Abstract
Charles Darwin believed that all traits of organisms have been honed to near perfection by natural selection. The empirical basis underlying Darwin’s conclusions consisted of numerous observations made by him and other naturalists on the exquisite adaptations of animals and plants to their natural habitats and on the impressive results of artificial selection. Darwin fully appreciated the importance of heredity but was unaware of the nature and, in fact, the very existence of genomes. A century and a half after the publication of the “Origin”, we have the opportunity to draw conclusions from the comparisons of hundreds of genome sequences from all walks of life. These comparisons suggest that the dominant mode of genome evolution is quite different from that of the phenotypic evolution. The genomes of vertebrates, those purported paragons of biological perfection, turned out to be veritable junkyards of selfish genetic elements where only a small fraction of the genetic material is dedicated to encoding biologically relevant information. In sharp contrast, genomes of microbes and viruses are incomparably more compact, with most of the genetic material assigned to distinct biological functions. However, even in these genomes, the specific genome organization (gene order) is poorly conserved. The results of comparative genomics lead to the conclusion that the genome architecture is not a straightforward result of continuous adaptation but rather is determined by the balance between the selection pressure, that is itself dependent on the effective population size and mutation rate, the level of recombination, and the activity of selfish elements. Although genes and, in many cases, multigene regions of genomes possess elaborate architectures that ensure regulation of expression, these arrangements are evolutionarily volatile and typically change substantially even on short evolutionary scales when gene sequences diverge minimally. Thus, the observed genome archtiectures are, mostly, products of neutral processes or epiphenomena of more general selective processes, such as selection for genome streamlining in successful lineages with large populations. Selection for specific gene arrangements (elements of genome architecture) seems only to modulate the results of these processes.
Introduction
Charles Darwin was the first to decipher some of the key features of biological evolution and to describe a general mechanism that had, at least, the potential to generate the remarkable diversity of existing life forms (Darwin, 1859). From the 21st century’s vantage point, it is almost unfathomable that Darwin was able to come up with his theory without having the idea of the genetic information encoding and replication, the concept that can be denoted the “genome principle”. On the pure force of logic, Darwin concluded that evolution could proceed only through the interplay of the deterministic process of heredity and the random process of heritable change. Under Darwin’s theory, the combination of these two factors yielded natural selection, the mighty force that gradually perfects the adaptation of organisms.
The elucidation of the genome principle through the pioneering genetic experimentation, primarily, by the Drosophila group led by Morgan (Morgan, 1926)and bold theorizing of Timofeev-Ressovsky, Delbruck, Pauling and others (Pauling and Delbruck, 1940, Timofeev-Ressovsky et al., 1935) culminated in the discovery of the DNA structure, the genetic code and the gene-product colinearity (see (Watson, 1963, Ycas, 1969) for early reviews), and vindicated Darwin’s vision by establishing the mechanisms of heredity and mutation. After the genome concept was established as a principle, a major, even if rarely spelled out, conundrum has emerged: is the genome an optimally organized “blueprint” for an organism that was shaped by Darwinian selection over eons of evolution, just like the organism’s phenotype, or a more or less random string of genes? Indications that the genome is unlikely to be an optimally designed instruction for an organism’s development appeared long before genome sequencing became feasible, in the form of the so-called C-value paradox, i.e., apparent large differences in genome sizes of organisms of comparable phenotypic complexity (Thomas, 1971).
In 1976, the first genome sequence of a life form, a bacteriophage, was reported (Fiers et al., 1976), and, since then, sequencing of thousands of genomes from viruses, bacteria, archaea, and eukaryotes has completely changed our understanding of genomes, their architectures, and the relationships between them. Genome architecture can be defined as the totality of non-random arrangements of functional elements (genes, regulatory regions etc) in the genome. A completely unorganized genome (a random string of genetic elements) potentially could be functional but the notion of architecture would not apply to it. Here I briefly review the emerging principles of genome architecture in different walks of life and argue that adaptive evolution of genome organization is not a viable concept. Instead, the genome architectures seem to be shaped by a complex gamut of forces, apparently, partially adaptive but largely neutral.
Overview of the principles of genome architecture: two types of genomes
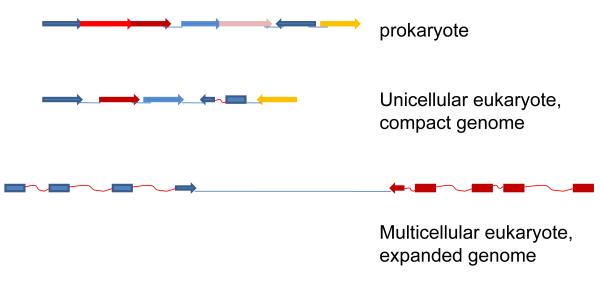
The genome layouts in different domains of life are fundamentally distinct (Figure 1). Viruses have relatively small genomes that are, typically, jam packed with genes, and overlapping genes are often used (Firth and Brown, 2006). Prokaryotes (archaea and bacteria) have compact genomes, albeit with larger intergenic regions than viruses, and very few (if any) long overlaps between genes (Lillo and Krakauer, 2007, Rogozin et al., 2002b, Rogozin et al., 2002c). Many prokaryotic genes are organized into cotranscribed groups, operons (Miller and Reznikoff, 1978, Salgado et al., 2000). Eukaryotes span an extremely wide range of genome sizes, from those well within the prokaryotic range to those that are orders of magnitude larger. They are all united by a distinctive gene architecture, the exon-intron organization whereby fragments of the protein-coding sequence of a gene (the exons) are separated by multiple non-coding regions, the introns, which are removed during splicing (Roy and Gilbert, 2006). Due to the presence of introns and long intergenic regions, the larger eukaryotic genomes are dramatically less compact than prokaryotic genomes (Fig. 2). In many unicellular eukaryotes, only a small fraction of the genes contain introns. However, the spliceosome is universally conserved in eukaryotes (Collins and Penny, 2005), and several independent evolutionary reconstructions have strongly suggested that ancestral eukaryotes had intron-rich genes, with many lineages undergoing extensive subsequent loss of introns (Carmel et al., 2007b, Csuros et al., 2008, Roy, 2006). These findings emphasize that exon-intron organization is a general architectural principle of eukaryotic genes, notwithstanding the paucity of introns in many eukaryotes. Another sharp difference between eukaryotic and prokaryotic genomes is that, typically, eukaryotes possess no operon organization (but see discussion of exceptions below).
