Differences in expression level of the effector caspases Drice and Dcp-1 and in their intrinsic abilities to induce apoptosis and to control the rate of cell death underlie the differential sensitivities of cells to apoptosis.
Abstract
Essentially, all metazoan cells can undergo apoptosis, but some cells are more sensitive than others to apoptotic stimuli. To date, it is unclear what determines the apoptotic potential of the cell. We set up an in vivo system for monitoring and comparing the activity levels of the two main effector caspases in Drosophila melanogaster, Drice and Dcp-1. Both caspases were activated by the apoptosome after irradiation. However, whereas each caspase alone could induce apoptosis, Drice was a more effective inducer of apoptosis than Dcp-1, which instead had a role in establishing the rate of cell death. These functional differences are attributed to their intrinsic properties rather than merely their tissue specificities. Significantly, the levels of the procaspases are directly proportional to their activity levels and play a key role in determining the cell’s sensitivity to apoptosis. Finally, we provide evidence for the existence of a cellular execution threshold of caspase activity, which must be reached to induce apoptosis.
Introduction
Proteases are a large group of enzymes that can cleave proteins during a multitude of physiological reactions in all organisms (Hooper, 2002). A unique group of cysteine proteases called caspases functions to execute the apoptotic cell death program (Yuan et al., 1993; Thornberry and Lazebnik, 1998; Song et al., 2000). Caspases are synthesized as inactive zymogens (or proenzymes) and work in a controlled proteolytic cascade to activate themselves and one another (Riedl and Shi, 2004; Salvesen and Riedl, 2008). Initiator caspases are generally activated through dimerization, facilitated at multiprotein complexes such as the apoptosome (cytochrome c-Apaf-1–caspase-9; Zou et al., 1997; Rodriguez and Lazebnik, 1999; Salvesen and Riedl, 2008). Once activated, initiator caspases cleave and activate effector caspases, such as caspase-3 and -7 (Boatright and Salvesen, 2003; Riedl and Shi, 2004), which in turn cleave a variety of cellular protein substrates, ultimately leading to apoptosis (Lüthi and Martin, 2007; Dix et al., 2008; Mahrus et al., 2008). Caspases are also regulated by inhibitory proteins, such as the inhibitor of apoptosis proteins, which can bind to and inhibit caspases in both insects and mammals (Goyal et al., 2000; Vaux and Silke, 2005; Orme and Meier, 2009). In addition to the efforts invested in revealing and understanding these global pathways of caspase regulation during apoptosis, other studies imply that caspase activation is not necessarily an all-or-nothing process (Hoeppner et al., 2001; Reddien et al., 2001; Ribeiro et al., 2007). Indeed, low, transient, or subcellularly restricted levels of caspase activity have been recently reported to promote vital cellular processes (Koto et al., 2009; Kaplan et al., 2010; Li et al., 2010; Schoenmann et al., 2010). Furthermore, different cells, under varying physiological and pathological conditions, often display substantial differences in their sensitivity to apoptotic stimuli, which may reflect distinct apoptotic potentials. However, it is still poorly understood why restrictive levels of caspase activity fail to induce apoptosis and what factors determine the apoptotic potential of the cell.
The Drosophila melanogaster genome encodes seven distinct caspases, out of which the initiator caspase-9 ortholog Dronc and the effector caspase-3 ortholog Drice are the major apoptotic caspases (Kumar, 2007; Cooper et al., 2009; Xu et al., 2009). Mutations in these caspases cause pleiotropic defects in developmental cell death and stress-induced apoptosis as well as the apoptosis-like process of spermatid individualization (Chew et al., 2004; Daish et al., 2004; Waldhuber et al., 2005; Xu et al., 2005, 2006; Arama et al., 2006; Muro et al., 2006). In contrast, only minor physiological roles in apoptosis have been demonstrated for the initiator-like atypical caspase Strica/Dream and the effector-like caspases Dcp-1 and Decay (Laundrie et al., 2003; Leulier et al., 2006; Muro et al., 2006; Xu et al., 2006; Baum et al., 2007; Denton et al., 2009). The caspase-8–like initiator Dredd appears not to be involved in cell death but rather in the innate immune response (Leulier et al., 2000; Stoven et al., 2003), whereas no role in apoptosis has been thus far demonstrated for the effector-like caspase Damm/Daydream (Harvey et al., 2001; Leulier et al., 2006). It is unclear why these caspases display different roles in apoptosis. For example, although Dcp-1 is highly homologous to Drice (67% identity; Song et al., 2000) and loss of dcp-1 can aggravate drice mutant phenotypes (Leulier et al., 2006; Muro et al., 2006; Xu et al., 2006), dcp-1 mutant flies are quite healthy, displaying only mild defects during starvation-induced autophagy and cell death in midoogenesis (Laundrie et al., 2003; Hou et al., 2008). Likewise, in mammals, knocking out caspase-3 causes decreased apoptosis and pleiotropic morphological defects, whereas caspase-7 knockout mice exhibit only mild antiapoptotic defects (Kuida et al., 1996; Woo et al., 1998; Houde et al., 2004). Whether these functional differences are the consequence of tissue specificity, distinct cellular levels of activity, or different execution efficiencies remains to be investigated.
Using a transgenic reporter of caspase-3–like (DEVDase) activity (Williams et al., 2006), we set up an in vivo system for monitoring and comparing the activity levels and execution efficiencies of the two main effector caspases in Drosophila, Drice and Dcp-1, during apoptosis. We show that after irradiation-induced apoptosis, Drice and Dcp-1 are the only DEVDases that become activated in wing imaginal discs (WDs), although Drice is more efficient than Dcp-1 in processing this reporter and an endogenous protein substrate. The apoptosome components, Dronc and its adaptor protein Ark (the Apaf-1 homolog), constitute the initiator activity required for Drice and Dcp-1 activation, which can be blocked by expression of caspase inhibitory proteins. Importantly, in a series of genetic and transgenesis studies, we demonstrate that whereas both Drice and Dcp-1 can induce apoptosis, Drice can execute apoptosis more effectively than Dcp-1. Dcp-1, on the other hand, functions to fine tune the rate of cell death, at least in part through further activation of Drice in a positive amplification loop. Building on the differential efficiencies of these caspases to induce apoptosis, we show that the apoptotic potential of the cell is significantly affected by the total level of caspase activity, which in turn is directly proportional to the levels of the procaspases and their ratios. Finally, we demonstrate that the total level of caspase activity must reach a critical threshold in order for the cell to undergo apoptosis; short of that threshold, apoptosis cannot occur, and the cell will recover.
Results
Detection of effector caspase–like DEVDase activity during apoptosis in vivo
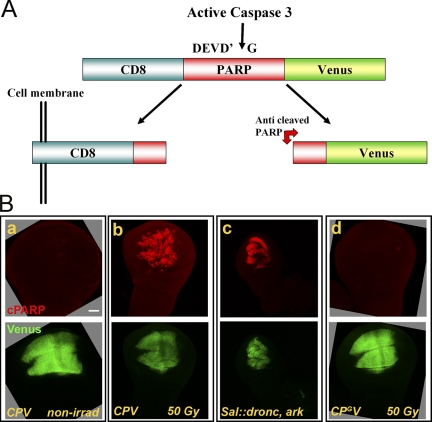
To monitor effector caspase activity in vivo, we took advantage of a genetic reporter previously designed for detection of caspase activity during the vital process of dendritic pruning in Drosophila sensory neurons (Williams et al., 2006; Schoenmann et al., 2010). This reporter, dubbed CD8::PARP::Venus (CPV), encodes an artificial effector caspase substrate composed of a 40-aa-long fragment of the human poly(ADP-ribose) polymerase (PARP) protein (including the caspase-3 consensus site DEVD) flanked by the extracellular/transmembrane domain of the mouse CD8 protein at the N-terminal side and the YFP Venus at the C terminus (Fig. 1 A). The activity is detected by staining with the anticleaved human PARP antibody, whereas the fluorescence of the Venus protein indicates which cells express the reporter. The CPV reporter has not been characterized during apoptosis, and the exact caspases (or other proteases) that may cleave it have not been determined. Therefore, we established a genetic assay for monitoring the CPV reporter during irradiation-induced apoptosis in Drosophila. The CPV was expressed in the pouch region of the WD using the spalt (sal)-Gal4 driver. Transgenic larvae were then γ-irradiated and subsequently allowed a 4-h recovery before their WDs were removed and stained to visualize cleaved human PARP. Whereas nonirradiated animals displayed almost no reporter processing activity, strong activity was detected after apoptosis induction (Fig. 1 B, a and b, respectively). Furthermore, coexpression of the apoptosome components Ark and Dronc, using the sal-Gal4 driver, also induced a high level of CPV processing activity (Fig. 1 B, c), consistent with the idea that this combination is a strong inducer of apoptosis (Shapiro et al., 2008).
Figure 1.
Detection of effector caspase activity in vivo. (A) A schematic representation of the CPV reporter of effector caspase activity. (B) CPV is a specific reporter of caspase-3–like (DEVDase) activity during apoptosis in vivo. The sal-Gal4 driver was used for expression of the UAS-dependent transgenes in the pouch region of the WD. WDs were dissected and stained with the anticleaved human PARP antibody (top row). The bottom row displays the fluorescence of the CPV reporter (Venus) in the corresponding discs. Third instar larvae containing the CPV reporter (a–c) or the mutant CPGV uncleavable control reporter (d) were either nonirradiated (nonirrad; a and c) or subjected to 50 Gy of γ-irradiation and allowed a 4-h recovery (b and d). (c) Coexpression of Dronc and Ark in the pouch region of the WD. The open boxes group images of the same disc detected in different fluorescent channels. Bar, 50 µm.
Apoptosis is associated with massive proteolytic activity (Crawford and Wells, 2011). To validate that CPV is cleaved only (immediately) after the DEVD consensus site, we introduced a point mutation to this reporter, which changes the invariably conserved aspartic acid residue to glycine (DEVD to DEVG). Transgenic flies expressing this modified construct (dubbed CPGV) were treated as described above for the visualization of CPV processing activity. However, this mutation completely abrogated processing of the reporter (Fig. 1 B, d). Therefore, the proteolytic cleavage of CPV during apoptosis reflects DEVDase activity only.
