by contamination of the primary tumour with events found in the metastases, absence of these events in the matched primary tumour makes reseeding much less likely (Fig. 2e). We hypothesize that events found only in one metastasis represent progression events acquired post-metastasis, and which could lead to localized progression of metastatic disease as is sometimes seen in human children. We observed highly clonal insertions in the primary tumour, including known medulloblastoma oncogenes such as Notch2, or Tert, which are not found in the matching metastases (Fig. 2f). This pattern could be explained through remobilization of the SB transposon in the metastatic tumour; however, no signs of the DNA footprint left after SB remobilization at these loci were observed (Supplemental Fig. S10)16. We suggest that these events, which may constitute driver events in the primary tumour, have arisen in the primary tumour after the metastases have disseminated (post-dispersion events). Although these known oncogenes represent attractive targets for therapy, their utility as targets for therapy may be limited if the target is not also found in the leptomeningeal compartment of the disease. Our data from two separate mouse lines supports a model in which medulloblastoma disseminates early from a restricted subclone of the primary tumour, and where the primary tumour and the matched metastases then undergo differential clonal selection and evolution. Failure to account for the differences between the primary and leptomeningeal compartments could lead to the failure of targeted therapies. Failure to study the leptomeningeal disease could result in systematically overlooking critical targets for therapy in this compartment (Fig. 2e).
Figure 2. Transposon driven metastatic medulloblastoma genetically differs from the primary tumour.
(a,b) Venn diagram showing the extent of overlap between gCISes in primary tumours and metastases on the Ptch +/− and Tp53mut backgrounds. (c–f) Insertion site, end point PCR to demonstrate relative clonality of insertions between samples. Three levels of input DNA were used for each sample as illustrated (1x, 5x, 25x). Insertions in the first column are from medulloblastoma #143 (c) Clonal events found in both primary tumour and matching metastases. (d) Insertions that are highly clonal in the metastases, but very sub-clonal in the matching primary tumour. (e) Insertions that are highly clonal in the metastases, but completely undetectable in the matching primary tumour. (f) Insertions that are highly clonal in the primary tumour, but completely undetectable in the matching metastases.
Examination of met-gCIS genes using GSEA demonstrates differences between the primary and metastatic disease, which importantly include enrichment for genes involved in the cytoskeleton among the metastases (Supplemental Table S12). Targets that are present in both compartments, and which are maintenance genes, will be optimal targets for therapy of both the primary and metastatic compartments, as exemplified by Pdgfra (Fig. 2c, Supplemental Tables S7, S9).
Pten, Akt2, Igf2, and Pik3r1 are all met-gCISes, implicating the PI3-kinase pathway in medulloblastoma progression. We injected the cerebella of Nestin-TVA mice 17 with either Shh virus alone, or Shh + Akt virus. Cerebellar injection of Shh alone resulted in medulloblastoma in 6/41 animals, compared to 20/42 animals injected with Shh + Akt (p=0.0018). Poignantly, while metastases were never seen with Shh virus alone (0/41), medulloblastoma metastases were seen in 9/42 animals injected with Shh + Akt (p=0.0024) (Supplemental Fig. S11). In vivo modeling validates PI3-kinase signaling and suggests that it can contribute to leptomeningeal dissemination of medulloblastoma.
Prior publications and clinical approaches to human medulloblastoma have largely assumed that the primary tumour and its matched metastases are highly similar 3,4. To test this assertion we formally reviewed all cases of medulloblastoma from the last decade at The Hospital for Sick Children, and identified 19 patients who had both bulk residual primary tumour post-surgery, and MRI visible metastases, both of which could be followed for response to treatment in the two compartments (Supplemental Fig. S12 and Supplemental Table S13). While it is possible that metastases might have received reduced radiotherapy than the primary tumor in a subset of patients, in 58% of overall cases (11/19) we observed a disparate response to therapy between the primary tumor and matched metastases (binomial test, p<2.2e-16). Identification of definitive differences in the clinical response to standard therapy between the primary and metastatic compartment awaits the completion of large, well controlled, prospective clinical trials.