Figure 1.
The distinct genome architectures of prokaryotes, unicellular eukaryotes and multicellular eukaryotes.
Genes are shown by rectangles with the arrowhead indicating the direction of transcription. Exons are shown by rectangles without arrowheads. Intergenic regions are shown by straight lines, and introns are shown by squiggly lines. The schematic is not to scale.
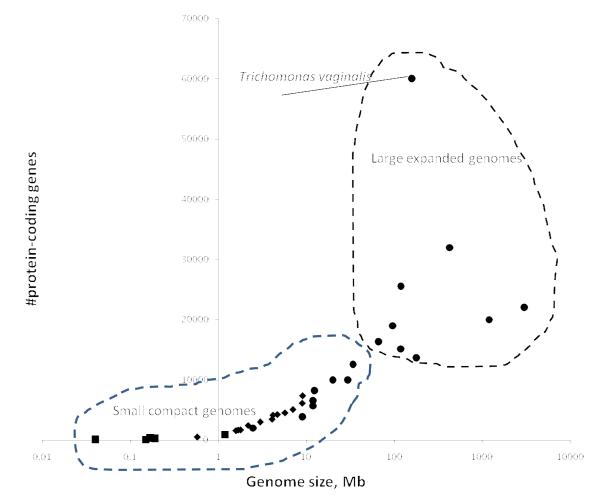
Figure 2.
Genome size versus the number of protein-coding genes for selected organisms from different divisions of life.
The plot is on a semi-logarithmic scale. Black squares, large DNA viruses; green diamonds, bacteria; red diamonds, archaea; blue circles, unicellular eukaryotes (including fungi); orange circles, multicellular eukaryotes (plants and animals). The data were from the NCBI Genome Project database (NCBI Genomes 2008).
Although the distinction between the principles of genome organization in viruses, prokaryotes, and eukaryotes is beyond doubt, the differences within each type of genomes go a long way towards blurring the boundaries. It seems like every conceivable “rule” of genome organization has its share of exceptions. The discovery of giant viruses and, conversely, of bacterial and archaeal parasites and symbionts with tiny genomes forever eliminated the separation of cellular and viral genomes by size (Koonin, 2005, Nakabachi et al., 2006, Raoult et al., 2004). Neither is there an appreciable difference in the gene density between the largest viral genomes and typical prokaryotic genomes (Iyer et al., 2006). Similarly, the genomes of many, albeit not all, unicellular eukaryotes are highly compact, almost “wall to wall” arrays of genes (with only a few tiny introns) that, in many respects, resemble the genomes of prokaryotes more than genomes of complex, multicellular eukaryotes (Lynch and Conery, 2003). Even the absence of introns in protein-coding genes of prokaryotes has been disproved: some archaeal open reading frames, are after all, interrupted by tiny introns (Watanabe et al., 2002). Conversely, the absence of operons in eukaryotes is not absolute either as two unrelated groups of eukaryotes, kinetoplastids and nematodes, possess a number of unique operons (Blumenthal, 2004).
As a generalization, it appears that genomes can be roughly partitioned into just two classes (Figure 2):
Compact, relatively small genomes of viruses, archaea, bacteria (typically, <10 Mb), and many unicellular eukaryotes (typically, <20 Mb). In these genomes, protein-coding and RNA-coding sequences occupy most of the genomic sequence.
Expansive, large genomes of multicellular and some unicellular eukaryotes (typically, >100 Mb). In these genomes, the majority of the nucleotide sequence is non-coding.
Of course, as always in biology, there are no sharp boundaries between the two types, with the genomes of certain unicellular eukaryotes such as Apicomplexa apparently being intermediate between “small” and “big” genomes. Nevertheless, the evolution of the two classes of genomes seems to be shaped by distinct forces as discussed below.
Genome architectures and their evolution
Prokaryotes: the malleable operonic organization, partially conserved gene neighborhoods, and the lack of synteny conservation
The operon, a group of co-transcribed and co-regulated genes, is one of the earliest and central concepts of bacterial genetics (Jacob and Monod, 1961). An enormous amount of variation on the simple theme of regulation by the Lac repressor developed by Jacob and Monod has been discovered over the nearly 50 years since the operon model was formulated. Nevertheless, the operon has stood the test of comparative genomics as the principle of organization of bacterial and archaeal genomes (Salgado et al., 2000, Wilson et al., 2007). Operons are much more strongly conserved during the evolution of bacterial and archaeal genomes than is large scale synteny (see below). Still, comparative analysis of gene order in bacteria and archaea reveals few operons that are shared by a broad range of organisms (Itoh et al., 1999, Wolf et al., 2001). As noticed early on, these highly conserved operons typically encode physically interacting proteins (Dandekar et al., 1998), a trend that is readily interpretable in terms of selection against the deleterious effects of imbalance between protein complex subunits (Papp et al., 2003). The most dramatic instantiation of this trend is the ribosomal superoperon that includes over 50 genes of ribosomal proteins that are found in different combinations and arrangements in all sequenced archaeal and bacterial genomes (Coenye and Vandamme, 2005, Wolf et al., 2001). Analysis of the ribosomal superoperon and other, smaller, groups of partially conserved operons led to the notion of an überoperon (Lathe et al., 2000) or a conserved gene neighborhood (Rogozin et al., 2002a), an array of overlapping, partially conserved (known or predicted) operons present in a collection of genomes. In addition to the ribosomal superoperon, notable examples of conserved neighborhoods are the group of predicted overlapping operons that encode subunits of the archaeal exosomal complex (Koonin et al., 2001) and the Cas genes that comprise an antivirus defense system (Haft et al., 2005, Makarova et al., 2006, Rogozin et al., 2002a) . The majority of genes in the überoperons encode proteins involved in the same process and/or complex but highly conserved arrangements including genes with seemingly unrelated functions exist as well, e.g., the common occurrence of the enolase gene in ribosomal neighborhoods or genes for proteasome subunits in the archaeal exosome neighborhood. The presence of these seemingly unrelated genes can be explained either by “gene sharing”, i.e., multiple functionalities of the respective proteins, or by “genomic hitchhiking”, a case when an operon combines genes without specific functional links but with similar requirements for expression (Rogozin et al., 2002a).
