Abstract
Identifying cellular reservoirs of human immunodeficiency virus type 1 (HIV-1) in patients on antiretroviral therapy (ART) is critical to finding a cure for HIV-1. In addition to resting CD4+ T cells, CD34+ hematopoietic progenitor cells have been proposed as another reservoir. We obtained bone marrow aspirates from 11 patients on ART who had undetectable plasma HIV-1 RNA. HIV-1 DNA was detected in CD4+ T cells from peripheral blood in all patients and from bone marrow cellular fractions containing T cells in most patients. We did not find HIV-1 DNA in highly purified CD34+ populations using either a sensitive real-time polymerase chain reaction assay or a coculture assay for replication-competent HIV-1.
Antiretroviral therapy (ART) stops replication of human immunodeficiency virus type 1 (HIV-1) virus and reduces plasma viremia to undetectable levels. Despite treatment, latent HIV-1 DNA is detected in resting CD4+ T cells; with therapy interruption, HIV-1 viremia rapidly rebounds to pretreatment levels (reviewed in [1]). Longitudinal studies confirm that due to the long-lived nature of resting CD4+ T cells, the size of this reservoir does not appreciably decrease with time on ART [2].
In addition to latent HIV-1 DNA in resting CD4+ T cells, most patients on ART have residual HIV-1 RNA in the plasma that can be detected with sensitive real-time polymerase chain reaction (PCR) assays [3]. HIV-1 sequence analysis and phylogenetic studies of residual viremia demonstrate that many sequences in the plasma are identical to sequences in resting CD4+ T cells. However, in some individuals, residual viremia contains predominant HIV-1 clones that are profoundly underrepresented in circulating CD4+ T cells [4]. This suggests that other significant HIV-1 reservoirs may exist in cells capable of proliferating after infection.
Hematopoietic progenitor cells (HPCs) are a logical candidate for a second reservoir due to expression of coreceptors required for HIV-1 entry, that is, CCR5 and CXCR4 [5]. Most studies of HPCs over the past 2 decades suggest that these cells do not harbor HIV-1 in vivo (reviewed in [6]). However, other studies found HIV-1 DNA in HPCs in patients infected with non–subtype B virus [7, 8]. A recent study was the first to address this in patients on ART with undetectable plasma HIV-1 [9]. The authors reported HIV-1 infection in CD34+ HPCs in 4 of 9 patients at a frequency of 2.5–40 copies of HIV-1 DNA/104 cells. The reasons for these different findings are unclear. Determining whether CD34+ cells constitute a second HIV-1 reservoir in patients on ART is critical to ongoing HIV-1 eradication efforts, which currently focus on latently infected CD4+ T cells as the barrier to curing HIV-1 infection. We conducted a study in ART-treated patients with undetectable plasma HIV-1 using rigorous purification techniques and a real-time PCR assay to look for HIV-1 DNA in CD34+ HPCs. In a subset of patients, we also performed an HPC/lymphocyte coculture assay, which is less sensitive for total HIV-1 DNA but highly specific for replication-competent HIV-1.
METHODS
Subjects
We studied HIV-1−infected adults on ART with a plasma HIV-1 level <50 copies/mL for at least 2 years. Isolated HIV-1 plasma RNA measurements between 50 and 200 copies/mL were considered viral “blips” and did not exclude participation. All patients gave informed consent according to a Johns Hopkins School of Medicine institutional review board.
Cell Purification
Study volunteers donated 40 mL of blood and 25–50 mL of bone marrow aspirated from the iliac crest. Bone marrow mononuclear cells (BMMCs) were obtained by Ficoll-Hypaque density gradient centrifugation. Cells were cultured in RPMI (Sigma-Aldrich) supplemented with 10% fetal bovine serum overnight. CD4+ T cells and CD34+ cells were purified within 24 hours by positive selection using a magnetic bead and column system (Miltenyi). The bead-enriched CD34+ population was labeled with phycoerythrin-conjugated anti-CD34 and fluorescein isothiocyanate−conjugated anti-CD3 monoclonal antibodies (BD Biosciences) and purified by fluorescence-activated cell sorting (FACS) on a MoFlo flow cytometer (Beckman Coulter). Viable CD34+ and CD3+ cells based on forward- and side-scatter properties were collected by FACS. Anti-CD3 was used to mark T cells because HPCs may express CD4 at low levels.