The effector caspases Drice and Dcp-1 are both activated during γ-irradiation–induced apoptosis
Apoptosis is mainly mediated by caspases, but other proteases may also be involved in this process (Schrader et al., 2010). To test whether CPV is cleaved by effector caspases during apoptosis, the effector caspase inhibitor baculovirus gene p35 and the Drosophila caspase inhibitor gene diap1 were each expressed in the pouch region of WDs. After irradiation, CPV processing activity was completely blocked in the presence of p35 or largely suppressed in the presence of Diap1, suggesting that effector caspases are indeed responsible for this activity (Fig. 2, A [a–c] and B). The residual activity in the Diap1-expressing WDs is attributed to the relatively short half-life of this protein (t1/2 = 30 min; Holley et al., 2002).
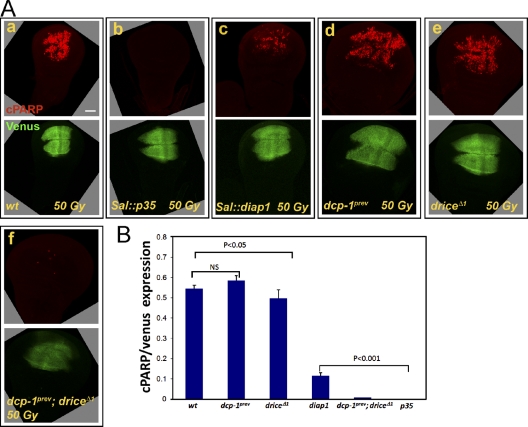
Figure 2.
Drice and Dcp-1 are both activated after irradiation-induced apoptosis. (A) CPV-expressing larvae of the indicated genotypes were treated as in Fig. 1 B. CPV processing activity was detected in wild-type (wt; a) and also in dcp-1 (d) and drice (e) mutants and was blocked upon expression of p35 (b) or in the dcp-1 and drice double mutants (f) or largely attenuated by expression of diap-1 (c). The open boxes group images of the same disc detected in different fluorescent channels. Bar, 50 µm. (B) Quantification of the relative caspase activity levels in the WDs described in A. Bars represent the mean cPARP staining area/reporter expression domain (represented by the Venus expression region). Error bars represent standard errors. P-values of the activity differences between various genotypes are indicated above the bars. All p-values were calculated using Fisher’s exact test.
Out of the four structurally related effector-like caspases in Drosophila, only Drice and Dcp-1 have been previously associated with apoptosis in vivo, although only drice mutants were shown to block irradiation-induced apoptosis in the WD (Muro et al., 2006; Xu et al., 2006). Therefore, we used a null allele of drice (driceΔ1; Fig. S1; see Fig. 6 E) to examine whether it is the only effector caspase responsible for the CPV processing activity in irradiated WDs. Unexpectedly, CPV processing activity persisted in WDs from irradiated drice mutants, suggesting the involvement of another effector caspase (Fig. 2, A [e] and B). Indeed, whereas a protein-null allele of dcp-1 (dcp-1prev; see Fig. 6 E) also failed to block the CPV processing activity in this system, this activity was almost completely abrogated in the double mutants (Fig. 2, A [d and f] and B). Therefore, Drice and Dcp-1 are both activated in WDs after irradiation-induced apoptosis. Notably, quantification of the processing activity in each of the mutants suggests that Dcp-1 activity is slightly lower than that of Drice (Fig. 2 B). However, comparative analysis of transcript expression by real-time quantitative PCR (RT-qPCR) indicates that both caspases are expressed at comparable levels in this tissue, suggesting that the Dcp-1 activity in this system may be less efficient at cleaving this reporter (Fig. S1).
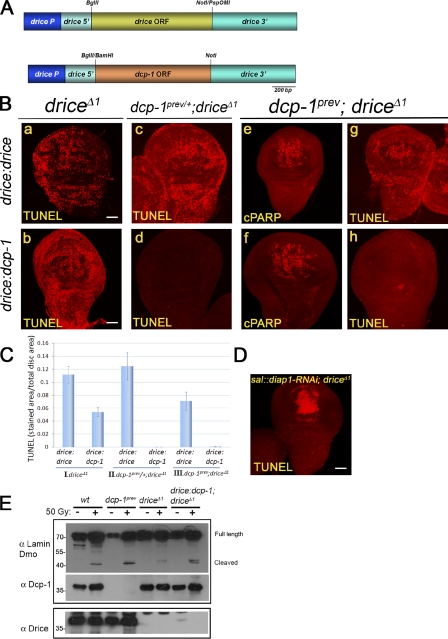
Figure 6.
Distinct execution efficiencies of Drice and Dcp-1. (A) A schematic structure of the constructs drice:drice (top) and drice:dcp-1 (bottom). (B) WDs from drice mutants (a and b), drice mutants that also carry one dcp-1 mutant allele (c and d), or drice and dcp-1 double mutant flies (e–h), which express either the transgenic Drice (a, c, e, and g) or transgenic Dcp-1 constructs (b, d, f, and h) under the promoter of drice and from the same genomic locus were either TUNEL labeled (a–d, g, and h) or stained to visualize the CPV processing activity (e and f). Flies were treated as in Fig. 1 B. Bars, 50 µm. (C) Quantification of the relative levels of cell death in WDs from transgenic flies expressing the Drice or Dcp-1 transgenes described in A and B in either of the following mutant backgrounds: drice homozygous mutants (I), drice homozygous and dcp-1 heterozygous mutants (II), and drice and dcp-1 double homozygous mutants (III). All calculations were performed as in Fig. 5 B. Error bars represent standard errors. (D) A representative image of a WD from drice mutants, which also expresses a diap1-targeting RNAi transgene in the pouch region. Flies were treated as in Fig. 1 B. Bar, 50 µm. (E) Drice and Dcp-1 display distinct efficiencies in cleaving lamin Dmo in vivo. Cleavage of the lamin Dmo protein was assessed by Western blotting of protein extracts from dissected WDs of the indicated genotypes using the anti–lamin Dmo antibody (top). The same membrane was reblotted with the anti–Dcp-1 antibody (middle). A similar membrane was blotted with anti-Drice antibody (bottom). Samples were either nonirradiated (−) or irradiated by a dose of 50 Gy and allowed to recover for 5–7 h (+). wt, wild type. Molecular mass is indicated in kilodaltons.
The apoptosome components Ark and Dronc are exclusively required for Drice and Dcp-1 activation after irradiation-induced apoptosis
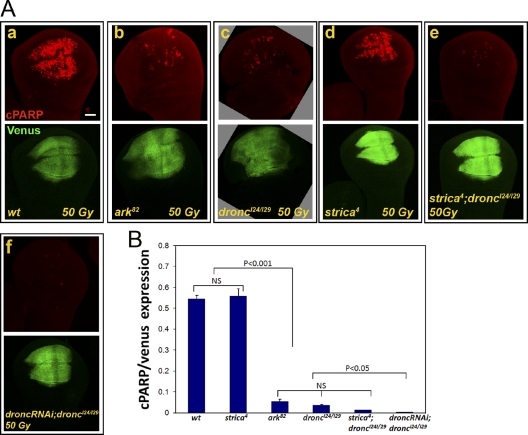
The apoptosome components Ark and Dronc are associated with most of the developmental and stress-induced apoptotic events in Drosophila. To examine whether these apoptosome components are also required for Drice and Dcp-1 activation in our system, CPV processing activity was monitored in WDs from ark82 and droncI24/I29 mutant flies. Indeed, most of the activity was strongly attenuated in these backgrounds, although some residual levels of activity remained (Fig. 3, A [a–c] and B). As both are null allelic backgrounds for their respective mutations, we asked whether this residual activity might be mediated by the initiator-like atypical caspase Strica, which has been suggested to be functionally redundant with Dronc in certain cellular paradigms (Baum et al., 2007). However, no significant attenuation of the activity was detected in the strica-null mutant (Fig. 3, A [d] and B). Moreover, WDs from dronc and strica double mutants exhibited similar levels of residual activity as those in the dronc mutant alone (Fig. 3, A [e] and B). An alternative possibility for the retained residual activity in the dronc and ark mutant backgrounds may be the persistence of maternal mRNA of these genes. Indeed, CPV processing activity was almost completely abrogated in WDs from droncI24/I29 mutants, which also express an RNAi construct against dronc (Fig. 3, A [f] and B). Therefore, the Ark-Dronc apoptosome is exclusively required for Drice and Dcp-1 activation after irradiation-induced apoptosis.
Figure 3.
The Ark-Dronc apoptosome is exclusively required for effector caspase activation after irradiation-induced apoptosis. (A) CPV-expressing larvae of the indicated genotypes were treated as in Fig. 1 B, and the figure panels are presented accordingly. WDs from wild-type (wt; a), ark (b), dronc (c), and strica (d)-null mutants, dronc and strica double mutants (e), and dronc mutants that also express an RNAi against dronc (f) are shown. The open boxes group images of the same disc detected in different fluorescent channels. Bar, 50 µm. (B) Quantification of the relative caspase activity levels in WDs described in A. All calculations were performed as in Fig. 2 B. Error bars represent standard errors.
Dcp-1 activity is insufficient to induce apoptosis in WDs after irradiation
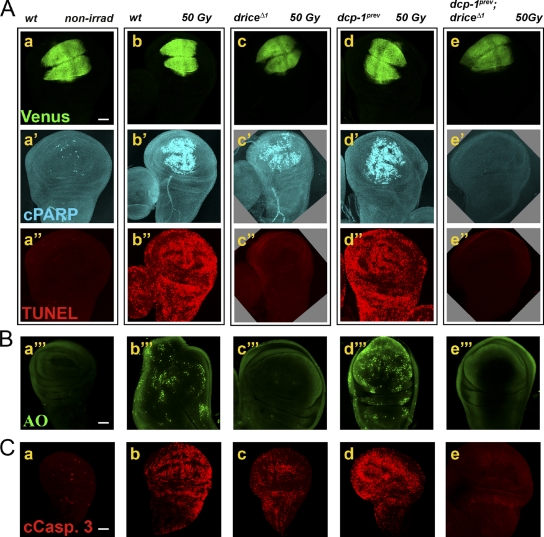
Previous works have suggested that irradiation-induced apoptosis in WDs is blocked in drice mutants (Muro et al., 2006; Xu et al., 2006). Therefore, we asked whether the Dcp-1 activity detected in WDs after irradiation is dispensable for cell death. To address this, WDs were first costained with anticleaved PARP (anti-cPARP) and TUNEL to visualize CPV processing activity and dying cells, respectively. In addition, discs of the same genotypes were also labeled with the vital dye acridine orange (AO), which is known to detect apoptotic corpses in Drosophila (Arama and Steller, 2006). 4 h after 50 Gray (Gy) irradiation, effector caspase activity and cell death were dramatically induced in WDs from wild-type and dcp-1 mutants (Fig. 4, A and B, a–a′′′, b–b′′′, and d–d′′′). In contrast, cell death was not induced in irradiated discs from drice mutants or drice and dcp-1 double mutants, although, as aforementioned, Dcp-1 activity could be readily detected in the drice (single) mutant (Fig. 4, A and B, c–c′′′ and e′–e′′′). Therefore, in the absence of Drice, Dcp-1 activity in WDs is insufficient to induce apoptosis after irradiation.