The majority of operons do not belong to complex, interconnected neighborhoods but instead are simple strings of 2 to 4 genes, with variations in their arrangement (Rogozin et al., 2002a, Tamames, 2001, Wolf et al., 2001). Identical, or similar, in terms of gene organization, operons are often found in highly diverse organisms and in different functional systems. A case in point are numerous metabolite transport operons that consist of similarly arranged genes encoding the transmembrane, ATPase, and periplasmic subunits of diverse permeases. The persistence of such common operons in diverse bacteria and archaea has been interpreted within the framework of the selfish operon concept, i.e., the notion that operons are maintained not so much because of the functional importance of coregulation of the constituent genes but owing to the selfish character of these compact genetic units that are prone to horizontal spread among prokaryotes (Lawrence, 1999, Lawrence, 1997, Lawrence and Roth, 1996) (see more on this concept below).
Comparative analysis of the arrangements of orthologous genes in archaeal and bacterial genomes revealed a relatively small fraction of conserved (predicted) operons and a much greater abundance of unique directons, i.e., strings of genes that are transcribed in the same direction and are separated by short intergenic regions (Salgado et al., 2000, Wolf et al., 2001). In benchmark analyses, directons have been shown to be highly accurate predictors of operons (Moreno-Hagelsieb and Collado-Vides, 2002). Thus, the local organization of archaeal and bacterial genomes seems to be governed by the operonic principle, with a small number of highly conserved operons and a much larger number of unique or rare ones.
Notably, although the great majority of the conserved gene pairs in prokaryotes are codirectional, in accordance with the operonic principle (Rogozin et al., 2002b), there is also significant conservation of divergent gene pairs which reflects coregulation by virtue of bidirectional transcription from symmetric promoters (Korbel et al., 2004). The degree of genome “operonization” widely differs among bacteria and archaea: some genomes, e.g., that of the hyperthermophilic bacterium Thermotoga maritima, are almost fully covered by (predicted) operons, whereas others, such as those of many Cyanobacteria, seem to contain few operons (Wolf et al., 2001). What determines the extent of operonization in an organism remains unclear although it stands to reason that this degree depends on the balance between the rates of gnome rearrangement that disrupts operons and horizontal gene transfer (HGT) that provides for survival and spread of operons (Lawrence, 1999, Lawrence, 1997, , 2003).
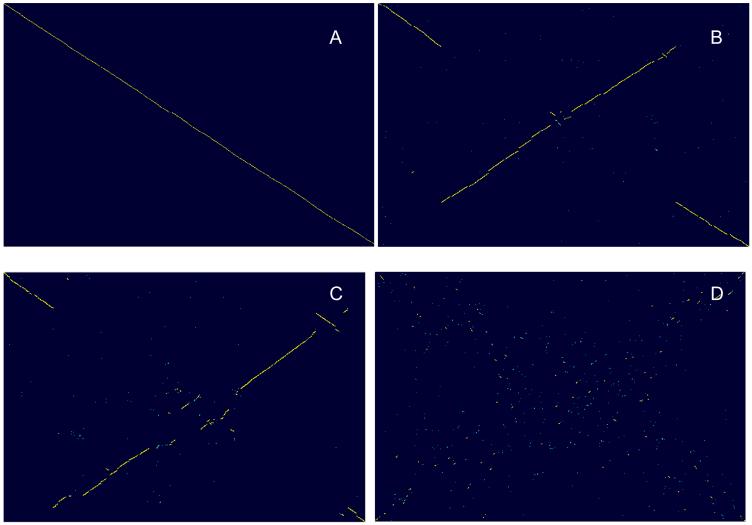
Comparisons of the first sequenced bacterial genomes revealed little conservation of gene order beyond the operonic scale (Dandekar et al., 1998, Itoh et al., 1999, Koonin et al., 1996, Mushegian and Koonin, 1996). The degree of gene order conservation between genomes can be conveniently visualized using a dot-plot where each point corresponds to a pair of orthologs. Examination of these plots reveals rapid divergence of gene order (Figure 3) so that, even between closely related bacteria, there are several breakpoints of synteny (Figure 3a), moderately diverged organisms show only a few extended colinear regions (Figure 3bc), whereas for any pair of relatively distant organisms, the plot looks like the starry sky (Figure 3d). Disruption of synteny during evolution of bacterial and archaeal genomes shows a clear and striking pattern, with an X-shape seen in the dot-plots. It appears most likely that the X-pattern is generated by symmetric chromosomal inversions around the origin of replication (Eisen et al., 2000). Such frequent inversions could be caused by the high frequency of recombination in replication forks that, in the circular chromosomes of bacteria and archaea, are typically located on both sides of and at the same distance from the origin site (Tillier and Collins, 2000). Together with small deletions and insertions, the symmetric inversions rapidly disrupt synteny during the evolution of prokaryotic genomes (Figure 3). Although extensive genome rearrangement is seen even between genomes in which the sequences of orthologous genes differ very little (Figure 3a), the rates of sequence evolution and genome rearrangement show a strong positive correlation (P. S. Novichkov, Y. I. Wolf, I. Dubchak, and EVK, unpublished results). This approximately clock-like decay of the genomic synteny suggests that genome rearrangement in prokaryotes is a largely neutral process that is affected by the same type of selective constraints that operate in sequence evolution.
Figure 3.
Comparison of gene orders in prokaryotic genomes.
In the genomic dot-plots, each point corresponds to a pair of orthologous genes identified as bidirectional best hits in BLASTP comparisons of the complete sets of protein sequences encoded in the corresponding genomes (Tatusov et al., 1997).