HIV-1 DNA Quantification
DNA was extracted immediately following cell sorting using a QIAamp DNA microkit (Qiagen). A single-step real-time PCR was used to quantify HIV-1 DNA in a 50-μL PCR reaction mix containing 25 μL of TaqMan Universal PCR Master Mix (Applied Biosystems), 20 μL of template DNA, and a primer-probe set designed to bind to a conserved region of gag: primers 6F (5′-CATGTTTTCAGCATTATCAGAAGGA-3′) and 84R (5′TGCTTGATGTCCCCCCACT-3′) (600 nm) and probe (5′FAM-CCACCCCACAAGATTTAAACACCATGCT) from a published single-copy assay to amplify HIV-1 RNA [3]. Amplifications were performed with an Applied Biosystems 7300 real-time PCR system. Thermocycling conditions were 95°C for 10 minutes, 50 cycles at 95°C for 15 seconds, and 60°C for 1 minute. HIV-1 copy number was determined by extrapolation from a standard curve generated from serial dilutions of a cell line harboring 1 HIV-1 genome per cell [10], which were spiked into genomic DNA from HIV-1–uninfected peripheral blood mononuclear cells (PBMCs) in order to accurately represent the in vivo situation of low HIV-1 copy number in a high background. The limit of detection of this assay was determined to be 2 copies of HIV-DNA in 105 cell equivalents of DNA, defined as the minimum copy number that could be detected >90% of the time. To determine if CD34+ cellular DNA in particular inhibited amplification of HIV-1, we measured the ability of our assay to detect 100, 10, and 5 HIV-1–positive cells spiked into 105 patient CD34+ cells. To quantify cell number, a separate reaction was performed to measure the human gene CCR5 using a published primer-probe set [11]. All reactions including negative controls with water and uninfected PBMCs were run in triplicate.
Culture Assay for Replication-Competent HIV-1
In patients from whom >106 CD34+ cells were obtained by FACS (n = 7), CD34+cells were cultured to amplify replication-competent HIV-1 RNA. Two 4-mL wells with CD34+ cells at 105 cells/mL were cultured in StemSpan (StemCell Technologies) with 100 ng/mL of stem cell factor, 100 ng/mL FMS-like tyrosine kinase–3 ligand, and 100 ng/mL thrombopoietin (R&D Systems) plus either (1) 100 ng recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) and 2.5 ng/mL tumor necrosis factor α (TNF-α; Biolegend) or (2) 10 ng/mL of phorbol-12-myristate-13-acetate (PMA) (Sigma-Aldrich) for 7 days as previously described [9]. Then 5 × 106 BMMCs and CD34-depleted cells were cultured under the same cytokine conditions (concentration 106 cells/mL). After 7 days, activated CD4+ lymphoblasts from uninfected donors were added as described in a resting CD4+ T-cell lymphocyte coculture assay [12]. Lymphoblasts were added at days 7, 14, and 21. Culture supernatants were collected on days 7, 14, 21, and 28 and tested for p24 antigen (PerkinElmer) and HIV-1 RNA (Roche Amplicor).
RESULTS
Patients
Patient characteristics are shown in Table 1. Median duration since HIV-1 diagnosis was 21 years and median duration of undetectable viremia on ART was 4.8 years. Patients had prior evidence of significant immunosuppression with a median CD4+ T-cell nadir of 212 cells/μL. Four of 11 participants had anemia or thrombocytopenia at the time of sampling. The majority (10 of 11) of patients started ART during chronic infection and 1 patient initiated ART within 2 months of acute HIV-1 infection.
Table 1.