Figure 4.
Inactivation of Drice, but not Dcp-1, completely blocks irradiation-induced cell death. (A) Visualization of the CPV expression domain (Venus), effector caspase activity (CPV processing), and cell death (TUNEL) in WDs from larvae treated as in Fig. 1 B. The specific genotypes indicated at the top relate to all the panels in a column. The first three columns are of the same imaginal disc. Note that caspase activity is confined to the reporter expression area, whereas cell death is detected in the entire WD. The open boxes group images of the same disc detected in different fluorescent channels. nonirrad, nonirradiated; wt, wild type. (B) WDs from irradiated larvae of the same genotypes as in A were stained with the vital dye AO to detect apoptotic corpses. (C) Similar WDs treated as in A and B and stained with the anticleaved caspase-3 antibody (cCasp. 3; lot no. 32; Cell Signaling Technology) to visualize the active forms of the effector caspases. Bars, 50 µm.
Because these experiments are performed by expressing relatively high levels of an artificial caspase substrate (the CPV reporter) within a large area of the WD, one concern is that this reporter may function as a competitive inhibitor of caspases and thus affect cell death. Therefore, we quantified and compared the levels of cell death induced in WDs from flies with the UAS-CPV construct alone (control) or flies containing the driver construct sal-Gal4 and the UAS-CPV. Importantly, the rates of cell death were almost identical between the control and CPV-expressing WDs at both 2 and 4 h after 50 Gy irradiation, suggesting that at least under these conditions, this reporter does not significantly affect the processing of endogenous death-associated substrates by caspases (Fig. S2).
It has been recently suggested that the anticleaved caspase-3 antibody (also known as CM1) may reflect Dronc activity in Drosophila, but the specific cleaved caspases identified by this antibody have not been determined (Fan and Bergmann, 2010). Furthermore, it has remained controversial whether this antibody can detect active Dcp-1 in vivo (Yu et al., 2002; Peterson et al., 2003; Muro et al., 2006). As the anticleaved caspase-3 antibody is widely used as a marker of caspase activation and apoptosis in Drosophila, we decided to examine four antibodies from two different sources raised against the same epitope (see also in Materials and methods). Interestingly, staining WDs with these antibodies revealed that they can recognize both active Drice (Fig. 4 C, d) and active Dcp-1 (Fig. 4 C, c) in vivo, and this staining was completely absent in the double mutants (Fig. 4 C, e). Nevertheless, the specificity of these antibodies toward active Dcp-1 varied among the different antibodies, which in all cases were much more specific toward active Drice (Fig. S3).
Dcp-1 activity fine tunes the rate of cell death after irradiation
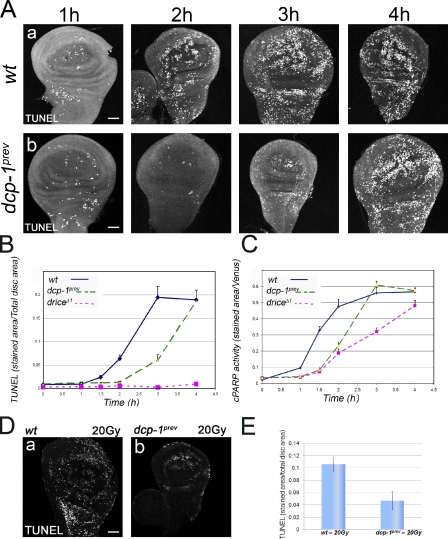
The idea that endogenous Dcp-1 activity cannot induce cell death in WDs after irradiation coupled with the fact that Drice activity alone is sufficient to induce apoptosis under the same conditions raised the question about the role of Dcp-1 in this system. One possibility is that Dcp-1 activity increases the total levels of effector activity in the cell, thus making cell death more efficient. To investigate this possibility, we quantified the levels of cell death in WDs from wild-type and dcp-1 and drice mutants at different time points after 50 Gy irradiation. In wild type, the number of dying cells started to accumulate ∼1.5 h after irradiation; this number reached its peak after 3 h and plateaued in the following hours (Fig. 5, A [a] and B). Interestingly, although the drice mutants displayed the expected complete block in cell death, the dcp-1 mutants displayed a significant delay of ∼1 h in the rate of cell death, reaching the wild-type plateau level 4 h after irradiation (Fig. 5, A [b] and B). To examine the correlation between the rates of cell death and caspase activity, we then quantified the levels of the CPV processing activities in these WDs. In accordance with the measured rates of cell death, both caspases became activated at ∼1.5 h after irradiation (Fig. 5 C). However, whereas Drice activity reached a peak and a plateau after 3 h, Dcp-1 activity reached that same plateau level ∼4 h after irradiation, suggesting that Dcp-1 may be less efficient at cleaving the CPV reporter than Drice or that Drice is more efficiently activated than Dcp-1 (Fig. 5 C). We conclude that whereas Drice activity is the trigger of cell death in this system, Dcp-1 activity fine tunes the rate of this process.
Figure 5.
dcp-1 mutants display reduced cell death rate. (A) Visualization of cell death by TUNEL labeling in WDs from wild-type (wt; a) and dcp-1 (b) mutants at the indicated time points after 50 Gy irradiation. Bars, 50 µm. (B) Quantification of the relative levels of cell death in WDs from wild-type, dcp-1, and drice mutants treated as in A. Cell death levels are calculated as the TUNEL-labeled area/total imaginal disc area. (C) Quantification of the relative caspase activity levels in WDs described in B. All calculations were performed as in Fig. 2 B. (D) Visualization of cell death by TUNEL labeling in WDs from wild-type (a) and dcp-1 (b) mutants at 4 h after 20 Gy irradiation. Bar, 50 µm. (E) Quantification of the relative levels of cell death in the WDs described in D. Cell death levels are calculated as in Fig. 2 B. (B, C, and E) Error bars represent standard errors.
One way by which Dcp-1 may contribute to the acceleration of Drice-triggered apoptosis may be through a positive amplification loop. According to this model, after apoptosis induction, the Ark-Dronc apoptosome activates Drice and Dcp-1, and, in turn, these effector caspases cleave death-associated substrates, whereas Dcp-1 may also cleave and further activate Drice. In support of this model, it has been previously shown that Dcp-1 can cleave Drice in a cell-free system, but not vice versa, similar to the way Dronc cleaves Drice (Hawkins et al., 2000; Song et al., 2000). To test this possibility in vivo, we performed Western blot analysis on protein extracts from wild-type, drice, and dcp-1 mutant adult flies using an anti-Drice antibody. Basal cleavage of Drice is detected in wild-type flies but not in the dcp-1 mutants, suggesting that Dcp-1 may promote cleavage of Drice also in vivo, albeit the significance of this cleavage (nonclassical p10 and p20) is unclear (Fig. S4 A). To test whether Dcp-1 may indeed promote amplification of Drice activation in vivo, we used the anticleaved caspase-3 antibody to quantify the levels of active Drice in WDs from wild-type and dcp-1 mutants at different time points after 50 Gy irradiation. Interestingly, a significant delay of ∼1 h is detected in the rate of Drice activation in the dcp-1 mutant versus wild type (Fig. S4 B). As the anticleaved caspase-3 antibody mostly detects active Drice in vivo (Fig. S3), this delay is mainly attributed to the lack of Drice activation by Dcp-1. Collectively, these findings provide evidence that Dcp-1 may function, at least in part, through activating Drice in a positive amplification loop.
The finding that Dcp-1 activity makes cell death more efficient prompted us to explore other conditions in which Drice activity alone may be suboptimal. By gradually decreasing the dose of γ-irradiation, the rate of cell death was reduced accordingly. At 4 h after an irradiation dose of 20 Gy, the rate of cell death in the wild type was reduced to about half that observed after a dose of 50 Gy (compare D [a] and E with B in Fig. 5). Under these conditions, the rate of cell death in the dcp-1 mutants (i.e., Drice activity only) was further reduced by 50% compared with wild type (Fig. 5, D and E). In contrast, the level of Drice activity alone at 4 h after 50 Gy irradiation could induce a similar level of cell death as that in wild type (albeit in a delay; Fig. 5 B). Therefore, the role of Dcp-1 activity in tuning the rate of cell death may be more significant when facing relatively weaker stresses.