(a) nearly complete colinearity with a few breakpoints: Borrelia afzelii strain PKo vs B. burgdorferi starin B31.
(b) moderate genome rearrangement in bacteria, X-shaped pattern indicative of inversion around the origin of replication: Xanthomonas axonopodis pv. citri strain 306 vs X. campestris pv. campestris strain 8004.
(c) moderate genome rearrangement in bacteria, X-shaped pattern indicative of inversion around the origin of replication: Pyrococcus horikoshii OT3 vs P. abyssi GE5
(d) Extensive genome rearrangement in bacteria: Streptococcus gordonii str. Challis substrain CH1vs S. pneumoniae D39.
Although gene order in prokaryotes is poorly conserved, there are discernible patterns of global genome architecture. Most prokaryotic genomes contain a single, bidirectional replication origin site that appears to be a special point in the genome with respect to the global genome architecture (Mott and Berger, 2007). By definition, a bidirectional origin is the switch point between the leading and lagging strands that in bacteria and archaea are replicated in different modes, continuous and discontinuous, respectively. In most prokaryotes, the leading and lagging strands show substantial asymmetries in nucleotide composition, gene orientation and gene content (Rocha, 2004). Typically, the leading strand is characterized by a greater density of genes than the lagging strand, and a substantial majority of the genes on the leading strand, especially, highly expressed and/or essential ones, e.g., those coding for ribosomal RNAs and proteins, are co-oriented with replication (Brewer, 1988, Nomura and Morgan, 1977, Rocha and Danchin, 2003a, 2003b). Usually, the patterns of gene distribution are explained by different versions of the polymerase collision model that postulates selection for minimizing the chance of head-on collision between the replicating DNA polymerase and the transcribing RNA polymerase that are both more likely and more damaging than codirectional collisions (Brewer, 1988, Nomura and Morgan, 1977, Rocha, 2004). The exact mechanisms that affect the overall layout of bacterial and archaeal chromosomes require much further analysis but the general conclusion seems clear that the mechanisms and rate of chromosomal replication are important factors that determine the genome architecture.
Eukaryotic genomes: what determines their architecture?
The distinctive feature of eukaryotic genomes that sharply separates them from prokaryotic genomes is the presence of spliceosomal introns that interrupt protein-coding genes. However, the content and density of introns differ dramatically, from 1-2 introns per genome in some unicellular eukaryotes (e.g., diplomonads) to a mean of 5-8 introns per gene in vertebrates (Logsdon, 1998, Rodriguez-Trelles et al., 2006, Roy and Gilbert, 2006). Most of the eukaryotes have relatively small introns (20-200 nucleotides) but some, e.g., plants also possess a fraction of long introns whereas in mammals the average length of intron is ~ 2 kb, and there are many extremely long introns (Gibbs et al., 2004). Considering this variance of intron densities and sizes, it is all the more notable that introns are, on average, well-conserved elements of the eukaryotic genome architecture. Indeed, for instance, among vertebrates or green plants, nearly all intron positions are conserved, and up to 30% of intron positions are conserved even between orthologous genes from animals and plants (Fedorov et al., 2002, Rodriguez-Trelles et al., 2006, Rogozin et al., 2003, Roy and Gilbert, 2006). The causes of such striking conservation of the positions of seemingly non-functional elements like introns remain unclear although multiple effects of introns on expression regulation have been demonstrated (Maniatis and Reed, 2002, Nott et al., 2003), in line with the possibility that, to some extent, introns are maintained by purifying selection (Carmel et al., 2007a). In addition to their effect on the expression of the “host” gene, some of the animal and plant introns harbor genes for small non-coding RNAs (Brown et al., 2008) or even protein-coding genes (Yu et al., 2005), adding an extra level of complexity to the genome architecture.
Comparison of gene orders between eukaryotic genomes reveals considerable conservation of synteny over long evolutionary spans (hundreds of millions of years), e.g., among vertebrates or insects. Indeed, approximately 50% of the orthologous genes in human and fish belong to conserved synteny blocks (Consortium., 2004). A detailed comparative analysis of 12 sequenced insect genomes reveals a nearly full range of synteny conservation, from 99% in different species of Drosophila to ~10% between flies and honeybee (Zdobnov and Bork, 2007). Remarkably, it has been convincingly shown that the rate of synteny loss during the evolution of animals is, at least, roughly, proportional to the rate of amino acid sequence divergence in orthologous proteins, so that at ~50% mean sequence divergence, all traces of ancestral gene order are lost (Zdobnov and Bork, 2007, Zdobnov et al., 2005). In full agreement with the results of prokaryotic genome analysis (see above), the approximately clock-like decay of synteny suggests that the change of gene order is a neutral, rather than an adaptive, process, that is partially constrained by purifying selection although, in the case of large eukaryotic genomes, it cannot be ruled out that these genome-wide observations obscure important differences between the driving forces of gene order evolution in different genome regions. . These observations indicate that, compared to prokaryotes, eukaryotes show a much slower decay of synteny: even at ~90% amino acid sequence identity, prokaryotes lose all synteny beyond the conserved operons (Figure 3). It seems likely that the mechanism of origin-centered inversion that is highly active in prokaryotes but absent in eukaryotes is, at least, in part, responsible for this dramatic difference in the rates of synteny decay.
As opposed to prokaryotes, where the operonic principle governs the local arrangement of genes in the genome, it seems certain that there is no such simple organizing principle in eukaryotes. The great majority of eukaryotic mRNAs are monocistronic, so there is no single, dominant mechanistic basis for clustering of functionally linked genes in eukaryotic genomes (but see below on more subtle causes that might still favor such clustering in some cases). The major exceptions include the genomes of kinetoplastids (trypanosomes and leishmania) in which the majority of genes are organized in operon-like units that are transcribed as polycistronic mRNAs. However, unlike the case of prokaryotes, the kinetoplastid transcripts are not translated directly but instead are processed into monocistronic mRNAs via a distinct process called trans-splicing (Clayton, 2002). The nematodes represent a less extreme case of eukaryotic operonization, with approximately 15% of the genes clustered in operons whose polycistronic transcripts are also processed via trans-splicing (Blumenthal and Gleason, 2003, Guiliano and Blaxter, 2006, Qian and Zhang, 2008). The operons in nematodes show considerable conservation even among distantly related species (Guiliano and Blaxter, 2006). However, the operons in nematodes and kinetoplastids are completely unrelated to each other or to the prokaryotic operons indicating that operons have independently evolved in at least two eukaryotic lineages.