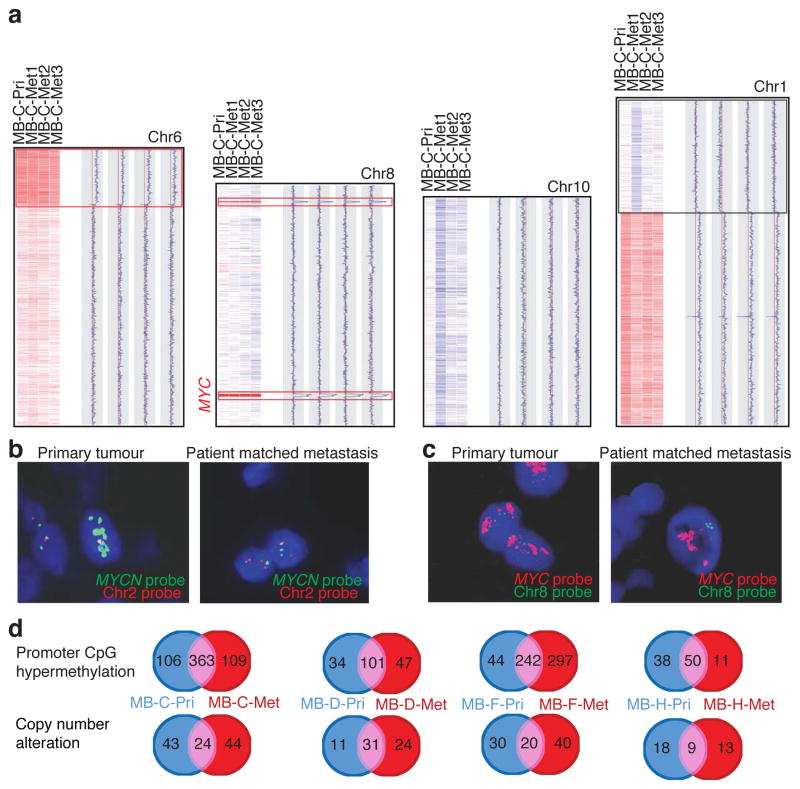
We examined seven matched primary/metastatic medulloblastomas for copy number aberrations (Fig. 3, Supplemental Figures S13, S14, Supplemental Tables S14, S15). In each case, the primary tumour and the matched metastases share complicated genetic events that highly support descent from a common transformed progenitor cell. Similar to our mouse data, in each case we see clonal genetic events in the metastatic tumour(s) that are not present in the matched primary tumour (Fig. 3. Supplemental Fig. S14). We also observe genetic events in the primary tumour that are absent from the matched metastasis, consistent with a ‘post-dispersion event’ (Fig. 3, Supplemental Fig. S14). Examination of a case with multiple leptomeningeal metastases demonstrates a deletion of chromosome 1p in only 1/3 metastases (Fig. 3a). This pattern of genetic events that are present only in a subset of metastases could be a mechanism for the emergence of therapy resistant metastatic clones.
Figure 3. Human medulloblastoma metastases are biologically distinct from their matched primary tumour.
(a) Copy number data from a primary medulloblastoma and three patient matched metastases, with chromosomal regions in red representing genetic gain (amplification), and blue denoting genetic loss (deletion). Examples of shared clonal events (red boxes), events limited to the metastases (blue boxes), and events limited to one but not all metastases (black box) are shown. (b) Interphase FISH shows amplification of MYCN in a primary tumour, but not the matched metastasis. (c) Interphase FISH for MYC demonstrates amplification in both the primary tumour and its matched metastases. (d) Venn diagrams depicting the degree of overlap and discordance in promoter CpG methylation events (above) and copy number alterations (below) in primary medulloblastomas and their matched metastasis.
We performed interphase FISH for the known medulloblastoma oncogenes MYCN and MYC on a collection of 17 paraffin embedded primary/metastatic pairs of human medulloblastoma 18–20. MYCN was amplified in 3 primary medulloblastomas, but not in the matching metastases (Fig. 3b, Supplemental Fig. S15). Conversely, MYC was amplified in 2 primary tumours and their matching metastases (Fig. 3c). These data are consistent with MYCN amplification being a ‘post-dispersion’ event, similar to examples in SB driven mouse medulloblastoma, and highly suggest that anti-MYCN therapeutics could lack efficacy in the metastatic compartment of human medulloblastoma. The possibility that MYCN amplicons are ‘lost’ overtime in the metastases cannot be excluded.