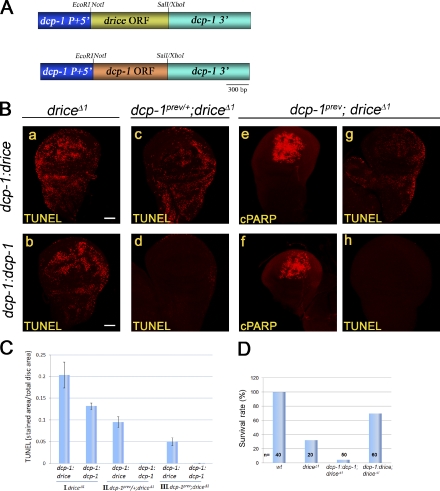
Dcp-1 can induce cell death but less efficiently than Drice
The fact that Dcp-1 activity cannot induce apoptosis in irradiated WDs raised the question of whether it is capable of inducing cell death at all. To answer this question, we generated transgenic fly lines expressing either the full-length drice or dcp-1 genes at identical levels. For this, the complete ORFs of drice and dcp-1 were each subcloned under the same regulatory regions (promoter 5′ and 3′ untranslated regions [UTRs]) of the drice gene, generating the drice:drice and drice:dcp-1 constructs, respectively (Fig. 6 A). Each construct was then targeted to the same genomic point using the ϕC31 integrase system, thus ensuring identical expression levels of these transgenes. By crossing these transgenes to the drice and dcp-1 double mutant, we validated that both can induce CPV processing activity after irradiation (Fig. 6 B, e and f). Next, we asked whether these transgenes could restore irradiation-induced cell death in the drice mutants (where Dcp-1 is the only endogenous source of effector activity), in drice mutants that also lack one allele of dcp-1, and in the double mutants. Interestingly, under these conditions, both transgenes were able to induce cell death in the drice mutants, as detected by TUNEL labeling (Fig. 6 B, a and b), but only drice:drice could induce cell death in the drice mutants that are also heterozygous for the dcp-1 allele (Fig. 6 B, c and d) and in the double mutants (Fig. 6 B, g and h). Of note, the same transgenes were also able to reverse the lethality of the drice mutant flies (see in the text for Fig. 7), indicating that the TUNEL-labeled WD cells are bona fide dying cells. Therefore, Dcp-1 has the potential to induce cell death by itself, but it is less efficient in doing so than Drice.
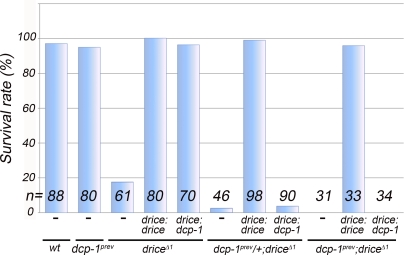
Figure 7.
Drice is a more efficient executioner than Dcp-1 during developmental cell death. Quantification of the survival rates from larva to adult after expression of the drice:drice or drice:dcp-1 transgenes in the following mutant combinations of drice and dcp-1: dcp-1prev, driceΔ1, dcp-1prev/+; driceΔ1, and dcp-1prev; driceΔ1. The survival rate is expressed as the percentage of third instar larvae that survived to adulthood. Data are derived from a single prolonged experiment. wt, wild type.
Procaspase levels and ratios affect cellular sensitivity to apoptosis by determining whether and at what rate caspase activity levels may reach a critical execution threshold
Next, we assessed the differences in the abilities of the two caspases to induce apoptosis by quantifying the levels of cell death induced by each transgene. Consistent with the idea that Drice is more efficient in inducing apoptosis than Dcp-1, drice:drice induced more than twice as many cell death events than drice:dcp-1 in the background of the drice mutant (Fig. 6 C, I). Further reduction of one or two copies of endogenous dcp-1 in this mutant abolished the ability of drice:dcp-1, but not drice:drice, to induce cell death, implying the existence of a threshold effect (Fig. 6 C, II). Finally, in the double mutant background, only drice:drice could induce cell death but in a more reduced rate than in the single drice mutant (Fig. 6 C, III).
In living cells, Diap1 binds to and inhibits the activation of both Drice and Dcp-1, whereas upon induction of apoptosis, Reaper family proteins bind to Diap1, relieving its inhibition of caspases (Ditzel et al., 2008; Steller, 2008). Therefore, we hypothesized that reducing the level of diap1 may lower the execution threshold of the cell. To test this, a diap1-RNAi transgene was expressed in the pouch region of the WD in the background of the drice mutant. After larval irradiation, these discs displayed dramatic induction of cell death in the pouch region but not in the surrounding tissue (Fig. 6 D). Therefore, reducing the levels of diap1 elevates the levels of the activated Dcp-1 in the cell, leading to apoptosis. Finally, to test the possibility that Diap1 may be a stronger inhibitor of Dcp-1 than Drice, the diap1-RNAi transgene was expressed in the pouch region of WDs in either the drice or dcp-1 mutants, and the WDs were tested for cell death induction as a result of spontaneous activation of the caspases (without irradiation). However, cell death was only detected in the pouch area of the dcp-1 mutant but not the drice mutant, which is consistent with the idea that the differences in the execution efficiencies of Drice and Dcp-1 are not the consequence of differential inhibition of these caspases by Diap1 (Fig. S5). Collectively, these results demonstrate that the levels of the proeffector caspases can tip the scales between cell survival and death and that it must go over a critical threshold to induce apoptosis.
Drice and Dcp-1 display distinct efficiencies in cleaving a protein substrate in vivo
A plausible reason for the relatively low efficiency of Dcp-1 to induce apoptosis is that it may cleave death-associated substrates less effectively than Drice. One such in vitro target of Drice and Dcp-1 is the Drosophila B-type lamin, lamin Dmo (Fraser and Evan, 1997; Song et al., 2000). To test whether Drice and Dcp-1 can also cleave this protein in vivo, Western blot analysis was performed on extracts prepared from wild-type, dcp-1, and drice mutant WDs both before and 5–7 h after irradiation. Using a specific antibody for lamin Dmo, cleavage of this protein was detected in all of the examined genotypes after irradiation, although this activity was dramatically reduced when drice was inactive (i.e., Dcp-1 activity only; Fig. 6 E). Furthermore, in accordance with the ability of transgenic Dcp-1 to rescue cell death after irradiation in the drice mutant, expression of transgenic Dcp-1 (on top of the endogenous Dcp-1 in the drice mutant) also increased the cleavage of lamin Dmo (Fig. 6 E). Therefore, Dcp-1 is less efficient than Drice at cleaving both the artificial CPV reporter and the death-associated substrate lamin Dmo in vivo.
Increasing Dcp-1 levels can compensate for the loss of drice during developmental cell death
Cell death plays a major role during metamorphosis of the fly by removing many larval structures, thus paving the way for the generation of new adult tissues. Indeed, whereas drice mutants survive to the third instar larval stage, most of them die during pupal development (Muro et al., 2006; Xu et al., 2006). As transgenic Dcp-1 could restore irradiation-induced cell death in the drice mutants (albeit in a reduced rate than transgenic Drice), we asked whether it can also compensate for the loss of Drice during pupal development. Therefore, we evaluated the ability of these transgenes to increase the rate of larva-to–adult fly survival in different combinations of drice and dcp-1 mutants. As shown in Figs. 7 and 8 C, only ∼20–30% of the drice mutants survived to adulthood, whereas the survival rate of the dcp-1 mutants was similar to that of wild type. Further loss of one or two copies of endogenous dcp-1 in the background of the drice mutants significantly reduced the survival rate or caused complete pupal lethality, respectively (Fig. 7). Importantly, all of the allelic combinations that restored irradiation-induced cell death, such as the transgenic Drice or Dcp-1 in the drice mutant background or transgenic Drice in the double mutants (Fig. 6 C), could also reverse pupal lethality and restore wild-type levels of survival, irrespective of the differences in the rates of cell death (Fig. 7). On the other hand, inability to restore irradiation-induced cell death (e.g., drice:dcp-1 in the double mutant background; Fig. 6 C) fully correlated with a failure of these transgenes to reverse fly lethality (Fig. 7). Therefore, Drice is also a more effective executioner caspase than Dcp-1 during developmental cell death.
Figure 8.
Rescue of cell death and survival by using the dcp-1 promoter are consistent with distinct efficiencies of the effector caspases and a threshold effect. (A) A schematic structure of the constructs dcp-1:drice (top) and dcp-1:dcp-1 (bottom). (B) WDs from drice mutants (a and b), drice mutants that also carry one dcp-1 mutant allele (c and d), or drice and dcp-1 double mutant flies (e–h), which express either the transgenic Drice (a, c, e, and g) or transgenic Dcp-1 constructs (b, d, f, and h) under the promoter of dcp-1 and from the same genomic loci were either TUNEL labeled (a–d, g, and h) or stained to visualize the CPV processing activity (e and f). Flies were treated as in Fig. 1 B. Bars, 50 µm. (C) Quantification of the relative levels of cell death in WDs from transgenic flies expressing the Drice or Dcp-1 transgenes described in A and B in either of the following mutant backgrounds: drice homozygous mutants (I), drice homozygous and dcp-1 heterozygous mutants (II), and drice and dcp-1 double homozygous mutants (III). All calculations were performed as in Fig. 5 B. Note that the transgenes in (a and b) were inserted into a similar genomic locus as those described in Fig. 6 (B and C) but a different genomic locus than the transgenes in c–g. Error bars represent standard errors. (D) Quantification of the survival rates from larva to adult after expression of the dcp-1:drice or dcp-1:dcp-1 transgenes in the background of drice homozygous mutants. Calculations were performed as in Fig. 7. Data are derived from a single prolonged experiment. wt, wild type.
Distinct tissue specificities of Drice and Dcp-1 also affect their differential requirements during developmental cell death
RT-qPCR analysis (Fig. S1) and microarray data and EST tissue information deposited in public domains suggested that drice expression levels are much higher than dcp-1 levels in almost all tissues investigated (unpublished data). To investigate the contribution of tissue specificities to the differences between the roles of the two caspases during developmental cell death, we generated new transgenes that contain the coding regions of drice and dcp-1, this time under the regulatory elements of dcp-1 (Fig. 8 A). Similar to the drice:drice and drice:dcp-1 transgenes and in agreement with the findings that the two caspases are expressed at similar levels in WDs (Fig. S1), the dcp-1:drice and dcp-1:dcp-1 transgenes were also able to restore irradiation-induced cell death in WDs of the drice mutants, with dcp-1:drice being more effective than dcp-1:dcp-1 (Fig. 8, B [a and b] and C). Furthermore, this similarity is extended to the double mutant backgrounds, showing that the dcp-1:drice transgene, but not the dcp-1:dcp-1, could restore irradiation-induced cell death in drice mutant flies that also contain either one or two copies of the dcp-1 mutant allele (Fig. 8, B [c–h] and C). Therefore, the fact that a different promoter gives almost identical results reinforces our findings that Dcp-1 is a less efficient inducer of cell death than Drice and further demonstrates the existence of a threshold effect.