Patient Characteristics
| Study ID (age in y, sex) | Current CD4 Count, cells/mm3 | CD4 Nadir, cells/mm3 | Years Since Diagnosis | Years on ART With HIV RNA <50 Copies/mLa | White Blood Cell Count, K/μLb | Hematocrit, %c | Platelet Count, K/μLd | No. of CD34+ Cells Assayed in PCR | Purity of CD34+ Cells, %e |
| BM1 (65, M) | 427 | 212 | 16 | 4.7 | 3.8 | 36.8 | 198 | 1.1 × 105 | 99.1 |
| BM2 (55, M) | 642 | 193 | 24 | 2.2 | 8 | 41.6 | 298 | 1.1 × 106 | 98.7 |
| BM3 (42, M) | 551 | 255 | 11 | 6 | 4.7 | 35.3 | 67 | 1.6 × 105 | >99.5 |
| BM4 (51, M) | 629 | 76 | 23 | 6.9 | 6.5 | 41 | 294 | 1.8 × 105 | >99.5 |
| BM6 (44, M) | 549 | 460 | 3 | 2.1 | 4 | 41 | 43 | 9.6 × 105 | >99.5 |
| BM7 (57, M) | 1168 | 386 | 28 | 9.3 | 7.4 | 43.4 | 223 | 6.9 × 105 | >99.5 |
| BM8 (52, M) | 356 | 200 | 13 | 3.6 | 4 | 43.5 | 153 | 5.1 × 105 | >99.5 |
| BM9 (52, M) | 491 | 44 | 26 | 2.5 | 6.3 | 45 | 113 | 3.7 × 105 | >99.5 |
| BM10 (45, M) | 784 | 231 | 22 | 5.8 | 6.7 | 42.9 | 164 | 6.4 × 105 | >99.5 |
| BM13 (49, M) | 972 | 264 | 11 | 6.4 | 6.8 | 43.3 | 161 | 7.9 × 105 | 99.2 |
| BM14 (46, M) | 689 | 212 | 21 | 7.3 | 5.6 | 46.6 | 212 | 2.2 × 105 | 99.3 |
Abbreviations: HIV, human immunodeficiency virus; PCR, polymerase chain reaction.
Isolated measurements between 50 and 200 copies/mL were considered clinically insignificant viral “blips” and were not included.
Normal range, 4.3–10.8 K/μL.
Normal range for men, 40%–54%.
Normal range, 150–450 K/μL.
Purity of CD34+ cells, expressed as the fraction of CD34+CD3− cells.
Purification
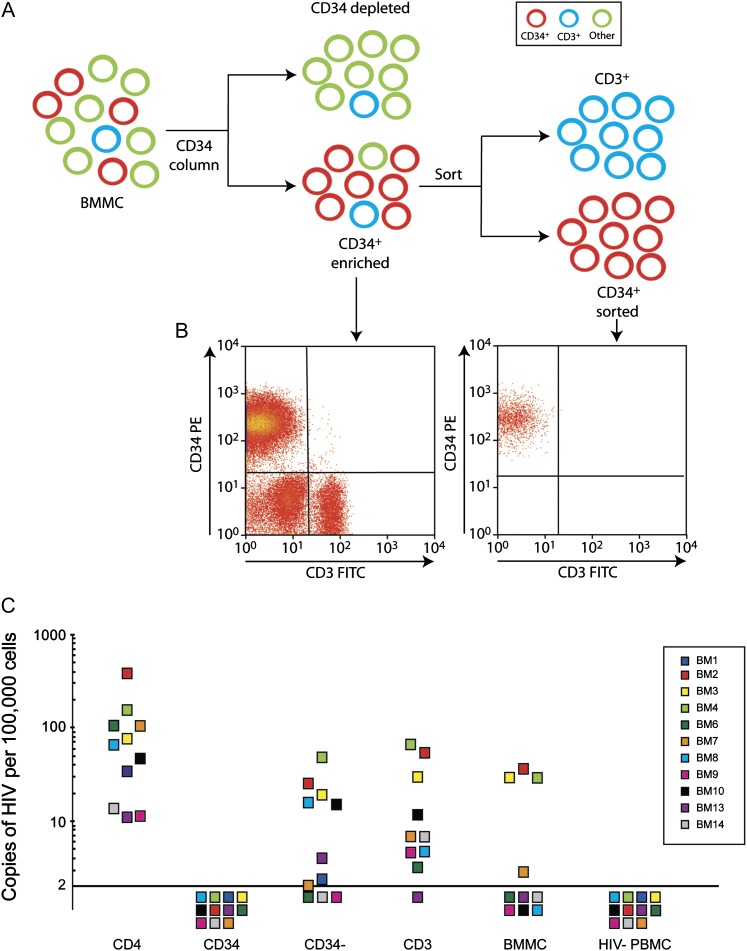
We isolated CD34+ HPCs using a 2-step process (Figure 1A). After bead enrichment, the average purity of CD34+ cells was 63% (range, 25%–87%). The average level of contaminating CD3+ T cells was 10% (range, 3%–17%). After FACS, the purity of the CD34+ cell population was >98.7% (Figure 1B), with contaminating CD3+ T cells <0.2% in all patients.
Figure 1.