We subsequently analyzed promoter CpG-methylation in these matched pairs and found that there was a great deal of discordance between the primary tumour and matched metastases (Fig. 3d, Supplemental Figures S13, S16, Supplemental Tables S16–17). Finally, we performed whole-exome sequencing on a limited set of matched primary/metastatic medulloblastomas, and found many single nucleotide variants (SNVs) that were restricted to one compartment or the other (Supplemental Fig. S13, Supplemental Table S18). Discordance of CNAs, promoter CpG methylation events, and SNVs between the primary tumour and its matched metastases supports a bicompartmental model for metastatic medulloblastoma. The mutational load in the human tumours (combination of CNAs, CpG methylation, and SNVs) compares favorably to the mutational load in our transposon driven mouse models (median number of gCISes=25 per tumour, Supplemental Table S19). Validation of individual CNAs restricted to the metastases, reveals that they can be detected in a very minor subclone of the primary tumour, in keeping with the relationship identified in the mouse model (Supplemental Fig. S17, Supplemental Table S20–21). Pathway analysis using DAVID to compare mouse gCISes with genes affected in human metastases identified only a single statistically significant shared signaling pathway – insulin signaling (p=0.027) (Supplemental Table S22). The known role for insulin receptor signaling in primary medulloblastoma21, and the data presented above on the role of Akt in metastatic medulloblastoma, might suggest prioritization of insulin signaling as a therapeutic target to be tested in clinical trials.
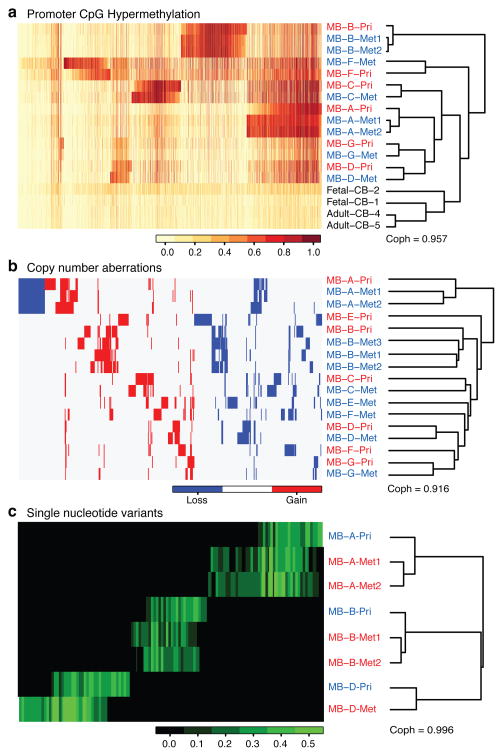
We performed unsupervised hierarchical clustering on CpG methylation data revealing that normal cerebellar controls cluster away from the medulloblastomas, while metastases clustered with their matching primary tumour (Fig. 4a). However, metastases cluster more closely to each other than they do to the matched primary tumour (z-test, p=0.0014, Supplemental Fig. S18). Unsupervised hierarchical clustering of CNA and exome SNV data reveals the same relationships (Fig. 4b,c). Evident within the exome data are many events that are shared only by patient matched metastases, as well as events restricted to the primary tumour, both of which are similar to the genetic patterns observed in the mouse. These three data sets support a model in which patient matched human medulloblastoma metastases are epigenetically, and genetically very similar to each other, but have substantially diverged from their primary tumour, resulting in two different disease compartments: primary and metastatic.
Figure 4. Human medulloblastoma metastases are genetically distinct from the primary tumour.