However, as opposed to drice:drice and drice:dcp-1 transgenes, the dcp-1:drice transgene only partially rescued pupal lethality of the drice mutants, whereas the dcp-1:dcp-1 transgene failed to rescue this lethality and instead further aggravated this phenotype (compare Fig. 8 D with Fig. 7). These results suggest that dcp-1 is either not expressed or expressed at very low levels in at least some of the tissues where Drice activity is required during pupal development. In addition, Dcp-1 may also be uniquely expressed in some other tissues where high Dcp-1 activity, but not Drice activity, is deleterious to the cells. Altogether, these results suggest that the differential requirement for Drice and Dcp-1 during developmental cell death is also the consequence of tissue specificities.
Discussion
The threshold effect of the effector caspase activity
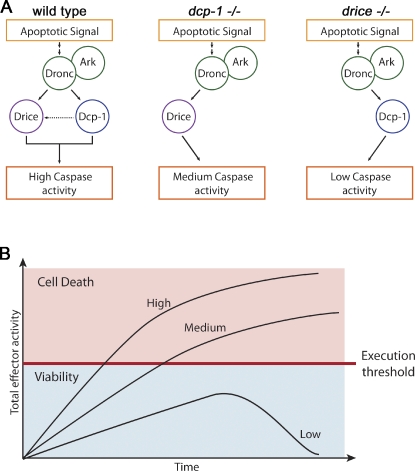
One of the most important questions in the fields of developmental apoptosis and cancer biology is what makes different cells more resistant or more sensitive to apoptotic stimuli. In the present study, we uncovered two factors important for establishing the overall apoptotic potential of the cell: the levels of the proeffector caspases and their execution efficiencies. We showed that the Drosophila effector caspases Drice and Dcp-1 differ in their competencies to induce apoptosis in vivo, with Drice being more effective than Dcp-1 (also illustrated in Fig. 9 A). Comparative transgenesis experiments indicate that these differences are the consequence of distinct intrinsic properties of the two caspases. Importantly, these experiments further demonstrate that cellular lethality is exerted only when caspase activity reaches a critical threshold. Short of that lethal level, cells fail to induce apoptosis, whereas surpassing that level increases the rate of cell death, thus further sensitizing the cells to apoptosis (see the model in Fig. 9 B and also below).
Figure 9.
Effector caspase activity and the cell apoptotic potential. (A) A schematic representation of the relative effector activity in wild-type and dcp-1 and drice mutants. A possible synergistic effect whereby Dcp-1 further activates Drice in a positive amplification loop is indicated with a dashed arrow. (B) Representative graphs of the corresponding different activities in A and their relations to the execution threshold point (red bar). When the effector activity is below that bar (viability zone; blue), cells can overcome the destructive activity of the caspases and survive, whereas above that bar (cell death zone; pink), cells commit to apoptosis, and the higher the level of effector activity, the more elevated the rate of cell death.
The levels of the proeffector caspases affect the sensitivity of the cell to apoptotic stimuli
Our experiments demonstrate that the level of activity of an effector caspase is directly proportionate to the level of expression of the respective inactive zymogene. Therefore, in addition to the established posttranslational regulatory mechanisms of caspases, which are crucial for the initiation of apoptosis, the expression levels of the proeffector caspases are an important factor in determining the sensitivity of the cells to apoptotic stimuli. This sensitivity is reflected in the rate by which the total level of effector activity meets the execution threshold of the cell. The higher the levels of the procaspases, the faster the cells reach that execution threshold level and the more sensitive the cells are to apoptotic stimuli (illustrated in Fig. 9 B). However, if the total levels of procaspases are too low, the activity rate may fail to reach that particular threshold, likely because the metabolic processes that normally sustain the cell overwhelm the catabolic processes, allowing for recovery and survival (Fig. 9 B). Interestingly, in an extreme case of apoptosis regulation at the level of procaspase expression, the onset of cell death in the tail-spiked cell of Caenorhabditis elegans was reported to be induced by an increase in the expression level of the caspase-encoding gene ced-3 (Maurer et al., 2007); however, whether or not this is caused by a threshold effect is still unclear.
How does the level of effector caspase activity affect the rate of cell death? The length of the time between the apoptotic stimulus and the point when the cell becomes apoptotic varies between different cell types and distinct apoptotic stimuli. It is believed that this time period mainly reflects the time it takes for the cells to process and transduce the apoptotic signal and eventually translate it to the action of the caspases (Pluquet and Hainaut, 2001; Hellwig et al., 2008). However, our work suggests that the levels of effector caspase activity also vary between different cells as a consequence of the levels and ratios of the two main proeffector caspases, and this factor plays an important role in determining the time length between the activation of caspases and the eventual death of the cell, where high levels of effector activity shorten that period, thus increasing the rate of cell death. A plausible explanation for this correlation is that in order for the cell to start dying, a critical mass of death-associated substrates must be cleaved by the effector caspases, such that the more caspase molecules available, the faster this mass is cleaved. To date, ∼1,000 cellular proteins are thought to be cleaved by effector caspases during apoptosis, many of which are believed to gain old or new activities (Lüthi and Martin, 2007; Dix et al., 2008; Mahrus et al., 2008). However, it is still unknown what caspase substrates are critical in apoptosis and what levels of cleaved substrates are required for induction of apoptosis. Future approaches to study that critical subset may further help in understanding how precisely the level of caspase activity affects the rate of cell death.
The Drosophila effector caspases differ in their abilities to induce apoptosis in vivo
We have presented several lines of evidence demonstrating that Drice is more efficient in inducing apoptosis than Dcp-1. First, despite the fact that both Drice and Dcp-1 are normally expressed at similar levels in the WD, only Drice activity could induce apoptosis after irradiation. Second, although when expressed under the drice regulatory regions both of the procaspase transgenes could restore irradiation-induced apoptosis and survival of the drice mutants, the rate of cell death was doubled with the Drice transgene. Moreover, only the Drice transgene could rescue irradiation-induced cell death and survival of the double mutants. Finally, similar differences in the efficiencies of the two caspases to induce apoptosis after irradiation were also observed when the procaspase transgenes were expressed under the dcp-1 regulatory regions.
What are the reasons for the differences in the in vivo execution efficiencies of Drice and Dcp-1? One idea is that Diap1 may inhibit Dcp-1 more efficiently than Drice. However, this possibility is unlikely, as reduction of the same endogenous levels of Diap1 led to spontaneous induction of cell death only in the dcp-1 mutant and not the drice mutant, demonstrating that despite similar levels of Diap1-free endogenous caspases, only activated Drice has the ability to induce apoptosis in this setup. Another possibility is that Dronc may cleave and activate Drice more efficiently than Dcp-1. In support of this idea, recombinant Dronc was reported to only poorly process Dcp-1, as compared with Drice, in a cell-free system (Hawkins et al., 2000). Consistently, cleavage of the endogenous Drice was more prominent than that of endogenous Dcp-1 in irradiated WDs (unpublished data). However, an alternative or additional mechanism is that Drice may be more efficient than Dcp-1 at cleaving some of the more critical death-associated substrates. Indeed, Drice was able to process both the CPV reporter and an endogenous death-associated substrate, lamin Dmo, more efficiently than Dcp-1 in irradiated WDs. Whereas both possibilities support a model in which Drice and Dcp-1 function in a similar manner to induce apoptosis (i.e., possessing overlapping repertoires of death-associated protein substrates) but with different efficiencies, our data additionally suggest that these caspases may also have some distinct protein substrates. The findings that when expressed under the regulatory regions of dcp-1 transgenic Drice partially rescued the survival of the drice mutants while the Dcp-1 transgene further increased their lethality imply that Dcp-1 may also cleave some distinct protein substrates. This is further supported by the notion that Dcp-1 can cleave Drice, whereas Drice cannot cleave Drice or Dcp-1 (see also below). Therefore, Drice and Dcp-1 may differ in their activation efficiencies by Dronc, their ability to effectively cleave similar critical death-associated substrates, and their repertoires of distinct substrates.
The coarse and fine modes of effector activity control
Both Drosophila and mammals possess major and minor effector caspases. Our work suggests that having both coarse (Drice; major) and fine (Dcp-1; minor) modes of effector activity control may allow the cells more flexibility in determining their apoptotic potential by setting their optimal execution threshold and rate of cell death. The finding that the rate of cell death in the absence of dcp-1 was dramatically reduced after a relatively weak stress (i.e., 20-Gy irradiation) as compared with a stronger stress (i.e., a 50-Gy dose) suggests that under conditions resembling the normal situation in the wild, the minor caspase plays a more prominent role in determining whether or not the effector activity will even meet the execution threshold. In higher stresses, however, there is no need for a cell to rely on Dcp-1 activity to reach the execution threshold, as its activity is obviated by the high activity of Drice. Interestingly, in addition to ced-3, which is essential for apoptosis in C. elegans (Yuan et al., 1993), the worm genome also contains other caspase genes, but no documented roles in apoptosis have been thus far assigned for these caspases (Shaham, 1998). Likewise, Drosophila and mammals also contain additional caspases, which do not appear to have a major role in apoptosis (Salvesen and Abrams, 2004). Therefore, it is attractive to consider that similar to Dcp-1, at least some of the additional caspases in these organisms may also function to fine tune the rate of cell death, providing maybe even a finer tuner.
Our RT-qPCR analysis suggests that the WD is an exceptional tissue in relation to the similar levels of dcp-1 and drice expression, as microarray data and EST tissue information deposited in public domains (FlyAtlas and FlyBase, respectively) suggest that drice expression levels are much higher than dcp-1 levels in almost all tissues examined (unpublished data). Thus, fine tuning the rate of cell death may be the main role of Dcp-1 in apoptosis. However, this is probably not the only role of Dcp-1, as this caspase appears to have more prominent roles in certain cellular paradigms. For example, Dcp-1 was reported to be essential for the sporadic and nutrient deprivation–induced germline cell death during midoogenesis, with Drice playing only a minor role, if any, in this system (Laundrie et al., 2003; Baum et al., 2007). Furthermore, genetic analyses revealed additional cellular paradigms in which the role of Dcp-1 in cell death is more prominent or that Dcp-1 inactivation significantly enhances drice mutant phenotypes (Leulier et al., 2006; Muro et al., 2006; Xu et al., 2006; Baum et al., 2007). Although it is largely unknown why different cells/tissues need to acquire distinct apoptotic potentials, it is not hard to envision that certain cells may need to be more resistant or more sensitive to apoptosis. For instance, cell types with low regeneration potential and with limited numbers, such as neurons, or cells with direct interface with the outside world and that are in higher risk of facing different stresses (e.g., eyes or sperm) may be more resistant to apoptosis and hence may express relatively low levels of Drice. On the other hand, dying cells that send signals to the surrounding tissue area (e.g., signals for compensatory proliferation) may need to express higher levels of Dcp-1 to fine tune their dying rate and thus control the level of the signal they send. Therefore, different cells in the organism may use distinct levels and ratios of Drice and Dcp-1 to optimize their sensitivity to apoptosis induction.