Beyond the unusual cases of operonization in kinetoplastids and nematodes, there are multiple, biologically important exceptions to the general lack of clustering of functionally related genes in eukaryotes. Typically, clusters of functionally related genes comprise tandem duplications Perhaps, the most spectacular of these are the clusters of Antennapedia (ANTP)-like homeobox genes (Hox, ParaHox, EHGBox, and NK clusters) that encode key regulators of animal development and seem to be, to a varying degree, conserved in all animals (Butts et al., 2008, Chourrout et al., 2006, Ferrier and Holland, 2001, Larroux et al., 2007). Evolutionary reconstructions based on comparative-genomic analysis indicate that the last common ancestor of the extant animals possessed a “Mega-cluster” of ANTP genes that subsequently independently and differentially deteriorated in different animal lineages(Butts et al., 2008, Ryan et al., 2006). The partial conservation of Hox and other ANTP gene clusters is likely to be maintained by purifying selection owing to the spatio-temporal colinearity whereby successive activation of genes in a cluster contributes to the progressive action along the anterior-posterior axis in the course of development (Ferrier and Minguillon, 2003, Monteiro and Ferrier, 2006). Among other notable clusters of duplicated and functionally similar genes are the thousands of vertebrate genes for olfactory receptors that are organized in multiple clusters (Niimura and Nei, 2005, , 2006), clusters of genes encoding various components of the vertebrate immune system (Hughes, 2006, Kelley and Trowsdale, 2005, Nei and Rooney, 2005), plant genes encoding proteins involved in pathogen response (Friedman and Baker, 2007), and many other, smaller clusters.
Beyond the relatively obvious clustering of paralogous genes, comparative genomics yielded many indications of non-random gene organization in eukaryotic genomes (Hurst et al., 2004, Michalak, 2008). Significant clustering has been observed among genes that can be considered related by a variety of criteria. Many reports have documented clustering of co-expressed genes. Thus, it has been shown that approximately 25% of the yeast genes that are expressed in the same stage of the mitotic cell cycle are clustered on chromosomes, i.e., at least one of the immediate neighbors in this set of control genes is expressed at the same stage (with <5% clustered genes expected by chance) (Cho et al., 1998). Similar findings have been reported for the nematode C. elegans, even after the contribution of operons was subtracted (Lercher et al., 2003, Roy et al., 2002). A comprehensive analysis of expression patterns among Drosophila genes indicated that ~20% of the genes form co-expression clusters (at least 10 times more than expected by chance) (Spellman and Rubin, 2002), and even more impressive clustering has been reported for genes that are expressed in the same tissue in both Drosophila (Boutanaev et al., 2002) and mammals(Bortoluzzi et al., 1998, Caron et al., 2001, Versteeg et al., 2003). However, the relationship between the observed clustering of coexpressed genes and clustering by gene function turned out to be complex. A significant but far from complete functional coherence was observed between clustered coexpressed genes in yeast (Cohen et al., 2000) and Arabidopsis (Williams and Bowles, 2004); by contrast, in animals, there seems to be little clustering of genes that are both co-expressed and belong to the same functional category (Fukuoka et al., 2004, Spellman and Rubin, 2002). A comparison of clusters of coexpressed genes in human and mouse genomes revealed significant conservation, in support of the functional significance of these clusters; however, the clusters encompassed less than 5% of the genes in each genome (Semon and Duret, 2006).
When eukaryotic genome architecture was analyzed explicitly from the functional perspective, it was shown that genes for enzymes of the same metabolic pathway are significantly (according to a scoring scheme developed to analyze the statistics of gene clusters) clustered in all analyzed genomes. The fraction of pathways that showed significant clustering varied widely, from approximately 98% in yeast to about 30% in Drosophila (Lee and Sonnhammer, 2003). Notably, however, this analysis found no coherence between gene clustering in different eukaryotic genomes, i.e., pathways that showed gene clustering in one genome were typically not clustered in others.
The mechanisms of co-expression of clustered genes in eukaryotic genomes, obviously, have to do with co-regulation of transcription and can be classified into local ones, such as the utilization of bidirectional promoters or common enhancers, and global ones, such as distinct chromatin structure that translates into similar expression patterns of genes in the corresponding region of a chromosome (Hurst et al., 2004). Although chromatin-level regulation is often considered important (Cremer and Cremer, 2001, Sproul et al., 2005), the small size of the majority of clusters of co-expressed genes suggests that, at least in mammals, local mechanisms of co-regulation could be decisive (Semon and Duret, 2006).
Evolutionary patterns and forces affecting genome architecture: chance and necessity
Probably, the single major conclusion from all the comparative analyses of genome organizations in prokaryotes and eukaryotes is the lack of uniformity and the plurality of evolution patterns, and underlying mechanisms. With regard to genome architecture, what is true of E. coli definitely does not apply to the elephant or even to the fly. The operonic principle of gene arrangement in prokaryotes is the only indisputably strong trend of genome organization but it only affects the short-range gene order. Demonstrable long-range trends definitely exist, such as the preferential positioning of prokaryotic genes on the leading strand or clustering of coexpressed genes in eukaryotes. However, all these trends are “statistical”, i.e., relatively weak, and also, highly variable even between genomes of relatively close organisms. In line with this lack of overriding trends in genome organization, synteny is not a trait that is generally conserved over long evolutionary distances (that is, such distances at which amino acid sequences of most proteins substantially diverge). Major exceptions, such as the partial conservation of the ribosomal superoperon in bacteria and archaea, and of the homeobox gene clusters in animals, are notable and can be attributed to functional constraints. However, these cases encompass only a small fraction of genes in genomes and only affect relatively short-range synteny. In prokaryotes, where inversions around the origin point are common, and so is HGT, complete deterioration of long-range synteny is often observed even between organisms that share nearly complete sets of highly conserved orthologous genes (Figure 3). Although, apparently owing to the absence of origin-centered inversions and low incidence of HGT, there is more synteny conservation in eukaryotes, almost none of it carries across phyla, and there definitely are no pan-eukaryotic gene clusters that would be comparable in their level of conservation to the ribosomal operons or ATPase operons in prokaryotes. Thus, in general, genome architecture is a highly variable, volatile feature of organisms.