(a) Profiling the methylation status of 27,578 CpG dinucleotides sites in the human genome in a collection of human matched primary and metastatic medulloblastomas, top 2000 genes are shown. Unsupervised hierarchical clustering by CpG methylation patterns demonstrates that patient matched metastases are more similar to each other than to their matched primary tumour. (b) Unsupervised clustering of regions of copy number gain and loss demonstrates that patient matched metastases are more similar to each other, than they are to their matched primary tumour. (c) Unsupervised hierarchical clustering of SNV data from whole exome sequencing demonstrates that patient matched metastases are more similar to each other than to the patient matched primary tumour. SNVs that are found only in the primary compartment, or only in both tumours in the metastatic compartment are evident.
Our data from two different mouse models with support by initial data from human medulloblastoma suggests that leptomeningeal metastases of medulloblastoma from a single human/mouse are genetically similar to each other, but highly divergent from the matched primary tumour, consistent with a bicompartmental model of the disease. Our results are consistent with a model in which metastases arise from a restricted subclone of the primary tumour through a process of clonal selection in both humans and mice. That metastases might arise from a pre-existing minor subclone of the primary tumour through clonal selection was suggested more than three decades ago, but remains a controversial hypothesis whose truth may vary from one disease to another 22–25. Failure to account for the divergent molecular pathology of the metastatic compartment may result in selection of therapeutic targets present in the primary tumour (which is more amenable to surgical control) and not the metastases, which are the more frequent cause of death.
Methods Summary
Generation of Math1SB11 transposase transgenic mouse
SB11 transposase cDNA was excised from pCMV-SB11 and ligated into J2QMath1 vector26,8.
Linker-mediated PCR and 454 deep sequencing
Bar-coded, linker-mediated PCR was performed as previously described 6. Sample preparation for 454 sequencing and subsequent procedures were performed as described 27.
Determination of gCISes: Gene-centric common insertion site analysis
A chi-square analysis was performed to determine if the number of observed integration events within each transcription unit in the SB driven medulloblastomas is significantly greater than expected given the number of TA dinucleotide sites within the gene relative to the number of TA sites in the genome, the number of integration sites within each tumour and the total number of tumours in each cohort. This gene-centric common insertion site (gCIS) analysis produced a p-value for each of the ~19,000 mouse RefSeq genes, and therefore Bonferroni correction was used to adjust for multiple-hypothesis testing.
Methods
Linker-mediated PCR and 454 deep sequencing
Genomic DNA was isolated and purified from mouse tissues with DNeasy Tissue and Blood kit (QIAGEN). The subsequent bar-coded linker-mediated PCR was performed as previously described. Sample preparation for 454 sequencing and subsequent procedures were performed as described 27.
PCR for SB-tagged fragments
Primers for amplifying SB transposon insertion sites were designed based on chromosomal location of each independent insertion site and its orientation to the transcription. The primers at IRDRL and IRDRR of the transposon are: 5′-CTGGGAATGTGATGAAAGAAATAAAA-3′ and 5′-TTGTGTCATGCACAAAGTAGATGT -3′, respectively. The input represents genomic DNA with SB transposition, which was illustrated by SB excision PCR that detect the transposon at post-transposition 6. Three points of input (1x, 5x, 25x) were used. Details about primer design for specific insertion sites and the PCR protocol are available upon request.
Review of Clinical Cases
We systematically reviewed all cases of medulloblastoma seen at the Hospital for Sick Children over the past ten years. Cases were identified that have both metastases and post-operative residual bulky disease at the primary site on the basis of postoperative imaging obtained within 72 hours of surgery. All radiology was reviewed by a senior neuro-oncologist (EB). Objective responses of both the primary tumor, and the metastatic disease were measured using the International Society for Pediatric Oncology (SIOP) criteria that are standard for clinical trials of pediatric brain tumors 28.
End-point PCR on human samples
For PCRs to confirm the deletion of CDKN2A locus at chromosome 9, a genome-walking approach (GenomeWalker Universal kit, Clonetech Cat#638904) was taken to locate the specific deletion region based on SNP coordinates. Primers flanking the deletion region are: Forward 5′-GCAATTAACCAAGACCACCCAATGGCAAG-3′ and Reverse 5′-GTAGCTATTGGGGAGGTTGAGAAGGAG-3′. Three points of input shown as ACTB (1x, 5x, 25x) were used. The PCR products were inserted into pCR2.1 TA cloning vector (Invitrogen), sequenced and blasted against human genome to confirm the deletion. For REXO1L1 deletion at chromosome 8, specific primers flanking the deletion region were designed based on SNP microarray results. The PCR products were TA-cloned and sequenced as described above. The primers are: Forward 5′-GGCTGACTCCCTTCT GATATAG-3′ and Reverse 5′-CAA TCA CTT ACA GTT ACT AGG CAC-3′. Details about the primer design and PCR protocols are available upon requested.