Drice and Dcp-1 synergize their activities during apoptosis
Our data also point to a synergistic effect of Drice and Dcp-1 on the overall effector activity levels in the cell. This effect is clearly detected when comparing the overall level of effector caspase activity in wild type (which reflects the combined activities of Drice and Dcp-1) and the sum of the single activity levels of Drice and Dcp-1 (measured in dcp-1 and drice mutant backgrounds, respectively). During the time window of 1–2 h after irradiation, wild-type activity levels were significantly higher than the simple summation of the levels in the mutants, implying the existence of a positive amplification loop of effector caspase activation. In a later window of time 3–4 h after irradiation, activity levels in wild type reached a peak and plateaued (Fig. 5 C). Because the levels of Drice activity alone also reached a similar peak and plateaued within the same time window, we attribute the formation of this plateau to either staining saturation or limited substrate availability but not the actual levels of caspase activity during that time frame. Interestingly, it has been previously shown that Dcp-1 can cleave Drice in a cell-free system but not vice versa, similar to the way Dronc cleaves Drice (Hawkins et al., 2000; Song et al., 2000). Likewise, we showed cleavage of Drice, but not Dcp-1, in extracts from wild-type adult flies, which was blocked in the dcp-1 mutants (Fig. S4 A). Moreover, monitoring the kinetics of Drice activation (using the anticleaved caspase-3 antibody) in wild-type and dcp-1 mutant WDs displayed a significant delay in the rate of Drice activation when Dcp-1 was lacking (Fig. S4 B). This delay cannot be attributed to merely the lack of staining of activated Dcp-1, as even if we were to assume that this antibody has identical specificities toward activated Drice and activated Dcp-1 (although, as aforementioned, this is not the case), it would be expected that without an amplification loop, the maximal contribution of activated Dcp-1 to the overall staining level shall not surpass that of activated Drice (i.e., a maximum of twofold increase). However, the lack of Dcp-1 caused much more than a twofold reduction in the levels of staining (i.e., 3–7 fold), indicating that this reduction is mainly a result of less activated Drice in the absence of Dcp-1. Altogether, these findings provide an indirect support to a model whereby Dcp-1 may, either directly or indirectly, promote further activation of Drice through a positive amplification loop (see also the model in Fig. 9 A). It remains to be determined whether this cleavage of Drice by Dcp-1 in vivo leads to activation of the former and whether this may account for the aforementioned synergistic effect.
In conclusion, we have demonstrated that cells can continue living in the presence of a considerable level of effector caspase activity, as long as this level does not reach a critical execution threshold. Current models of the conserved core apoptotic machinery between Drosophila and mammals commonly illustrate the effector caspase activation step as an all-or-nothing process (Fuchs and Steller, 2011). However, recent studies of caspase-dependent nonapoptotic cellular processes suggest that to avoid excessive caspase activation and apoptosis, effector caspase activity may be low or restricted in space and time in these cells (Feinstein-Rotkopf and Arama, 2009; Kaplan et al., 2010; Li et al., 2010). Furthermore, it is well established that cancer cells can escape cellular lethality by manipulating a variety of steps and components in the apoptotic machinery, ultimately affecting the activity levels of the effector caspases (Johnstone et al., 2002; Hanahan and Weinberg, 2011). The current study provides insight into why low or restricted caspase activity levels fail to induce apoptosis (i.e., the threshold effect) and suggests that the critical step of caspase activation is more complex than has been previously appreciated.
Materials and methods
Fly strains and expression vectors
Flies containing the sal-gal4 driver (provided by K. Basler, University of Zurich, Zurich, Switzerland) were crossed to flies with the reporter transgene CPV (Williams et al., 2006), and the progeny were used as wild-type controls. The fly mutant alleles used in this study are droncI24 and droncI29 (Xu et al., 2005), driceΔ1 (Muro et al., 2006), dcp-1prev (Laundrie et al., 2003), ark82 (Akdemir et al., 2006), and strica4 (Baum et al., 2007). The UAS-CPV and sal-gal4 transgenes were recombined to the drice and dronc mutant alleles. The following UAS fly lines were also used: UAS-p35 (provided by H. Steller, Howard Hughes Medical Institute, Strang Laboratory of Cancer Research, The Rockefeller University, New York, NY), UAS-pro-dronc and UAS-ark (provided by H.D. Ryoo, New York University School of Medicine, New York, NY), and UAS-diap1 (provided by T. Volk, Weizmann Institute of Science, Rehovot, Israel). The dronc-RNAi line was obtained from the Vienna Drosophila RNAi Center (transformant ID no. 23035), and diap1-RNAi was a gift from P. Meier (Institute of Cancer Research, London, England, UK).
To generate the UAS-CD8::parpDEVG::venus (UAS-CPGV) construct, the CD8::parp::venus was subcloned into pBluescript II SK plasmid, and then a point mutation was introduced using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) with the primers forward 5′-GGCGATGAGGTGGGTGGAGTGGATGAA-3′ and reverse 5′-TTCATCCACTCCACCCACCTCATCGC-3′, which changed the adenine residue (A722) into guanine, leading to a change of the conserved aspartic acid at position P1 into glycine.
The rescue transgenic lines drice:drice and drice:dcp-1 were generated as follows: first, a 564-bp fragment from the drice genome encompassing its promoter and 5′ UTR was PCR amplified using the forward primer 5′-GGCAATTGCCTCTTTGAGAGTGTGACCG-3′ and reverse primer 5′-CCAAGATCTGGCTAAGTTCTCTCCTTGAG-3′ with added MfeI and BglII restriction sites, respectively. Second, a 708-bp fragment of drice 3′ UTR was also amplified by genomic PCR using the forward primer 5′-CCGGGCGGCCGCTGGCTAATGGTATGGATCAA-3′ and reverse primer 5′-GCGGTACCAGGGTCAACAGCAAACAGCCAA-3′ with added NotI and Acc65I restriction sites, respectively. Both the promoter/5′ UTR and 3′ UTR fragments were respectively cloned in a sequential order into the EcoRI + BglII and NotI + Acc65I sites of the pattB plasmid (a gift from J. Bischof, University of Zurich, Zurich, Switzerland), giving rise to the pattB-drice-5′-3′ plasmid. The drice coding region was amplified from a cDNA clone (a gift from H. Steller) using the forward primer 5′-CCAAGATCTATGGACGCCACTAACAATGG-3′ and reverse primer 5′-GGGGGGCCCTCAAACCCGTCCGGCTGGTG-3′ with added BglII and PspOMI restriction sites and was subsequently respectively cloned into the BglII and NotI restriction sites of the pattB-drice-5′-3′ plasmid. The dcp-1 coding region was amplified from a cDNA clone (a gift from H. Steller) using the forward primer 5′-CGGGGATCCATGACCGACGAGTGCGTAAC-3′ and reverse primer 5′-CGGCGCGGCCGCCTAGCCAGCCTTATTGCCGT-3′ with added BglII and NotI restriction sites and was subsequently respectively cloned into the BamHI and NotI restriction sites of the pattB-drice-5′-3′ plasmid. The two transgenic fly lines, drice:drice and drice:dcp-1, were generated using the ϕC31-mediated site-specific transgenesis technique, which allows insertion of transgenes into known sites of the Drosophila genome (Bischof et al., 2007; Fish et al., 2007). Specifically, these transgenes were inserted into the attP18 site on the X chromosome. Transcriptional expression of the transgenes was confirmed by RT-qPCR analysis on RNA from WDs of the two transgenic fly lines.
The rescue transgenic lines dcp-1:drice and dcp-1:dcp-1 were generated as follows: first, a 703-bp fragment from the dcp-1 genome encompassing its promoter and 5′ UTR was amplified using the forward primer 5′-AACAGATCTCTGTTTTTAATGTTAGATTAG-3′ and reverse primer 5′-GCGAATTCCTTGCGCCCCTTTTCTTGC-3′ with added BglII and EcoRI restriction sites, respectively. Second, a 1,090-bp fragment of dcp-1 3′ UTR was also amplified by genomic PCR using the forward primer 5′-AACTCGAGGAAGAGATCTCCCTTCGAAG-3′ and reverse primer 5′-AATCTAGAGTAAGCGGCTCCATCCATGGG-3′ with added XhoI and XbaI restriction sites, respectively. Both the promoter/5′ UTR and 3′ UTR fragments were respectively cloned in a sequential order into the BamHI + EcoRI and XhoI + XbaI sites of the pattB plasmid, giving rise to the pattB-dcp-1-5′-3′ plasmid. The dcp-1 and drice coding regions were each amplified from the cDNA clones as described above but this time using primers with added NotI and SalI restriction sites. Each fragment was then cloned into the NotI and XhoI restriction sites in the pattB-dcp-1-5′-3′ plasmid. These transgenes were inserted into the attP40 site on chromosome 2L using the ϕC31 system as before.
Immunofluorescence staining, TUNEL, and AO labeling and antibodies
Third instar larvae were subjected to γ-irradiation (50- or 20-Gy doses) and allowed to recover for several hours at room temperature, and their WDs were dissected and stained for either cleaved human PARP, cleaved caspase-3, TUNEL (see below), or AO (see below) using standard protocols (Arama et al., 2003; Arama and Steller, 2006). The corresponding antibodies used were rabbit polyclonal anticleaved human PARP (1:500, Ab2317; Abcam), rabbit polyclonal anticleaved caspase-3 (1:75, Asp175; Cell Signaling Technology), and rabbit polyclonal anticleaved caspase-3 (1:500, Ab13847; Abcam).