What are, then, the evolutionary forces that shape genome architecture? Of course, there are multiple ones. Clearly, genome organization is neither random – no genome is simply an arbitrary string of genes - nor a fully optimized design selected to encode the optimal phenotype. The principal explanatory framework for understanding evolution of genome organization can be drawn from the population-genetic theory of evolution of genomic complexity that was recently expounded by Lynch (Lynch, 2007, Lynch and Conery, 2003). The theory maintains that genetic changes leading to an increase of complexity such as gene duplications or intron insertions are slightly deleterious and can be fixed only when purifying selection in a population is weak. Therefore, substantial genome complexification is possible only during population bottlenecks, given that the strength of purifying selection is proportional to the effective population size. Under this concept, genomic complexity is not adaptive but is brought about by neutral population-genetic processes under conditions when purifying selection is ineffective. Complexification starts off as a “genomic syndrome” although complex features subsequently become subject to adaptive selection. By contrast, in “highly successful”, large populations, purifying selection is intense, so that the prevailing mode of evolution in these prokaryotes is genome contraction. Most of the prokaryotic genomes and genomes of many unicellular eukaryotes do not pass the “complexification threshold”, the result being compact, streamlined genomes with a relatively small number of genes, short intergenic regions, and few selfish elements. By contrast, the genomes of multicellular eukaryotes are beyond the threshold, so fixation of multiple duplications as well as proliferation of transposable elements (TEs), the latter also facilitated by sex (Lynch, 2007), become possible.
Of course, all these trends are far from being hard principles, and there are bacterial genomes with more than 12,000 genes (Schneiker et al., 2007) as well as genomes of unicellular eukaryotes (e.g., Chlamydomonas (Merchant et al., 2007)or Trichomonas (Carlton et al., 2007)) that are at least as complex by any criteria as the genomes of multicellular animals or plants. Furthermore, some prokaryotic genomes (e.g., the crenarchaeon Sulfolobus solfataricus (She et al., 2001)) and genomes of unicellular eukaryotes (e.g., Trichomonas vaginalis (Carlton et al., 2007)) are among those with the highest content of TEs. Apparently, the evolution of even these, relatively small genomes depends on the balance between the pressure of purifying selection, itself dependent on the population size and mutation rate, the intensity of recombination processes, and the activity of selfish genetic elements.
Where in the evolution of genome architecture can we see clear imprints of selection, in particular, positive selection? It seems that selection is an important factor in the evolution of operons. Operons can easily form by chance, in a completely neutral fashion, through genome compactification (streamlining) which leads to the formation of tightly spaced strings of codirectional genes, directons (Salgado et al., 2000, Wolf et al., 2001). Those of the randomly assembled operons that consist of functionally linked genes provide a selective advantage to their carriers owing to the possibility of co-expression and co-regulation, so such operons are fixed in evolution and often become widespread via HGT. This view of operon evolution incorporates the selfish operon hypothesis according to which operons are maintained as selfish elements via HGT (Lawrence, 1999, Lawrence, 1997, , 2003) but also includes a distinct effect of positive selection that is amplified by HGT. So operons can be reasonably viewed as partially selfish elements whose survival depends both on their selective value for the carrier organisms and on random HGT.
The role of HGT in the persistence of operons is indirectly but, in my view, strongly supported by the fact that no strings of genes homologous to prokaryotic operons are detectable in eukaryotic genomes (Y.I. Wolf and EVK, unpublished results). Regardless of the exact scenario for the origin of eukaryotes, the genome of the last common ancestor of the extant eukaryotes must have acquired diverse operons, at least, as part of the DNA transferred from the mitochondrial endosymbiont, and possibly, also from the archaeal (under the symbiotic hypotheses of eukaryotic origin (Embley and Martin, 2006, Martin and Koonin, 2006)) or protoeukaryotic (under the archaezoan or related hypotheses (Kurland et al., 2006, Poole and Penny, 2007)) host. The lack of any traces of such inherited operons in eukaryotic genomes suggests a ratchet-type scenario of operon elimination: once an operon is gone, in the absence of appreciable HGT, the loss is virtually irreversible.
Conversely, reconstruction of the evolutionary dynamics of operons in nematodes yielded a “easy come, slow go” scenario, with the rate of gain substantially exceeding the rate of loss (Qian and Zhang, 2008). Thus, it appears that operons that are randomly created by recombination are subsequently maintained by purifying selection.
In multicellular eukaryotes, the relatively small population size and relatively low characteristic mutation rates translate into comparatively weak purifying selection, so that various degrees of genome enlargement and complexification become possible. Hence the formation of large clusters of tandemly duplicated genes, a feature that can be viewed as an increase in genome ordering. However, the counter trend is also apparent, namely, the increased activity of transposable elements that leads to an increase in genomic disorder. In vertebrates, this mobilization of transposable elements is particularly dramatic so that the genomes consist mostly of TE-derived sequences (Makalowski, 2000). In an already familiar pattern that is a crucial part of the neutral paradigm of the evolution of genomic complexity (Lynch, 2007), the TEs comprise an important source for recruitment (exaptation) of new regulatory and, possibly, even structural sequences (Jordan et al., 2003, Thornburg et al., 2006).