Chromosomal mapping of gCISes
Chromosomal maps of gCISes-associated genes were obtained from UCSC Mouse Genome Browser (assembly in July 2007). Each insertion site of a specific CIS was mapped to the gene with the same orientation as transcription (arrow in green) or inverse (arrow in red) to the direction of transcription.
Human medulloblastoma tumour specimens
All tumour specimens were obtained in accordance with the Research Ethics Board at the Hospital for Sick Children (Toronto, Canada). Surgically-resected, fresh-frozen samples were obtained from the Co-operative Human Tissue Network (Columbus, OH), and the Brain Tumor Tissue Bank (London, Canada).
SB remobilization
Potential SB insertion sites either at Fubp1, Mnat1 and Igf2 in primary tumours from mouse#143, #14 and 112, or at Ptges, Aof1 and Notch2 in the matched spine mets were tested for remobilization. Primers were designed to amplify each insertion site to produce a proximate 300bp with the insertion site in the middle. PCR products were either sequenced directly or after being TA-cloned. The resulted sequences were examined for ‘scars’ from potential remobilization. As positive controls for the ‘scars’, primers were used to amplify T2Onc transposon in each samples8. The products were sequenced and examined for the ‘scars’ as described above. Details about the primer design and PCR protocols are available upon requested.
Hierarchical clustering
Agglomerative hierarchical clustering analyses were performed in the R statistical programming environment (v2.13). The average linkage method was used in all cases. As different data types were used in the various analyses, the metric used for clustering differed among analyses. Manhattan distance metric was used for the copy number data, since the data was encoded as {−1, 0, 1}. The magnitudes of the copy number aberrations (CNAs) were not considered, due to multitude of confounding factors including tumour heterogeneity and ploidy. Kendall correlation was used for the single nucleotide variant (SNV) frequency data, since the data distributions were not normal. Pearson correlation was used for the methylation data, which were normally distributed.
Identification of CpG Hypermethylation Events
Human genomic DNA was isolated from matching primary and metastatic medulloblastomas obtained from John Hopkins University, Virginia Commonwealth University and New York University. Zymogen’s EZ DNA Methylation kit was employed for bisulfite-converting 500ng of each sample. The recovered DNA was profiled on Illumina HumanMethylation27 BeadChips at TCAG. Subsequently 27,578 CpG dinucleotides spanning 14,495 genes were analyzed. Probe signal intensity was corrected by using Illumina BeadStudio 3.2.0 software. The background normalization and differential methylation analyses were performed against fetal cerebella by Illumina Custom Error Model. Cancer-specific DNA hypermethylation events were defined as those with a 30% increase in methylation in at least one medulloblastoma samples, relative to an average methylation level (less than 50%) in normal fetal and adult cerebellum samples. An unsupervised clustering using Euclidian hierarchical clustering metrics was then performed on 2503 data points that were filtered for cancer-specific hypermethylation events. CpG methylation data is available from GEO under GSE 34356.
Bisulfite sequencing CpG promoter methylation
Representative examples of primary- and metastatic-specific methylation events were identified from normalized Illumina Hg27 methylation data. Bisulfite PCR (BSP) primers were designed using Sequenom Epidesigner tool (http://www.epidesigner.com/) encompassing a genomic region flanking the Illumina Hg27 gene-specific probe. 500ng of primary and corresponding metastatic DNA were bisulfite converted using EZ DNA Methylation Kit (Zymo Research). Following PCR optimization, 10ng of bisulfite converted DNA was used to amplify genomic regions of interest. Amplicons were subcloned into pCR2.1-TOPO (Invitrogen) and plasmid DNA from 10–12 colonies was extracted using Purelink Quick Plasmid Miniprep Kit (Invitrogen). Sequencing was performed at the TCAG using the M13 Reverse primer (5′-CAG GAA ACA GCT ATG AC). Details about the primer design and PCR protocols are available upon requested.