Analysis of human immunodeficiency virus type 1 (HIV-1) DNA in CD34+ hematopoietic progenitor cells. A, Purification methods used. CD34+ cells were first enriched from bone marrow mononuclear cells (BMMCs) using anti-CD34–conjugated magnetic beads. Highly pure populations were then obtained by flow-cytometric cell sorting using anti-CD34 and anti-CD3 antibodies. B, Representative flow cytometric analysis corresponding to CD34+-enriched vs CD34+-sorted cell populations from (A); phycoerythrin (PE) and fluorescein isothiocyanate (FITC). C, HIV-1 DNA copies/100 000 cells in peripheral CD4+ T cells (CD4+), sorted CD34+ cells, bone marrow depleted of CD34+ cells (CD34-depleted), CD3+ cells sorted from enriched CD34+ cells (CD3+), unfractionated BMMCs, and peripheral blood mononuclear cells from uninfected donors (HIV-PBMCs). Each color square represents a different patient designated by BM (bone marrow) and study number. The corresponding color HIV- PBMCs are the negative controls for the specific patient experiment. For patient 1, only sorted CD34+ and CD34-depleted fractions were tested.
HIV-1 DNA Quantification
To measure HIV-1 DNA, we used a real-time PCR assay with a limit of detection of 2 copies of HIV-1 DNA/105 cells. We compared HIV-1 copy numbers in sorted CD34+ cells to copy numbers in peripheral CD4+ T cells and other bone marrow fractions: CD34-depleted cells, sorted CD3+ cells from the bead-enriched CD34+ cells, and BMMCs. Table 1 lists the number of CD34+ cells assayed per patient (median, 5.5 × 105 cells; range, 1.1 × 105 to 1.1 × 106).
The copy number of HIV-1 per 105 cells is shown in Figure 1C. HIV-1 DNA was detected in CD4+ T cells in 11 of 11 patients, with a geometric mean of 64 copies/105 cells. In many patients, HIV-1 DNA was detected in the other bone marrow fractions. In 11 of 11 patients, no HIV-1 DNA was detected in sorted CD34+ cells. Lack of signal was not due to PCR inhibition because the CCR5 gene was amplified in all samples. In addition, an internal positive control using 100, 10, and 5 HIV-1–positive cells spiked into 105 sorted CD34+ patient cells confirmed that small amounts of HIV-1 could be detected in these cells (data not shown).
CD34+ HPC/Lymphocyte Coculture
To assess whether production of HIV-1 is induced with ex vivo differentiation of CD34+ HPCs, we cultured cells from 7 patients with cytokines and added activated CD4+ lymphoblasts as target cells for HIV-1 replication. In resting CD4 T cells, this coculture assay detects replication-competent virus released from a single infected cell [2]. In 7 of 7 patients, neither HIV-1 antigen nor HIV-1 RNA was detected in culture supernatants from CD34+ cells over 28 days. We cultured BMMCs and CD34-depleted cells under the same conditions and detected replication-competent HIV-1 in 3 of 7 patients (2 in BMMC in GM-CSF/TNF-α and PMA, respectively, and 1 in CD34-depleted cells in GM-CSF and TNF-α).
DISCUSSION
We have found that in patients undergoing ART, HIV-1 DNA can be frequently detected in bone marrow fractions that contain mature T cells but not in highly pure populations of CD34+ HPCs. Our quantitative PCR assay detected HIV-1 in CD4+ T cell in 11 of 11 patients but found no evidence of HIV-1 in CD34+ cells with a sensitivity down to 2 copies of HIV-1 DNA/105 cells and a median of 5.5 × 105 CD34+ cells assayed per patient. Because it has been reported that latent infection of CD34+ cells can be reversed by cytokine-directed differentiation, we cultured bone marrow fractions from a subset of 7 patients with uninfected lymphocytes. After culture for 1 month, we detected replication-competent HIV-1 in CD34+ cells from 0 of 7 patients and in bone marrow fractions containing T cells in 3 of 7 patients. This assay only detects replication-competent HIV-1 and thus is less sensitive for total HIV-1 DNA.
This study was limited by the small sample size and sensitivity of our PCR assay. We had >90% power to detect infection in 20% of patients. We can conclude that in the majority of HIV-1 subtype-B–infected patients on ART, the frequency of HIV-infected CD34+ is <0.002%. These conclusions extend a large body of clinical and translational data reported over the past 2 decades, indicating that CD34+ cells do not constitute a significant reservoir for HIV-1 in vivo (reviewed in [6]).