To what extent gene clustering in eukaryotes is affected by selection and what the targets of this potential selection are remain widely open questions. As such, co-expression of adjacent genes cannot be considered evidence of selection because, when genes are located in the same chromatin domain, up- or down-regulation of one gene can accidentally cause a concordant change in the expression of the other owing to the effect of chromatin remodeling (Spellman and Rubin, 2002). Such co-expression does not necessarily confer any benefits on the organism and might not be subject to selection (Hurst et al., 2004). Clustering of genes that are directly functionally associated, such as enzymes in the same pathway (Lee and Sonnhammer, 2003), is hard to explain without invoking selection. However, the lack of significant evolutionary conservation of such clusters is surprising and suggests that either the selective pressure that leads to fixation and persistence of these clusters is quite weak, or that relative importance of clustering (and the ensuing co-regulation) changes rapidly in the course of evolution (or both).
Conclusions
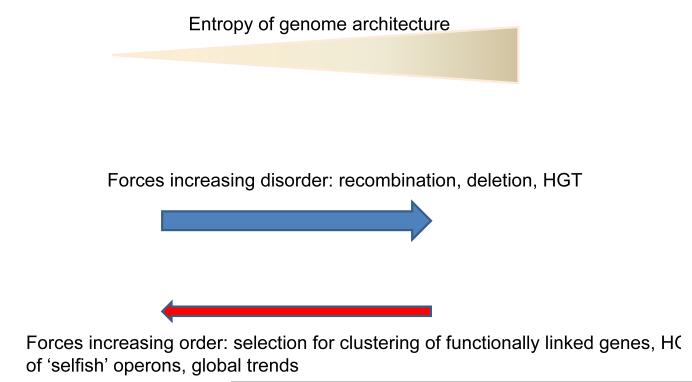
The evolution of genome architecture appears to be defined by a dynamic balance between forces that enhance disorder, primarily, various forms of intragenomic and intergenomic recombination including HGT, and the ordering effects of selection (Figure 4). The result is a complex genomescape that encompasses a variety of non-random features, particularly, local ones, that emerged with participation of selection, but is far removed from an optimally designed architectural blueprint for an organism. On the whole, the large-scale organization of genomes appears to be, mostly, random and, indeed, evolves rapidly (at least, compared to protein sequences) and in an approximately clock-like manner. Thus, in a sense, the title of this article is not fully appropriate, as there is no such thing as global genome architecture although elaborate local architectural features are shaped by selection and play crucial roles in the functioning of all genomes.
Figure 4.
The evolutionary forces affecting genome architecture
Acknowledgements
I thank Pavel Novichkov for providing the data for Figure 3. The author’s research is supported by the DHHS (National Library of Medicine) intramural funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- NCBI genomes 2008 < http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi>.
- Blumenthal T. Operons in eukaryotes. Brief Funct Genomic Proteomic. 2004;3:199–211. doi: 10.1093/bfgp/3.3.199. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Gleason KS. Caenorhabditis elegans operons: form and function. Nat Rev Genet. 2003;4:112–20. doi: 10.1038/nrg995. [DOI] [PubMed] [Google Scholar]
- Bortoluzzi S, Rampoldi L, Simionati B, Zimbello R, Barbon A, d’Alessi F, et al. A comprehensive, high-resolution genomic transcript map of human skeletal muscle. Genome Res. 1998;8:817–25. doi: 10.1101/gr.8.8.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–9. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–86. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Brown JW, Marshall DF, Echeverria M. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008;13:335–42. doi: 10.1016/j.tplants.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Butts T, Holland PW, Ferrier DE. The Urbilaterian Super-Hox cluster. Trends Genet. 2008;24:259–62. doi: 10.1016/j.tig.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–12. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel L, Rogozin IB, Wolf YI, Koonin EV. Patterns of intron gain and conservation in eukaryotic genes. BMC Evol Biol. 2007a;7:192. doi: 10.1186/1471-2148-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007b;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–92. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Chourrout D, Delsuc F, Chourrout P, Edvardsen RB, Rentzsch F, Renfer E, et al. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442:684–7. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- Clayton CE. Life without transcriptional control? From fly to man and back again. Embo J. 2002;21:1881–8. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Vandamme P. Organisation of the S10, spc and alpha ribosomal protein gene clusters in prokaryotic genomes. FEMS Microbiol Lett. 2005;242:117–26. doi: 10.1016/j.femsle.2004.10.050. [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–6. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- Collins L, Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol. 2005;22:1053–66. doi: 10.1093/molbev/msi091. [DOI] [PubMed] [Google Scholar]
- Consortium. ICGS Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Csuros M, Rogozin IB, Koonin EV. Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach. Mol Biol Evol. 2008;25:903–11. doi: 10.1093/molbev/msn039. [DOI] [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–8. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Heidelberg JF, White O, Salzberg SL. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-research0011. RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–30. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Merican AF, Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc Natl Acad Sci U S A. 2002;99:16128–33. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier DE, Holland PW. Ancient origin of the Hox gene cluster. Nat Rev Genet. 2001;2:33–8. doi: 10.1038/35047605. [DOI] [PubMed] [Google Scholar]
- Ferrier DE, Minguillon C. Evolution of the Hox/ParaHox gene clusters. Int J Dev Biol. 2003;47:605–11. [PubMed] [Google Scholar]
- Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–7. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Firth AE, Brown CM. Detecting overlapping coding sequences in virus genomes. BMC Bioinformatics. 2006;7:75. doi: 10.1186/1471-2105-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AR, Baker BJ. The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007;17:493–9. doi: 10.1016/j.gde.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Fukuoka Y, Inaoka H, Kohane IS. Inter-species differences of co-expression of neighboring genes in eukaryotic genomes. BMC Genomics. 2004;5:4. doi: 10.1186/1471-2164-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Guiliano DB, Blaxter ML. Operon conservation and the evolution of trans-splicing in the phylum Nematoda. PLoS Genet. 2006;2:e198. doi: 10.1371/journal.pgen.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Evolutionary relationships of vertebrate NACHT domain-containing proteins. Immunogenetics. 2006;58:785–91. doi: 10.1007/s00251-006-0148-8. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Itoh T, Takemoto K, Mori H, Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol Biol Evol. 1999;16:332–46. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–84. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Kelley J, Trowsdale J. Features of MHC and NK gene clusters. Transpl Immunol. 2005;14:129–34. doi: 10.1016/j.trim.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Virology: Gulliver among the Lilliputians. Curr Biol. 2005;15:R167–9. doi: 10.1016/j.cub.2005.02.042. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR, Rudd KE. Sequencing and analysis of bacterial genomes. Curr Biol. 1996;6:404–16. doi: 10.1016/s0960-9822(02)00508-0. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–52. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 2004;22:911–7. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–4. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS, Degnan BM. The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol. 2007;17:706–10. doi: 10.1016/j.cub.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lathe WC, 3rd, Snel B, Bork P. Gene context conservation of a higher order than operons. Trends Biochem Sci. 2000;25:474–9. doi: 10.1016/s0968-0004(00)01663-7. [DOI] [PubMed] [Google Scholar]