Alignment and variant calling for whole exome sequencing
Standard manufacturer protocols were used to perform target capture with the Illumina TruSeq exome enrichment kit and sequencing of 100 bp paired end reads on Illumina Hiseq. Approximately 10 Gb of sequence were generated for each subject such that >90% of the coding bases of the exome defined by the consensus coding sequence (CCDS) project were covered by at least 10 reads. Adaptor sequences and quality trimmed reads were removed by using the Fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and then a custom script was used to ensure that only read pairs with both mates present were subsequently used. Reads were aligned to hg19 with BWA1, and duplicate reads were marked using Picard (http://picard.sourceforge.net/) and excluded from downstream analyses. Single nucleotide variants (SNVs) and short insertions and deletions (indels) were called using samtools (http://samtools.sourceforge.net/) pileup and varFilter2 with the base alignment quality (BAQ) adjustment disabled, and quality filtered to require at least 20% of reads supporting the variant call. Variants were annotated using both Annovar3 and custom scripts to identify whether they affected protein coding sequence, and whether they had previously been seen in dbSNP131, the 1000 genomes pilot release (Nov. 2010), or in approximately 160 exomes previously sequenced at our center.
SNV analysis of whole Exome sequencing data
For clustering analysis, a SNV frequency matrix was constructed by calculating frequencies from the read counts of the reference and the alternative nucleotide. The matrix was not standardized (i.e. converted to z-scores) prior to clustering, since the absolute SNV frequencies were of interest.
For Venn analysis, the samples were grouped into primary-metastasis sets, and the filtered SNVs were used to identify SNVs that are enriched in one sample compared to all other samples of the same set, as determined by the hypergeometric test (p-value threshold = 0.05). For sets consisting of three or more samples (A, B, and C), a SNV was considered to be enriched in samples A and B if the SNV was enriched in A as compared to C alone and also enriched in B as compared to C alone. SNVs that were not enriched in any sample or subsets of samples were considered to be common SNVs. Many of these common SNVs likely represented germline SNVs specific to the patient.
Analysis of CpG promoter methylation data
The similarities between patient-matched metastatic and primary tumour samples and between patient-matched metastatic tumour samples were determined by using Pearson correlation analysis. As Pearson’s r values are not normally distributed, they were standardized by Fisher’s z transformation. Subsequently, the correlations between metastatic samples and the matched primary samples were compared to the correlations between the patient-matched metastatic samples, using the paired heteroscedastic Student’s t test. Clustering analysis was performed as described above. The methylation matrix was not standardized prior to clustering, as doing so would discard critical information regarding the differences in overall methylation profiles among samples or average methylation among CpG promoters.
The stability of the CpG hypermethylation profile clusters was assessed using three methods. First, the clustering analysis was run for different numbers of CpG hypermethylation sites that vary most widely among samples. The partitions generated by each clustering run is compared to the reference partitions generated by original clustering based on the 1000 most variable hypermethylated CpG islands using the Jaccard similarity index. The same analysis was applied to a set of 100 background hypermethylation data matrices in which the sites are permuted within and independently for each sample. Second, the clustering analysis was performed for random sub-samples of 1000 sites, for 1000 repeat runs. In each run, the resulting cluster was compared to the original cluster using the Jaccard index. Analysis on the original data matrix was compared to a set of 100 background matrices, permuted as described above. Third, the cluster stability was further assessed by bootstrap re-sampling of the samples using the Pvclust R package (v1.2).