For TUNEL labeling, WDs were dissected and fixed for 20 min at room temperature in 4% PFA in PBS. The samples were then rinsed in PBS, washed twice, 10 min per wash, in 1× BSS (5× BSS: 270 mM NaCl, 200 mM KCl, 37 mM MgSO4, 12 mM CaCl2, 2 mM H2O, 24 mM tricine, 1.8% glucose, and 8.5% sucrose), and washed three times, 5 min per wash, with PBTw (0.1% Tween 20 in PBS). The samples were refixed in 4% PFA for 20 min, washed in PBTw five times, 5 min per wash, incubated in equilibration buffer (ApopTag kit; Millipore) for 1 h, and incubated again in reaction buffer (TdT enzyme; ratio 7:3; ApopTag kit) at 37°C overnight. On the next day, the TdT reaction mix was replaced with stop buffer (diluted 1:34 in dH2O; ApopTag kit) and incubated at 37°C for 3–4 h. Then, the samples were washed three times, 5 min per wash, blocked in BTN solution (1× BSS, 0.3% Triton X-100, and 5% normal goat serum) at room temperature for 1 h, and incubated with antidigoxigenin antibody solution (diluted 47:53 in blocking solution; ApopTag kit) overnight in the dark at 4°C. On the following day, the samples were washed four times in 1× BSS, 20 min per wash, and mounted in Fluoromount-G (SouthernBiotech).
For AO staining, WDs were incubated in a 0.6-µg/ml solution of AO (diluted in PBS) for 5 min, washed briefly in PBS, and mounted in a drop of PBS. Images were taken on a confocal microscope (LSM510 Meta Inverted Axio Observer; Carl Zeiss) using an EC Plan Neofluar 20×/0.50 M27 lens. Fluoromount-G was used as the imaging medium. All images were captured using the LSM510 operating software (Carl Zeiss).
Detection of effector caspase activity in vivo using the CPV reporter
In living cells, the reporter is localized to the plasma membrane via its CD8 domain. Upon cleavage by caspases, Venus is released to the cytoplasm, and, in principle, this process can be revealed by live imaging in large-diameter cells. However, in tissues of small-diameter cells, it is more difficult to distinguish between the membranal and cytoplasmic Venus, and, thus, the detection requires further staining of the tissue with an anti-cPARP antibody, which specifically detects the cleaved C-terminal fragment of human PARP.
Quantification of images
Quantification of staining was performed by measuring the area of the positively stained pixels (i.e., cPARP or TUNEL) and dividing it in the area of the Venus expression for cPARP or the total disc area for TUNEL using the ImageJ program (National Institutes of Health; Abramoff et al., 2004). Statistical analysis was performed using a one-way analysis of variance test followed by Fisher’s protected least significant difference posttest for multiple comparisons using the StatView Program (Abacus Concepts). For each experiment, 12–20 WDs were analyzed for each genotype or time point. Significance level was considered as either P < 0.05 or P < 0.001, as indicated above the bars in the figures.
Quantification of drice and dcp-1 mRNA expression levels
WDs from wild-type and mutants flies were dissected, and total RNA was extracted using the RNeasy Micro Kit (QIAGEN). Reverse transcription was performed with 1 µg of total RNA. First-strand synthesis used Oligo(dT) primers (Promega) and SuperScript II Reverse Transcriptase (Promega) in the presence of RNase-free DNase to eliminate DNA contamination and Protector RNase inhibitor (Roche). Measurements were normalized to mitochondrial large-ribosomal RNA (MtlrRNA1: forward 5′-AAAAAGATTGCGACCTCGAT-3′ and reverse 5′-AAACCAACCTGGCTTACACC-3′) or RP49 (Dmel/RpL32: forward 5′-GACCATCCGCCCAGCATAC-3′ and reverse 5′-CCATTTGTGCGACAGCTTAGC-3′). The primers for drice mRNA were forward 5′-CCACTAACAATGGAGAATCCGCC-3′ and reverse 5′-GCCGCTGCTACCCGCTCCTC-3′. The primers for dcp-1 mRNA were forward 5′-ACCGACGAGTGCGTAACCAGAA-3′ and reverse 5′-ACAAGAGACTCCGGCGTACAGC-3′. Products were amplified by the KAPA SYBR FAST quantitative PCR kit (Kapa Biosystems) using the LightCycler 480 quantitative PCR machine (Roche). Results of expression levels were calculated as the mean from three independent experiments with at least two biological repeats with two duplicates each.
Survival assay
Third instar larvae of each genotype were collected in vials with fresh food at 25°C. Eclosing adults were counted, and fly viability was determined as the number of eclosed adults divided in the total larvae collected. Survival rate was normalized to wild-type levels, and the larvae were collected during a period of 3 wk.
Western blotting
50 WDs from larvae with the desired genotype were dissected in PBS and collected into cold lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% NP-40, and a protease inhibitor mix). Then, these WDs were homogenized and incubated for 15 min on ice and centrifuged for 20 min, and the supernatant was collected and mixed with a sample buffer. Gel running and subsequent blotting was performed using standard methods, and the blots were incubated with a guinea pig anti–Dcp-1 antibody (1:1,000; a gift from P. Meier), a mouse anti–lamin Dmo antibody (1:500; a gift from Y. Gruenbaum, The Hebrew University of Jerusalem, Jerusalem, Israel), or a rabbit anti-Drice antibody (1:2,000; a gift from P. Friesen, University of Wisconsin-Madison, Madison, WI).
Online supplemental material
Fig. S1 shows that drice and dcp-1 transcripts are expressed at comparable levels in WDs. Fig. S2 shows that the CPV reporter does not affect the rate of cell death. Fig. S3 shows that different anticleaved caspase-3 (CM1) antibodies detect both active Drice and Dcp-1, although they are more specific toward active Drice. Fig. S4 shows that Dcp-1 may further activate Drice in a positive amplification loop. Fig. S5 shows that Diap1 does not inhibit Drice and Dcp-1 with different efficiencies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201107133/DC1.
Acknowledgments
We are grateful to J.M. Abrams, K. Basler, A. Bergmann, P. Friesen, Y. Gruenbaum, B. Hay, K. McCall, P. Meier, H.D. Ryoo, H. Steller, T. Volk, and D.W. Williams for providing additional stocks and reagents, S. Kredo-Russo for helping with the RT-qPCR, V. Kalchenko for helping with the ImageJ program, Y. Kaplan for helping with coimmunoprecipitation experiments, and S. Pietrokovski and E. Schejter for useful discussions. We thank the Arama laboratory members for encouragement and advice, O. Reiner and L. Ravid for critically reading the manuscript, and S. Brown for English editing.
This research was supported in part by the Israel Science Foundation (grant no. 308/09) and Minerva Foundation, with funding from the German Federal Ministry of Education and Research, Israel Cancer Research Fund, Israel Cancer Association, Yeda-Sela Center for Basic Research, and the M.D. Moross Institute for Cancer Research. E. Arama is also supported by grants from the Y. Leon Benoziyo Institute for Molecular Medicine, the Jeanne and Joseph Nissim Center for Life Sciences Research, and Lord Mitchell. E. Arama is the Incumbent of the Corinne S. Koshland Career Development Chair.
Footnotes
Abbreviations used in this paper:
- AO
- acridine orange
- cPARP
- cleaved PARP
- PARP
- poly(ADP-ribose) polymerase
- RT-qPCR
- real-time quantitative PCR
- UTR
- untranslated region
- WD
- wing imaginal disc
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. 2004. Image processing with ImageJ. Biophotonics International. 11:36–42 [Google Scholar]
- Akdemir F., Farkas R., Chen P., Juhasz G., Medved’ová L., Sass M., Wang L., Wang X., Chittaranjan S., Gorski S.M., et al. 2006. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 133:1457–1465 10.1242/dev.02332 [DOI] [PubMed] [Google Scholar]
- Arama E., Steller H. 2006. Detection of apoptosis by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling and acridine orange in Drosophila embryos and adult male gonads. Nat. Protoc. 1:1725–1731 10.1038/nprot.2006.235 [DOI] [PubMed] [Google Scholar]
- Arama E., Agapite J., Steller H. 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell. 4:687–697 10.1016/S1534-5807(03)00120-5 [DOI] [PubMed] [Google Scholar]
- Arama E., Bader M., Srivastava M., Bergmann A., Steller H. 2006. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 25:232–243 10.1038/sj.emboj.7600920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J.S., Arama E., Steller H., McCall K. 2007. The Drosophila caspases Strica and Dronc function redundantly in programmed cell death during oogenesis. Cell Death Differ. 14:1508–1517 10.1038/sj.cdd.4402155 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 104:3312–3317 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright K.M., Salvesen G.S. 2003. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15:725–731 10.1016/j.ceb.2003.10.009 [DOI] [PubMed] [Google Scholar]
- Chew S.K., Akdemir F., Chen P., Lu W.J., Mills K., Daish T., Kumar S., Rodriguez A., Abrams J.M. 2004. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev. Cell. 7:897–907 10.1016/j.devcel.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Cooper D.M., Granville D.J., Lowenberger C. 2009. The insect caspases. Apoptosis. 14:247–256 10.1007/s10495-009-0322-1 [DOI] [PubMed] [Google Scholar]
- Crawford E.D., Wells J.A. 2011. Caspase substrates and cellular remodeling. Annu. Rev. Biochem. 80:1055–1087 10.1146/annurev-biochem-061809-121639 [DOI] [PubMed] [Google Scholar]
- Daish T.J., Mills K., Kumar S. 2004. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell. 7:909–915 10.1016/j.devcel.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Denton D., Shravage B., Simin R., Mills K., Berry D.L., Baehrecke E.H., Kumar S. 2009. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 19:1741–1746 10.1016/j.cub.2009.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M., Broemer M., Tenev T., Bolduc C., Lee T.V., Rigbolt K.T., Elliott R., Zvelebil M., Blagoev B., Bergmann A., Meier P. 2008. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol. Cell. 32:540–553 10.1016/j.molcel.2008.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix M.M., Simon G.M., Cravatt B.F. 2008. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 134:679–691 10.1016/j.cell.2008.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Bergmann A. 2010. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 17:534–539 10.1038/cdd.2009.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein-Rotkopf Y., Arama E. 2009. Can’t live without them, can live with them: Roles of caspases during vital cellular processes. Apoptosis. 14:980–995 10.1007/s10495-009-0346-6 [DOI] [PubMed] [Google Scholar]
- Fish M.P., Groth A.C., Calos M.P., Nusse R. 2007. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. Nat. Protoc. 2:2325–2331 10.1038/nprot.2007.328 [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Evan G.I. 1997. Identification of a Drosophila melanogaster ICE/CED-3-related protease, drICE. EMBO J. 16:2805–2813 10.1093/emboj/16.10.2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y., Steller H. 2011. Programmed cell death in animal development and disease. Cell. 147:742–758 10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L., McCall K., Agapite J., Hartwieg E., Steller H. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589–597 10.1093/emboj/19.4.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2011. Hallmarks of cancer: The next generation. Cell. 144:646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Harvey N.L., Daish T., Mills K., Dorstyn L., Quinn L.M., Read S.H., Richardson H., Kumar S. 2001. Characterization of the Drosophila caspase, DAMM. J. Biol. Chem. 276:25342–25350 10.1074/jbc.M009444200 [DOI] [PubMed] [Google Scholar]
- Hawkins C.J., Yoo S.J., Peterson E.P., Wang S.L., Vernooy S.Y., Hay B.A. 2000. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 275:27084–27093 [DOI] [PubMed] [Google Scholar]
- Hellwig C.T., Kohler B.F., Lehtivarjo A.K., Dussmann H., Courtney M.J., Prehn J.H., Rehm M. 2008. Real time analysis of tumor necrosis factor-related apoptosis-inducing ligand/cycloheximide-induced caspase activities during apoptosis initiation. J. Biol. Chem. 283:21676–21685 10.1074/jbc.M802889200 [DOI] [PubMed] [Google Scholar]
- Hoeppner D.J., Hengartner M.O., Schnabel R. 2001. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature. 412:202–206 10.1038/35084103 [DOI] [PubMed] [Google Scholar]
- Holley C.L., Olson M.R., Colón-Ramos D.A., Kornbluth S. 2002. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat. Cell Biol. 4:439–444 10.1038/ncb798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, N.M., editor. 2002. Proteases in Biology and Medicine. Portland Press, London. 201 pp.