- Lawrence J. Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr Opin Genet Dev. 1999;9:642–8. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- Lawrence JG. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–9. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- Lawrence JG. Gene organization: selection, selfishness, and serendipity. Annu Rev Microbiol. 2003;57:419–40. doi: 10.1146/annurev.micro.57.030502.090816. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Roth JR. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–60. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Sonnhammer EL. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–82. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 2003;13:238–43. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo F, Krakauer DC. A statistical analysis of the three-fold evolution of genomic compression through frame overlaps in prokaryotes. Biol Direct. 2007;2:22. doi: 10.1186/1745-6150-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon JM., Jr The recent origins of spliceosomal introns revisited. Curr Opin Genet Dev. 1998;8:637–48. doi: 10.1016/s0959-437x(98)80031-2. [DOI] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8597–604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–4. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Makalowski W. Genomic scrap yard: how genomes utilize all that junk. Gene. 2000;259:61–7. doi: 10.1016/s0378-1119(00)00436-4. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentation. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–50. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–8. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Monteiro AS, Ferrier DE. Hox genes are not always Colinear. Int J Biol Sci. 2006;2:95–103. doi: 10.7150/ijbs.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18(Suppl 1):S329–36. doi: 10.1093/bioinformatics/18.suppl_1.s329. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–54. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. Gene order is not conserved in bacterial evolution. Trends Genet. 1996;12:289–90. doi: 10.1016/0168-9525(96)20006-x. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–52. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005;346:13–21. doi: 10.1016/j.gene.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51:505–17. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Morgan EA. Genetics of bacterial ribosomes. Annu Rev Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. Rna. 2003;9:607–17. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–7. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Pauling L, Delbruck M. The nature of the intermolecular forces operative in biological processes. Science. 1940;92:77–79. doi: 10.1126/science.92.2378.77. [DOI] [PubMed] [Google Scholar]
- Poole A, Penny D. Eukaryote evolution: engulfed by speculation. Nature. 2007;447:913. doi: 10.1038/447913a. [DOI] [PubMed] [Google Scholar]
- Qian W, Zhang J. Evolutionary dynamics of nematode operons: easy come, slow go. Genome Res. 2008;18:412–21. doi: 10.1101/gr.7112608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, et al. The 1. 2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–50. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–27. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat Genet. 2003a;34:377–8. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 2003b;31:6570–7. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarro R, Ayala FJ. Origin and Evolution of Spliceosomal Introns. Annu Rev Genet. 2006 doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Makarova KS, Murvai J, Czabarka E, Wolf YI, Tatusov RL, et al. Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res. 2002a;30:2212–23. doi: 10.1093/nar/30.10.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Makarova KS, Natale DA, Spiridonov AN, Tatusov RL, Wolf YI, et al. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res. 2002b;30:4264–71. doi: 10.1093/nar/gkf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Spiridonov AN, Sorokin AV, Wolf YI, Jordan IK, Tatusov RL, et al. Purifying and directional selection in overlapping prokaryotic genes. Trends Genet. 2002c;18:228–32. doi: 10.1016/s0168-9525(02)02649-5. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 2003;13:1512–7. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–9. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Roy SW. Intron-rich ancestors. Trends Genet. 2006;22:468–71. doi: 10.1016/j.tig.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–21. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006;7:R64. doi: 10.1186/gb-2006-7-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci U S A. 2000;97:6652–7. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–9. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- Semon M, Duret L. Evolutionary origin and maintenance of coexpressed gene clusters in mammals. Mol Biol Evol. 2006;23:1715–23. doi: 10.1093/molbev/msl034. [DOI] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001;98:7835–40. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–81. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- Tamames J. Evolution of gene order conservation in prokaryotes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0020. RESEARCH0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thomas CA., Jr The genetic organization of chromosomes. Annu Rev Genet. 1971;5:237–56. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- Thornburg BG, Gotea V, Makalowski W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006;365:104–10. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Tillier ER, Collins RA. Genome rearrangement by replication-directed translocation. Nat Genet. 2000;26:195–7. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- Timofeev-Ressovsky NW, Zimmer KG, Delbruck M. Uber die Natur der Genmutation und der Genstruktur. Nachr. Ges. Wiss. Gottingen, Math.-Phys. Kl. 1935;6:190–245. [Google Scholar]
- Versteeg R, van Schaik BD, van Batenburg MF, Roos M, Monajemi R, Caron H, et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yokobori S, Inaba T, Yamagishi A, Oshima T, Kawarabayasi Y, et al. Introns in protein-coding genes in Archaea. FEBS Lett. 2002;510:27–30. doi: 10.1016/s0014-5793(01)03219-7. [DOI] [PubMed] [Google Scholar]
- Watson JD. Involvement of RNA in the synthesis of proteins. Science. 1963;140:17–26. doi: 10.1126/science.140.3562.17. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Bowles DJ. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14:1060–7. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol Life Sci. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- Yu P, Ma D, Xu M. Nested genes in the human genome. Genomics. 2005;86:414–22. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, von Mering C, Letunic I, Bork P. Consistency of genome-based methods in measuring Metazoan evolution. FEBS Lett. 2005;579:3355–61. doi: 10.1016/j.febslet.2005.04.006. [DOI] [PubMed] [Google Scholar]