The only prior study to examine this issue in ART-treated patients with undetectable plasma HIV-1 RNA levels found PCR evidence of HIV-1 DNA in CD34+ cells in 4 of 9 patients [9]. There are several potential explanations for the conflicting data. This could be the result of CD34+ cells being infected at a level below the limit of detection of our assay and/or be due to insufficient sampling of cells per patient. However, our reported sensitivity is comparable to the prior report and the number of cells assayed per patient is larger. We hypothesize that contamination with HIV-1–infected mature T cells could produce false-positive results in bead-enriched HPCs. The purity of the bead-enriched CD34+ cells in patients on ART studied by Carter et al [9] ranged from 30% to 97% (median 85%) and was 30%, 57%, 83%, and 90% in the 4 patients with detectable HIV-1 DNA. This purity is typical of column-enrichment methods, and we had a similar range of purity (25%–87%) after CD34+ bead enrichment. We did not perform PCR on the CD34-enriched cells in order to maximize our yield from FACS. As an alternative control, we tested other bone marrow fractions known to contain CD4+ T cells and detected HIV-1 in a significant proportion of patients.
Lack of HIV-1 detection in our cohort could be explained by HIV-1 coreceptor tropism. CD34+ cells are heterogeneous and include rare hematopoietic stem cells as well as committed myeloid and lymphoid progenitors. These cells vary in their expression of CCR5 and CXCR4 used for HIV-1 entry. More primitive cells express CXCR4 but not CCR5 [5]. An in vitro study by Carter et al [13] showed that HIV-1 vectors pseudotyped with X4 envelope infect progenitor cells more efficiently than vectors pseudotyped with R5 envelope. However, whether this occurs in vivo is less clear. We do not know the tropism of viruses present in the majority of patients in our cohort. In vivo studies that focus on patients with different frequencies of X4 virus may clarify this issue.
Notes
Acknowledgments.
We thank Joel Blankson and Andrew Redd for their careful reading of the manuscript. We thank Patricia Rennie for her help in coordinating all the patient visits. Finally, we acknowledge the generous contribution of the study participants, without whom this research would not be possible.
Financial support.
This work was supported by the National Institutes of Health (NIH), National Cancer Institute (grants P30CA06973 and P01CA1539627); Howard Hughes Medical Institute and NIH; and National Institute of Allergy and Infectious Diseases (grant AI43222 to R. F. S).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Haggerty CM, Pitt E, Siliciano RF. The latent reservoir for HIV-1 in resting CD4+ T cells and other viral reservoirs during chronic infection: insights from treatment and treatment-interruption trials. Curr Opin HIV AIDS. 2006;1:62–8. doi: 10.1097/01.COH.0000191897.78309.70. [DOI] [PubMed] [Google Scholar]
- 2.Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 3.Palmer S, Wiegand AP, Maldarelli F, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;10:4531–6. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey JR, Sedaghat AR, Kieffer TL, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–57. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiz ME, Cicala C, Arthos J, et al. Peripheral blood-derived CD34+ progenitor cells: CXC chemokine receptor 4 and CC chemokine receptor 5 expression and infection by HIV. J Immun. 1998;161:4169–76. [PubMed] [Google Scholar]
- 6.McNamara LA, Collins KL. Hematopoietic stem/precursors cells as HIV reservoirs. Curr Opin HIV AIDS. 2011;6:43–8. doi: 10.1097/COH.0b013e32834086b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley SK, Kessler S, Justement JS, et al. CD34+ bone marrow cells are infected with HIV in a subset of seropositive individuals. J Immun. 1992;149:689–97. [PubMed] [Google Scholar]
- 8.Redd AD, Avalos A, Essex M. Infection of hematopoietic progenitor cells by HIV-1 subtype C, and its association with anemia in southern Africa. Blood. 2007;110:3143–9. doi: 10.1182/blood-2007-04-086314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter CC, Onafuwa-Nuga A, McNamara LA, et al. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med. 2010;16:446–51. doi: 10.1038/nm.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Y, Lin Y, Wenfeng A, et al. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Micro. 2008;4:134–46. doi: 10.1016/j.chom.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malnati MS, Scarlatti G, Gatto F, et al. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Prot. 2008;7:1240–8. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- 12.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantification of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 13.Carter CC, McNamara LA, Onafuwa-Nuga A, et al. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe. 2011;9:223–34. doi: 10.1016/j.chom.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]