- Hou Y.C., Chittaranjan S., Barbosa S.G., McCall K., Gorski S.M. 2008. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182:1127–1139 10.1083/jcb.200712091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde C., Banks K.G., Coulombe N., Rasper D., Grimm E., Roy S., Simpson E.M., Nicholson D.W. 2004. Caspase-7 expanded function and intrinsic expression level underlies strain-specific brain phenotype of caspase-3-null mice. J. Neurosci. 24:9977–9984 10.1523/JNEUROSCI.3356-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone R.W., Ruefli A.A., Lowe S.W. 2002. Apoptosis: A link between cancer genetics and chemotherapy. Cell. 108:153–164 10.1016/S0092-8674(02)00625-6 [DOI] [PubMed] [Google Scholar]
- Kaplan Y., Gibbs-Bar L., Kalifa Y., Feinstein-Rotkopf Y., Arama E. 2010. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev. Cell. 19:160–173 10.1016/j.devcel.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Koto A., Kuranaga E., Miura M. 2009. Temporal regulation of Drosophila IAP1 determines caspase functions in sensory organ development. J. Cell Biol. 187:219–231 10.1083/jcb.200905110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K., Zheng T.S., Na S., Kuan C., Yang D., Karasuyama H., Rakic P., Flavell R.A. 1996. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 384:368–372 10.1038/384368a0 [DOI] [PubMed] [Google Scholar]
- Kumar S. 2007. Caspase function in programmed cell death. Cell Death Differ. 14:32–43 10.1038/sj.cdd.4402060 [DOI] [PubMed] [Google Scholar]
- Laundrie B., Peterson J.S., Baum J.S., Chang J.C., Fileppo D., Thompson S.R., McCall K. 2003. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 165:1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F., Rodriguez A., Khush R.S., Abrams J.M., Lemaitre B. 2000. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 1:353–358 10.1093/embo-reports/kvd073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F., Ribeiro P.S., Palmer E., Tenev T., Takahashi K., Robertson D., Zachariou A., Pichaud F., Ueda R., Meier P. 2006. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. 13:1663–1674 10.1038/sj.cdd.4401868 [DOI] [PubMed] [Google Scholar]
- Li Z., Jo J., Jia J.M., Lo S.C., Whitcomb D.J., Jiao S., Cho K., Sheng M. 2010. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell. 141:859–871 10.1016/j.cell.2010.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi A.U., Martin S.J. 2007. The CASBAH: A searchable database of caspase substrates. Cell Death Differ. 14:641–650 10.1038/sj.cdd.4402103 [DOI] [PubMed] [Google Scholar]
- Mahrus S., Trinidad J.C., Barkan D.T., Sali A., Burlingame A.L., Wells J.A. 2008. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 134:866–876 10.1016/j.cell.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer C.W., Chiorazzi M., Shaham S. 2007. Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development. 134:1357–1368 10.1242/dev.02818 [DOI] [PubMed] [Google Scholar]
- Muro I., Berry D.L., Huh J.R., Chen C.H., Huang H., Yoo S.J., Guo M., Baehrecke E.H., Hay B.A. 2006. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development. 133:3305–3315 10.1242/dev.02495 [DOI] [PubMed] [Google Scholar]
- Orme M., Meier P. 2009. Inhibitor of apoptosis proteins in Drosophila: Gatekeepers of death. Apoptosis. 14:950–960 10.1007/s10495-009-0358-2 [DOI] [PubMed] [Google Scholar]
- Peterson J.S., Barkett M., McCall K. 2003. Stage-specific regulation of caspase activity in drosophila oogenesis. Dev. Biol. 260:113–123 10.1016/S0012-1606(03)00240-9 [DOI] [PubMed] [Google Scholar]
- Pluquet O., Hainaut P. 2001. Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett. 174:1–15 10.1016/S0304-3835(01)00698-X [DOI] [PubMed] [Google Scholar]
- Reddien P.W., Cameron S., Horvitz H.R. 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature. 412:198–202 10.1038/35084096 [DOI] [PubMed] [Google Scholar]
- Ribeiro P.S., Kuranaga E., Tenev T., Leulier F., Miura M., Meier P. 2007. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J. Cell Biol. 179:1467–1480 10.1083/jcb.200706027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl S.J., Shi Y. 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 5:897–907 10.1038/nrm1496 [DOI] [PubMed] [Google Scholar]
- Rodriguez J., Lazebnik Y. 1999. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 13:3179–3184 10.1101/gad.13.24.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen G.S., Abrams J.M. 2004. Caspase activation - stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 23:2774–2784 10.1038/sj.onc.1207522 [DOI] [PubMed] [Google Scholar]
- Salvesen G.S., Riedl S.J. 2008. Caspase mechanisms. Adv. Exp. Med. Biol. 615:13–23 10.1007/978-1-4020-6554-5_2 [DOI] [PubMed] [Google Scholar]
- Schoenmann Z., Assa-Kunik E., Tiomny S., Minis A., Haklai-Topper L., Arama E., Yaron A. 2010. Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J. Neurosci. 30:6375–6386 10.1523/JNEUROSCI.0922-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader K., Huai J., Jöckel L., Oberle C., Borner C. 2010. Non-caspase proteases: Triggers or amplifiers of apoptosis? Cell. Mol. Life Sci. 67:1607–1618 10.1007/s00018-010-0287-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S. 1998. Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J. Biol. Chem. 273:35109–35117 10.1074/jbc.273.52.35109 [DOI] [PubMed] [Google Scholar]
- Shapiro P.J., Hsu H.H., Jung H., Robbins E.S., Ryoo H.D. 2008. Regulation of the Drosophila apoptosome through feedback inhibition. Nat. Cell Biol. 10:1440–1446 10.1038/ncb1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Guan B., Bergman A., Nicholson D.W., Thornberry N.A., Peterson E.P., Steller H. 2000. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Mol. Cell. Biol. 20:2907–2914 10.1128/MCB.20.8.2907-2914.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. 2008. Regulation of apoptosis in Drosophila. Cell Death Differ. 15:1132–1138 10.1038/cdd.2008.50 [DOI] [PubMed] [Google Scholar]
- Stoven S., Silverman N., Junell A., Hedengren-Olcott M., Erturk D., Engstrom Y., Maniatis T., Hultmark D. 2003. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc. Natl. Acad. Sci. USA. 100:5991–5996 10.1073/pnas.1035902100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry N.A., Lazebnik Y. 1998. Caspases: Enemies within. Science. 281:1312–1316 10.1126/science.281.5381.1312 [DOI] [PubMed] [Google Scholar]
- Vaux D.L., Silke J. 2005. IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 6:287–297 10.1038/nrm1621 [DOI] [PubMed] [Google Scholar]
- Waldhuber M., Emoto K., Petritsch C. 2005. The Drosophila caspase DRONC is required for metamorphosis and cell death in response to irradiation and developmental signals. Mech. Dev. 122:914–927 10.1016/j.mod.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Williams D.W., Kondo S., Krzyzanowska A., Hiromi Y., Truman J.W. 2006. Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9:1234–1236 10.1038/nn1774 [DOI] [PubMed] [Google Scholar]
- Woo M., Hakem R., Soengas M.S., Duncan G.S., Shahinian A., Kägi D., Hakem A., McCurrach M., Khoo W., Kaufman S.A., et al. 1998. Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 12:806–819 10.1101/gad.12.6.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Li Y., Arcaro M., Lackey M., Bergmann A. 2005. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 132:2125–2134 10.1242/dev.01790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Wang Y., Willecke R., Chen Z., Ding T., Bergmann A. 2006. The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ. 13:1697–1706 10.1038/sj.cdd.4401920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Woodfield S.E., Lee T.V., Fan Y., Antonio C., Bergmann A. 2009. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin). 3:78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.Y., Yoo S.J., Yang L., Zapata C., Srinivasan A., Hay B.A., Baker N.E. 2002. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 129:3269–3278 [DOI] [PubMed] [Google Scholar]
- Yuan J., Shaham S., Ledoux S., Ellis H.M., Horvitz H.R. 1993. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 75:641–652 10.1016/0092-8674(93)90485-9 [DOI] [PubMed] [Google Scholar]
- Zou H., Henzel W.J., Liu X., Lutschg A., Wang X. 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 90:405–413 10.1016/S0092-8674(00)80501-2 [DOI] [PubMed] [Google Scholar]