Supplementary Material
Acknowledgments
M.D.T. holds a CIHR Clinician-Scientist Phase II Award, was a Sontag Foundation Distinguished Scholar, and is supported by grants from the National Institutes of Health (R01CA148699), the Pediatric Brain Tumour Foundation, the Canadian Cancer Society, and Brainchild. X.W. was supported by a fellowship from the American Brain Tumour Association in tribute to Tracy Greenwood. L.G. was supported by a fellowship from the Davis M. Ferguson Fund from The American Brain Tumour Association. A. Dubuc was supported by a Vanier Doctoral Fellowship from the CIHR. LSC was supported by K01CA122183 and a Kimmel Scholar award from the Kimmel foundation. CGE was supported by a grant from the NIH (NS055089). We thank Susan Archer for technical writing assistance.
Footnotes
Author Contributions: MDT, XW, PAN, LC, A Dupuy, and DL conceived the research and planned the experiments. XW, PAN, A Dubuc, DS conducted the vast majority of the experiments under the guidance of MDT. CE, TVM, DZ, JC, TM, PC, XNL, TM, VPC, YJC, and WAW provided human tumour materials. All authors contributed experimental expertise and participated in the writing of the manuscript. A Dupuy, DJHS, TES, SWS, KA, JK, ALS, DL, and LC provided biostatistical and bioinformatic expertise. EB provided the clinical data and analysis. DWF did the Akt experiments. NJ, JS, and JM did the exome sequencing. HW, SFP, and AK performed immunostaining of human medulloblastoma tissue microarrays and did FISH for MYCN and MYC. SC performed the pathological analysis of mouse brain tumours.
Author Information: The authors declare no competing financial interests.
References
- 1.Gajjar A, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813–820. doi: 10.1016/S1470-2045(06)70867-1. S1470-2045(06)70867-1 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Mabbott DJ, et al. Serial evaluation of academic and behavioral outcome after treatment with cranial radiation in childhood. J Clin Oncol. 2005;23:2256–2263. doi: 10.1200/JCO.2005.01.158. 23/10/2256 [pii] [DOI] [PubMed] [Google Scholar]
- 3.MacDonald TJ, et al. Expression profiling of medulloblastoma:PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet. 2001;29:143–152. doi: 10.1038/ng731. ng731 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. ng1060 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 6.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. nature03681 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. nature03691 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- 9.Wetmore C, Eberhart DE, Curran T. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res. 2001;61:513–516. [PubMed] [Google Scholar]
- 10.Brett BT, et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PLoS One. 2011;6:e24668. doi: 10.1371/journal.pone.0024668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. ng.336 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10(Unit 10):11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. nrg2101 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Norton L, Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12:875–878. doi: 10.1038/nm0806-875. nm0806-875 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao G, Pedone CA, Coffin CM, Holland EC, Fults DW. c-Myc enhances sonic hedgehog-induced medulloblastoma formation from nestin-expressing neural progenitors in mice. Neoplasia. 2003;5:198–204. doi: 10.1016/S1476-5586(03)80052-0. NO_DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swartling FJ, et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 2010;24:1059–1072. doi: 10.1101/gad.1907510. 24/10/1059 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfister S, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27:1627–1636. doi: 10.1200/JCO.2008.17.9432. JCO.2008.17.9432 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Korshunov A, et al. Accumulation of genomic aberrations during clinical progression of medulloblastoma. Acta Neuropathol. 2008;116:383–390. doi: 10.1007/s00401-008-0422-y. [DOI] [PubMed] [Google Scholar]
- 21.Hahn H, et al. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. The Journal of biological chemistry. 2000;275:28341–28344. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- 22.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 23.Scheel C, Onder T, Karnoub A, Weinberg RA. Adaptation versus selection: the origins of metastatic behavior. Cancer Res. 2007;67:11476–11479. doi: 10.1158/0008-5472.CAN-07-1653. discussion 11479-11480, 67/24/11476 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Talmadge JE. Clonal selection of metastasis within the life history of a tumor. Cancer Res. 2007;67:11471–11475. doi: 10.1158/0008-5472.CAN-07-2496. 67/24/11471 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. 0008-5472.CAN-10-1040 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geurts AM, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. S1525001603000996 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. 1163040 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnekow AK. Recommendations of the Brain Tumor Subcommittee for the reporting of trials. SIOP Brain Tumor Subcommittee. International Society of Pediatric Oncology. Medical and pediatric oncology. 1995;24:104–108. doi: 10.1002/mpo.2950240209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.